Human versus Artificial Intelligence: Validation of a Deep Learning Model for Retinal Layer and Fluid Segmentation in Optical Coherence Tomography Images from Patients with Age-Related Macular Degeneration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Setting
2.3. Study Population
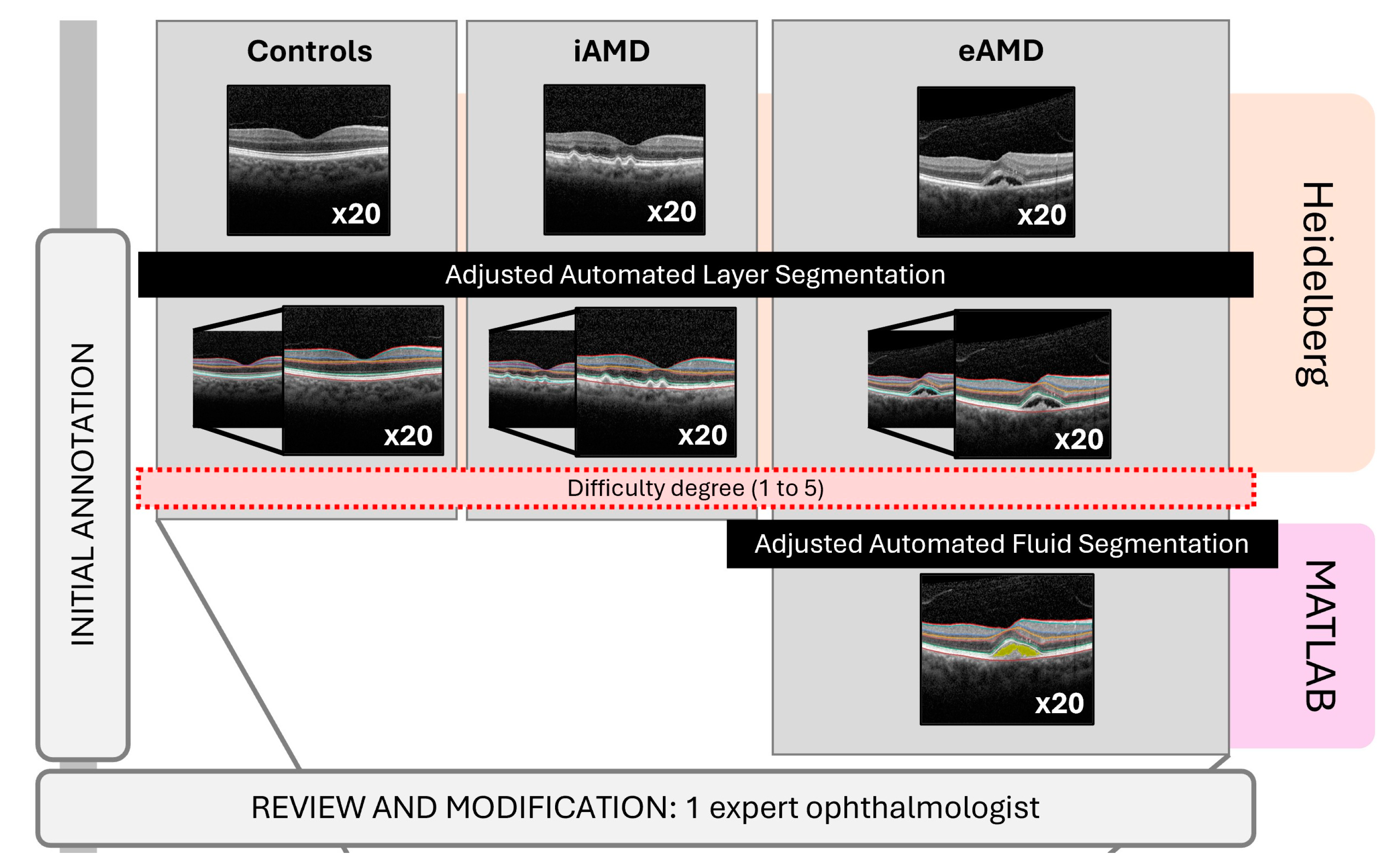
2.4. Identification and Image Data Collection
2.5. Algorithm Description
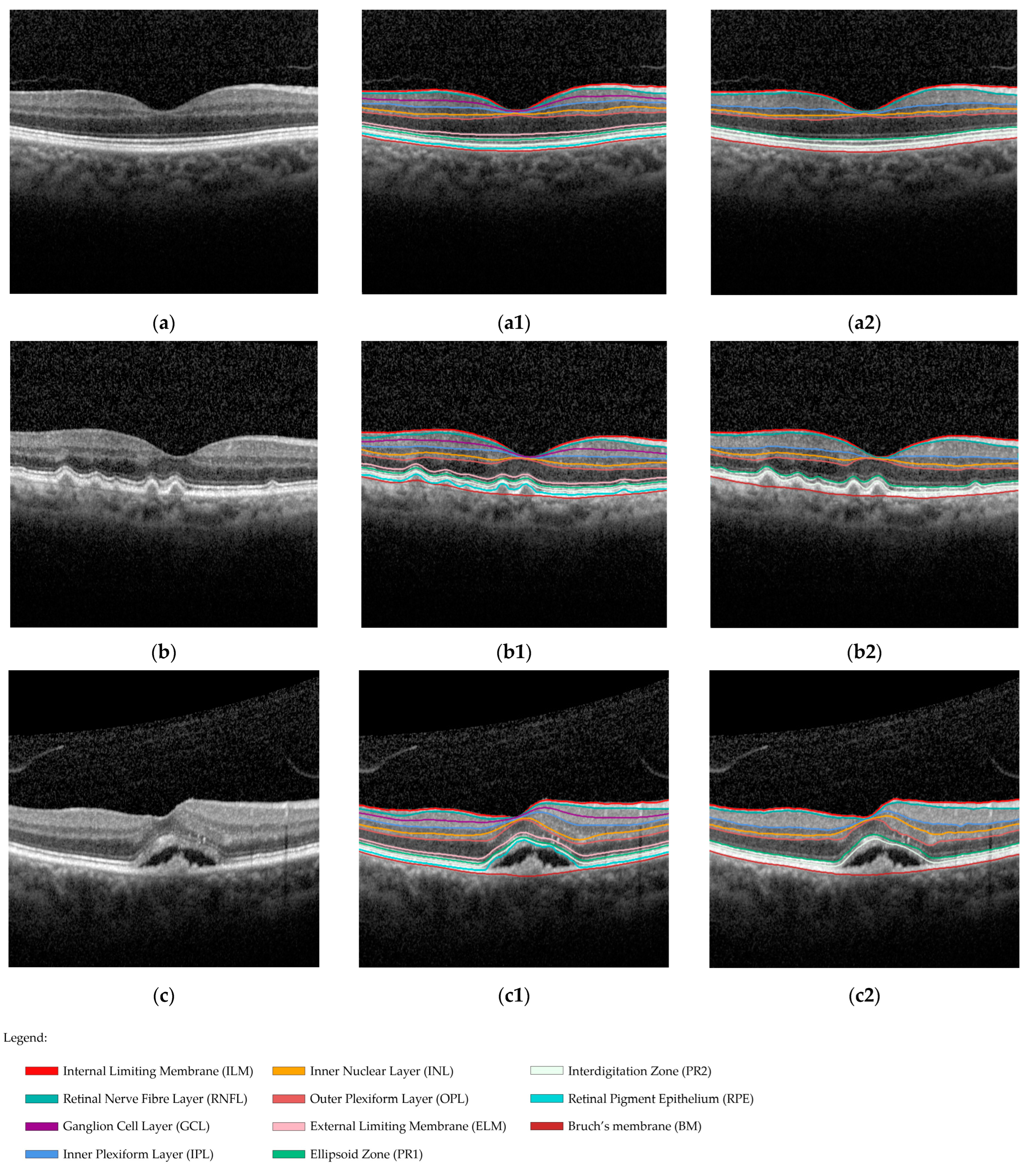
2.5.1. Reference Standard
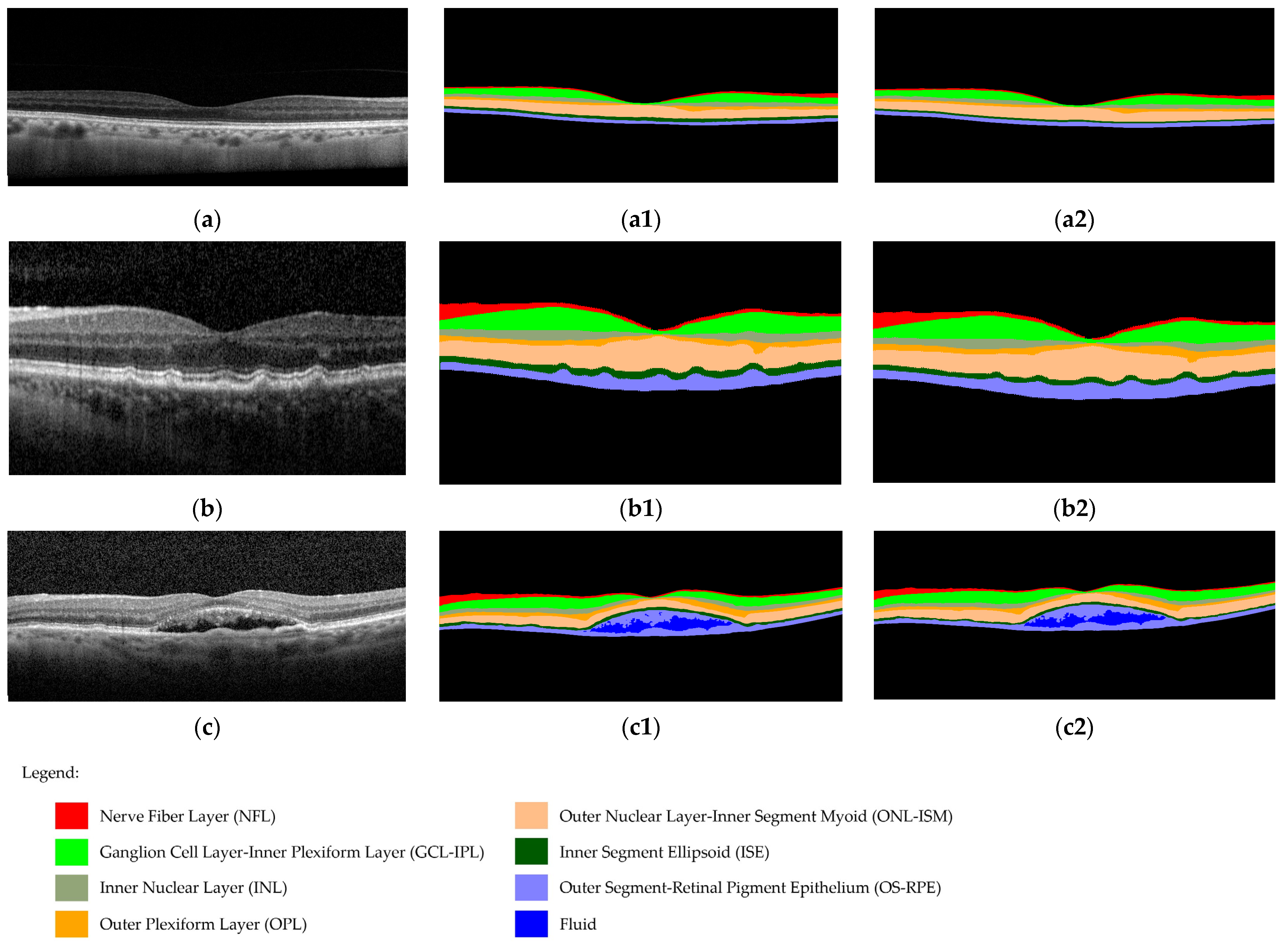
2.5.2. DL Software
2.6. Image Annotations
2.7. Outcomes
2.8. Statistical Analysis
3. Results
3.1. Performance Evaluation of the DL Software
3.2. Accessing DL Software’s Accuracy
3.2.1. Retinal Layer Thickness Segmentation
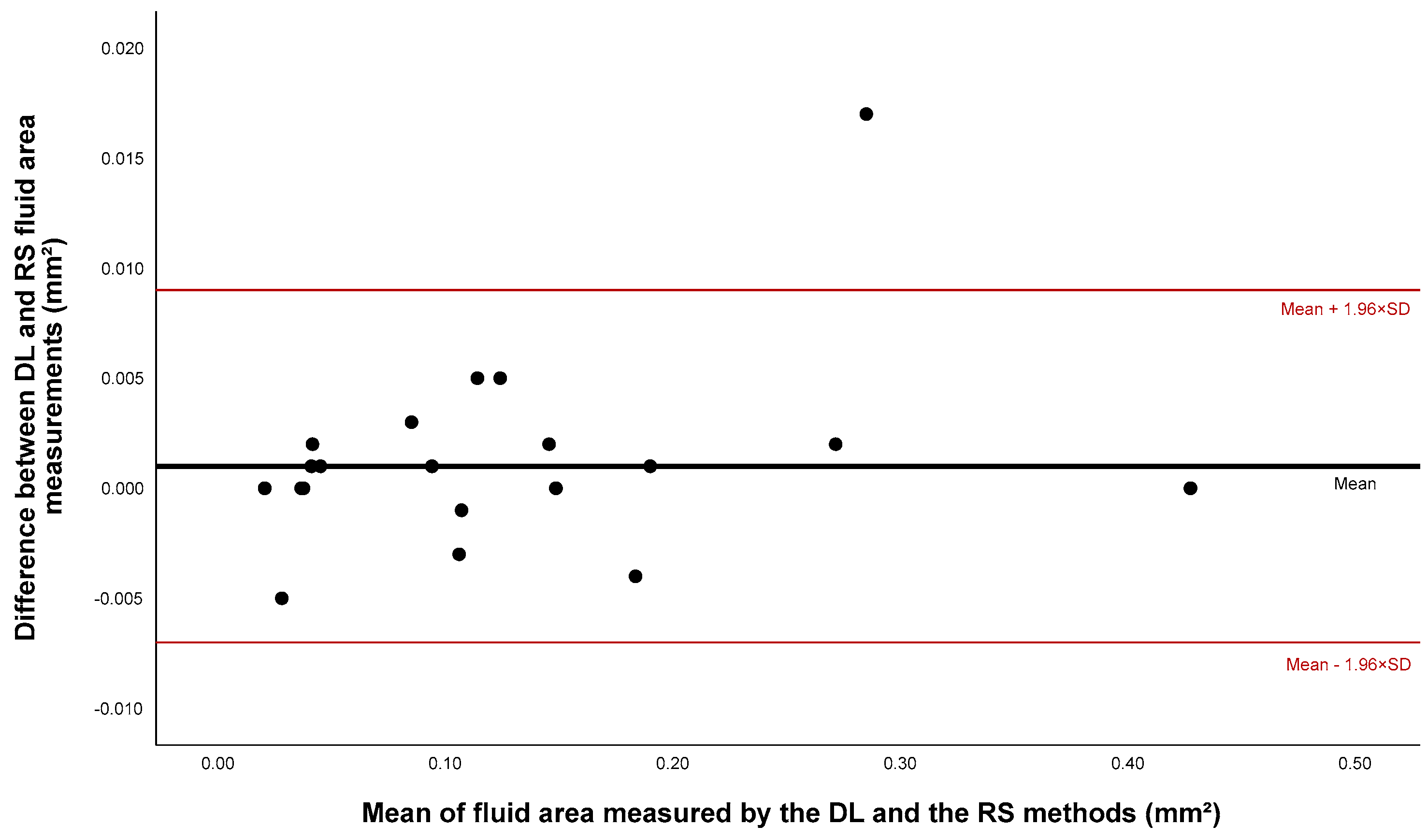
3.2.2. Fluid Segmentation
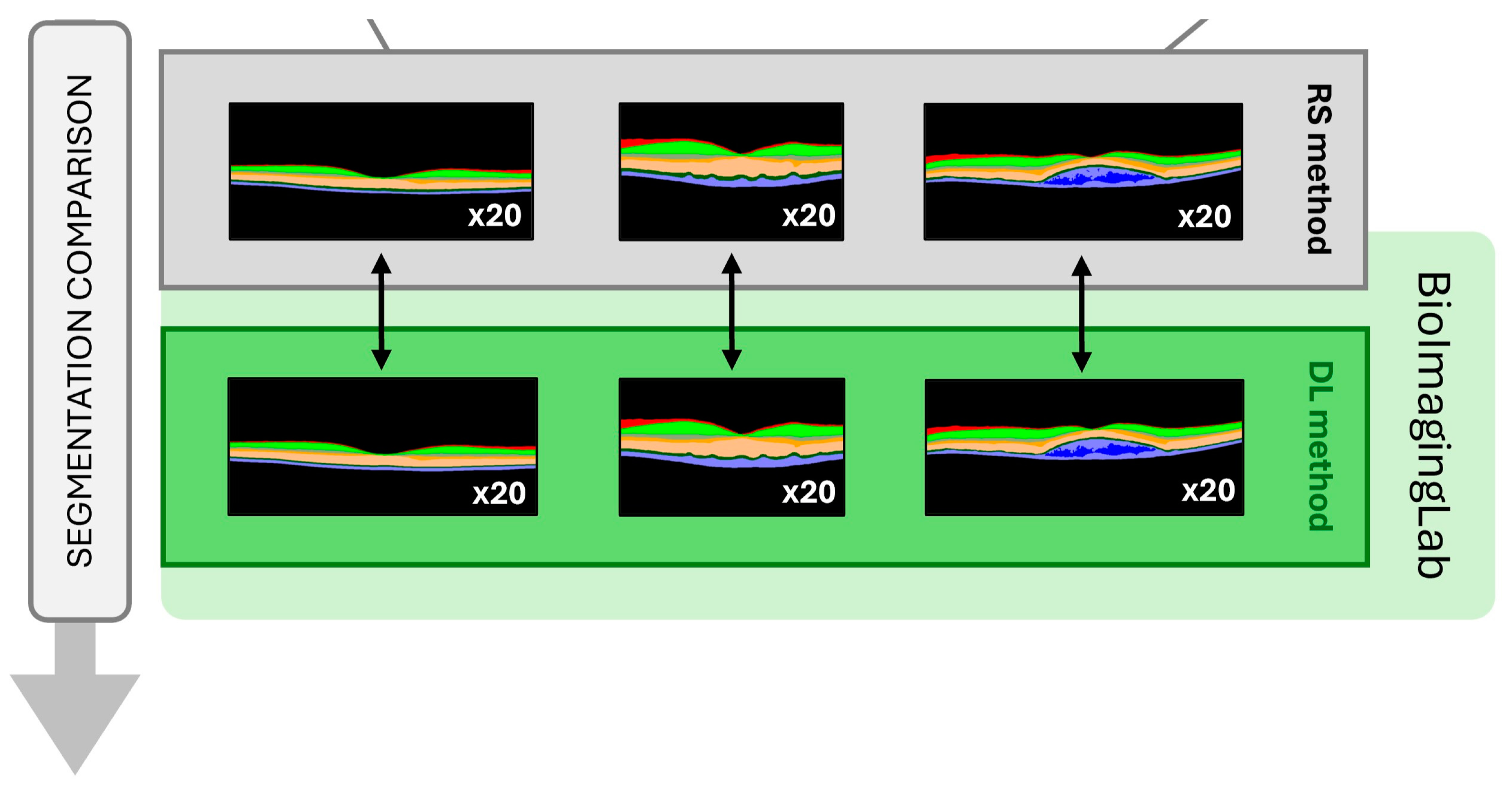
3.3. Disagreement between Methods in Layers’ Segmentation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Causes of blindness and vision impairment in 2020 and trends over 30 years, and prevalence of avoidable blindness in relation to VISION 2020: The Right to Sight: An analysis for the Global Burden of Disease Study. Lancet Glob. Health 2021, 9, e144–e160. [CrossRef]
- Bourne, R.R.A.; Jonas, J.B.; Bron, A.M.; Cicinelli, M.V.; Das, A.; Flaxman, S.R.; Friedman, D.S.; Keeffe, J.E.; Kempen, J.H.; Leasher, J.; et al. Prevalence and causes of vision loss in high-income countries and in Eastern and Central Europe in 2015: Magnitude, temporal trends and projections. Br. J. Ophthalmol. 2018, 102, 575–585. [Google Scholar] [CrossRef]
- Wong, W.L.; Su, X.; Li, X.; Cheung, C.M.; Klein, R.; Cheng, C.Y.; Wong, T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Glob. Health 2014, 2, e106–e116. [Google Scholar] [CrossRef]
- Jabbehdari, S.; Handa, J.T. Oxidative stress as a therapeutic target for the prevention and treatment of early age-related macular degeneration. Surv. Ophthalmol. 2021, 66, 423–440. [Google Scholar] [CrossRef]
- Chen, M.; Xu, H. Parainflammation, chronic inflammation, and age-related macular degeneration. J. Leukoc. Biol. 2015, 98, 713–725. [Google Scholar] [CrossRef]
- Mullins, R.F.; Schoo, D.P.; Sohn, E.H.; Flamme-Wiese, M.J.; Workamelahu, G.; Johnston, R.M.; Wang, K.; Tucker, B.A.; Stone, E.M. The membrane attack complex in aging human choriocapillaris: Relationship to macular degeneration and choroidal thinning. Am. J. Pathol. 2014, 184, 3142–3153. [Google Scholar] [CrossRef]
- Fritsche, L.G.; Igl, W.; Bailey, J.N.; Grassmann, F.; Sengupta, S.; Bragg-Gresham, J.L.; Burdon, K.P.; Hebbring, S.J.; Wen, C.; Gorski, M.; et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2016, 48, 134–143. [Google Scholar] [CrossRef]
- Heesterbeek, T.J.; Lorés-Motta, L.; Hoyng, C.B.; Lechanteur, Y.T.E.; den Hollander, A.I. Risk factors for progression of age-related macular degeneration. Ophthalmic Physiol. Opt. 2020, 40, 140–170. [Google Scholar] [CrossRef]
- Curcio, C.A. Soft Drusen in Age-Related Macular Degeneration: Biology and Targeting Via the Oil Spill Strategies. Investig. Ophthalmol. Vis. Sci. 2018, 59, AMD160–AMD181. [Google Scholar] [CrossRef]
- Ferris, F.L., 3rd; Wilkinson, C.P.; Bird, A.; Chakravarthy, U.; Chew, E.; Csaky, K.; Sadda, S.R. Clinical classification of age-related macular degeneration. Ophthalmology 2013, 120, 844–851. [Google Scholar] [CrossRef]
- Elsharkawy, M.; Elrazzaz, M.; Ghazal, M.; Alhalabi, M.; Soliman, A.; Mahmoud, A.; El-Daydamony, E.; Atwan, A.; Thanos, A.; Sandhu, H.S.; et al. Role of Optical Coherence Tomography Imaging in Predicting Progression of Age-Related Macular Disease: A Survey. Diagnostics 2021, 11, 2313. [Google Scholar] [CrossRef] [PubMed]
- Schlanitz, F.G.; Baumann, B.; Kundi, M.; Sacu, S.; Baratsits, M.; Scheschy, U.; Shahlaee, A.; Mittermüller, T.J.; Montuoro, A.; Roberts, P.; et al. Drusen volume development over time and its relevance to the course of age-related macular degeneration. Br. J. Ophthalmol. 2017, 101, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Waldstein, S.M.; Vogl, W.D.; Bogunovic, H.; Sadeghipour, A.; Riedl, S.; Schmidt-Erfurth, U. Characterization of Drusen and Hyperreflective Foci as Biomarkers for Disease Progression in Age-Related Macular Degeneration Using Artificial Intelligence in Optical Coherence Tomography. JAMA Ophthalmol. 2020, 138, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Metrangolo, C.; Donati, S.; Mazzola, M.; Fontanel, L.; Messina, W.; D‘Alterio, G.; Rubino, M.; Radice, P.; Premi, E.; Azzolini, C. OCT Biomarkers in Neovascular Age-Related Macular Degeneration: A Narrative Review. J. Ophthalmol. 2021, 2021, 9994098. [Google Scholar] [CrossRef] [PubMed]
- Lai, T.T.; Hsieh, Y.T.; Yang, C.M.; Ho, T.C.; Yang, C.H. Biomarkers of optical coherence tomography in evaluating the treatment outcomes of neovascular age-related macular degeneration: A real-world study. Sci. Rep. 2019, 9, 529. [Google Scholar] [CrossRef] [PubMed]
- Dahrouj, M.; Miller, J.B. Artificial Intelligence (AI) and Retinal Optical Coherence Tomography (OCT). Semin. Ophthalmol. 2021, 36, 341–345. [Google Scholar] [CrossRef]
- Leng, X.; Shi, R.; Wu, Y.; Zhu, S.; Cai, X.; Lu, X.; Liu, R. Deep learning for detection of age-related macular degeneration: A systematic review and meta-analysis of diagnostic test accuracy studies. PLoS ONE 2023, 18, e0284060. [Google Scholar] [CrossRef] [PubMed]
- Bhuiyan, A.; Wong, T.Y.; Ting, D.S.W.; Govindaiah, A.; Souied, E.H.; Smith, R.T. Artificial Intelligence to Stratify Severity of Age-Related Macular Degeneration (AMD) and Predict Risk of Progression to Late AMD. Transl. Vis. Sci. Technol. 2020, 9, 25. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Erfurth, U.; Reiter, G.S.; Riedl, S.; Seeböck, P.; Vogl, W.D.; Blodi, B.A.; Domalpally, A.; Fawzi, A.; Jia, Y.; Sarraf, D.; et al. AI-based monitoring of retinal fluid in disease activity and under therapy. Prog. Retin. Eye Res. 2022, 86, 100972. [Google Scholar] [CrossRef]
- Yim, J.; Chopra, R.; Spitz, T.; Winkens, J.; Obika, A.; Kelly, C.; Askham, H.; Lukic, M.; Huemer, J.; Fasler, K.; et al. Predicting conversion to wet age-related macular degeneration using deep learning. Nat. Med. 2020, 26, 892–899. [Google Scholar] [CrossRef]
- Melo, T.; Carneiro, Â.; Campilho, A.; Mendonça, A.M. Retinal layer and fluid segmentation in optical coherence tomography images using a hierarchical framework. J. Med. Imaging 2023, 10, 014006. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yan, S.; Lu, N.; Yang, D.; Lv, H.; Wang, S.; Zhu, X.; Zhao, Y.; Wang, Y.; Ma, Z.; et al. Automated retinal boundary segmentation of optical coherence tomography images using an improved Canny operator. Sci. Rep. 2022, 12, 1412. [Google Scholar] [CrossRef] [PubMed]
- Schlegl, T.; Waldstein, S.M.; Bogunovic, H.; Endstraßer, F.; Sadeghipour, A.; Philip, A.M.; Podkowinski, D.; Gerendas, B.S.; Langs, G.; Schmidt-Erfurth, U. Fully Automated Detection and Quantification of Macular Fluid in OCT Using Deep Learning. Ophthalmology 2018, 125, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Camacho, P.; Dutra-Medeiros, M.; Salgueiro, L.; Sadio, S.; Rosa, P.C. Manual Segmentation of 12 Layers of the Retina and Choroid through SD-OCT in Intermediate AMD: Repeatability and Reproducibility. J. Ophthalmic Vis. Res. 2021, 16, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, O.; Tennant, M.; Grewal, P.; Rubin, U.; Seamone, M. Artificial intelligence and machine learning in ophthalmology: A review. Indian. J. Ophthalmol. 2023, 71, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Acton, J.H.; Smith, R.T.; Hood, D.C.; Greenstein, V.C. Relationship between retinal layer thickness and the visual field in early age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2012, 53, 7618–7624. [Google Scholar] [CrossRef]
- Bjerager, J.; Schneider, M.; Potapenko, I.; van Dijk, E.H.C.; Faber, C.; Grauslund, J.; Pfau, K.; Huemer, J.; Muttuvelu, D.V.; Rasmussen, M.L.R.; et al. Diagnostic Accuracy of the Amsler Grid Test for Detecting Neovascular Age-Related Macular Degeneration: A Systematic Review and Meta-analysis. JAMA Ophthalmol. 2023, 141, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Finger, R.P.; Schmitz-Valckenberg, S.; Schmid, M.; Rubin, G.S.; Dunbar, H.; Tufail, A.; Crabb, D.P.; Binns, A.; Sánchez, C.I.; Margaron, P.; et al. MACUSTAR: Development and Clinical Validation of Functional, Structural, and Patient-Reported Endpoints in Intermediate Age-Related Macular Degeneration. Ophthalmologica 2019, 241, 61–72. [Google Scholar] [CrossRef]
- Taylor, D.J.; Hobby, A.E.; Binns, A.M.; Crabb, D.P. How does age-related macular degeneration affect real-world visual ability and quality of life? A systematic review. BMJ Open 2016, 6, e011504. [Google Scholar] [CrossRef]
- Chapman, N.A.; Jacobs, R.J.; Braakhuis, A.J. Role of diet and food intake in age-related macular degeneration: A systematic review. Clin. Exp. Ophthalmol. 2019, 47, 106–127. [Google Scholar] [CrossRef]
- Chew, E.Y. Complement inhibitors for the treatment of geographic atrophy. Lancet 2023, 402, 1396–1398. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.M.; Kaiser, P.K.; Michels, M.; Soubrane, G.; Heier, J.S.; Kim, R.Y.; Sy, J.P.; Schneider, S. Ranibizumab versus verteporfin for neovascular age-related macular degeneration. N. Engl. J. Med. 2006, 355, 1432–1444. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.F.; Maguire, M.G.; Ying, G.S.; Grunwald, J.E.; Fine, S.L.; Jaffe, G.J. Ranibizumab and bevacizumab for neovascular age-related macular degeneration. N. Engl. J. Med. 2011, 364, 1897–1908. [Google Scholar] [CrossRef] [PubMed]
- Alex, V.; Motevasseli, T.; Freeman, W.R.; Jayamon, J.A.; Bartsch, D.G.; Borooah, S. Assessing the validity of a cross-platform retinal image segmentation tool in normal and diseased retina. Sci. Rep. 2021, 11, 21784. [Google Scholar] [CrossRef] [PubMed]
- Pawloff, M.; Gerendas, B.S.; Deak, G.; Bogunovic, H.; Gruber, A.; Schmidt-Erfurth, U. Performance of retinal fluid monitoring in OCT imaging by automated deep learning versus human expert grading in neovascular AMD. Eye 2023, 37, 3793–3800. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, I.; de Sisternes, L.; Hallak, J.A.; Leng, T.; Osborne, A.; Rosenfeld, P.J.; Gregori, G.; Durbin, M.; Rubin, D. Prediction of age-related macular degeneration disease using a sequential deep learning approach on longitudinal SD-OCT imaging biomarkers. Sci. Rep. 2020, 10, 15434. [Google Scholar] [CrossRef]
- Russakoff, D.B.; Lamin, A.; Oakley, J.D.; Dubis, A.M.; Sivaprasad, S. Deep Learning for Prediction of AMD Progression: A Pilot Study. Investig. Ophthalmol. Vis. Sci. 2019, 60, 712–722. [Google Scholar] [CrossRef]
- Midena, E.; Toto, L.; Frizziero, L.; Covello, G.; Torresin, T.; Midena, G.; Danieli, L.; Pilotto, E.; Figus, M.; Mariotti, C.; et al. Validation of an Automated Artificial Intelligence Algorithm for the Quantification of Major OCT Parameters in Diabetic Macular Edema. J. Clin. Med. 2023, 12, 2134. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.; Han, J.; Yoon, J.; Park, J.I.; Hwang, J.S.; Han, J.M.; Park, K.H.; Hwang, D.D. Assessing central serous chorioretinopathy with deep learning and multiple optical coherence tomography images. Sci. Rep. 2022, 12, 1831. [Google Scholar] [CrossRef]
- Jin, K.; Yan, Y.; Wang, S.; Yang, C.; Chen, M.; Liu, X.; Terasaki, H.; Yeo, T.H.; Singh, N.G.; Wang, Y.; et al. iERM: An Interpretable Deep Learning System to Classify Epiretinal Membrane for Different Optical Coherence Tomography Devices: A Multi-Center Analysis. J. Clin. Med. 2023, 12, 400. [Google Scholar] [CrossRef]
- Mariottoni, E.B.; Datta, S.; Shigueoka, L.S.; Jammal, A.A.; Tavares, I.M.; Henao, R.; Carin, L.; Medeiros, F.A. Deep Learning-Assisted Detection of Glaucoma Progression in Spectral-Domain OCT. Ophthalmol. Glaucoma 2023, 6, 228–238. [Google Scholar] [CrossRef] [PubMed]

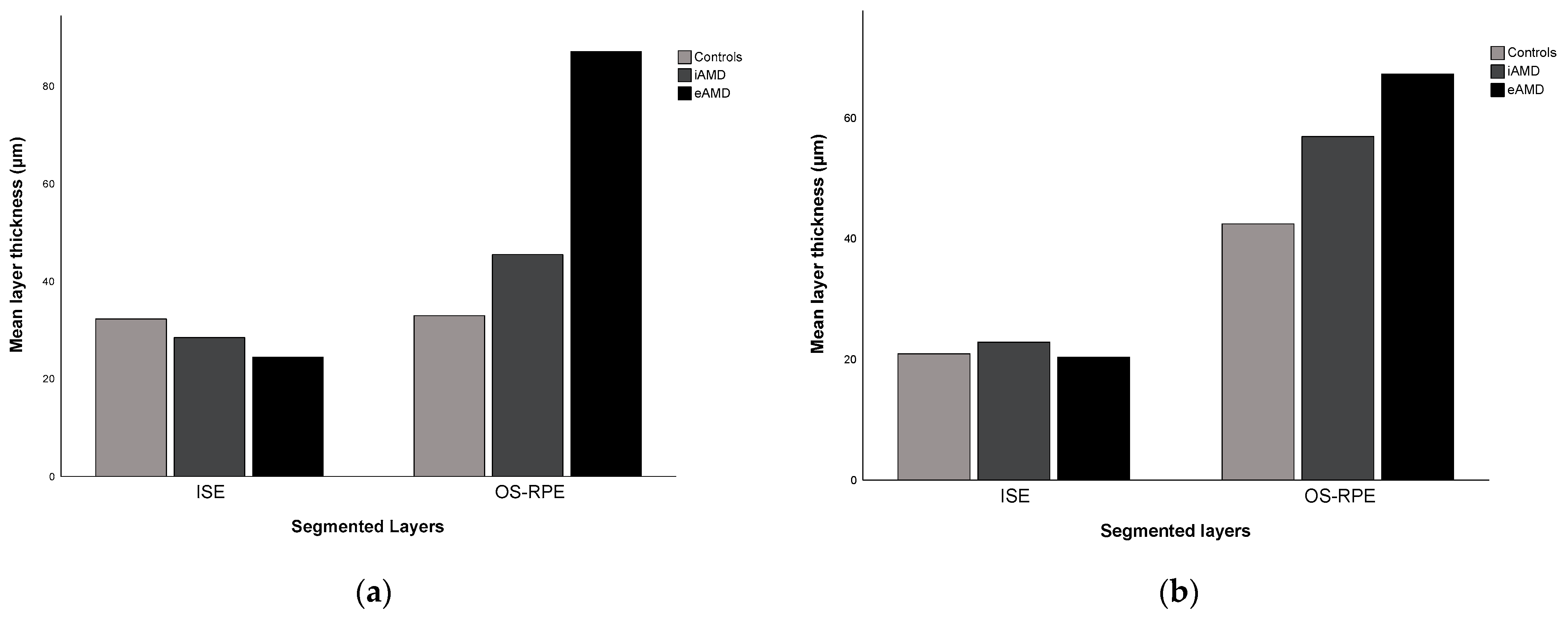
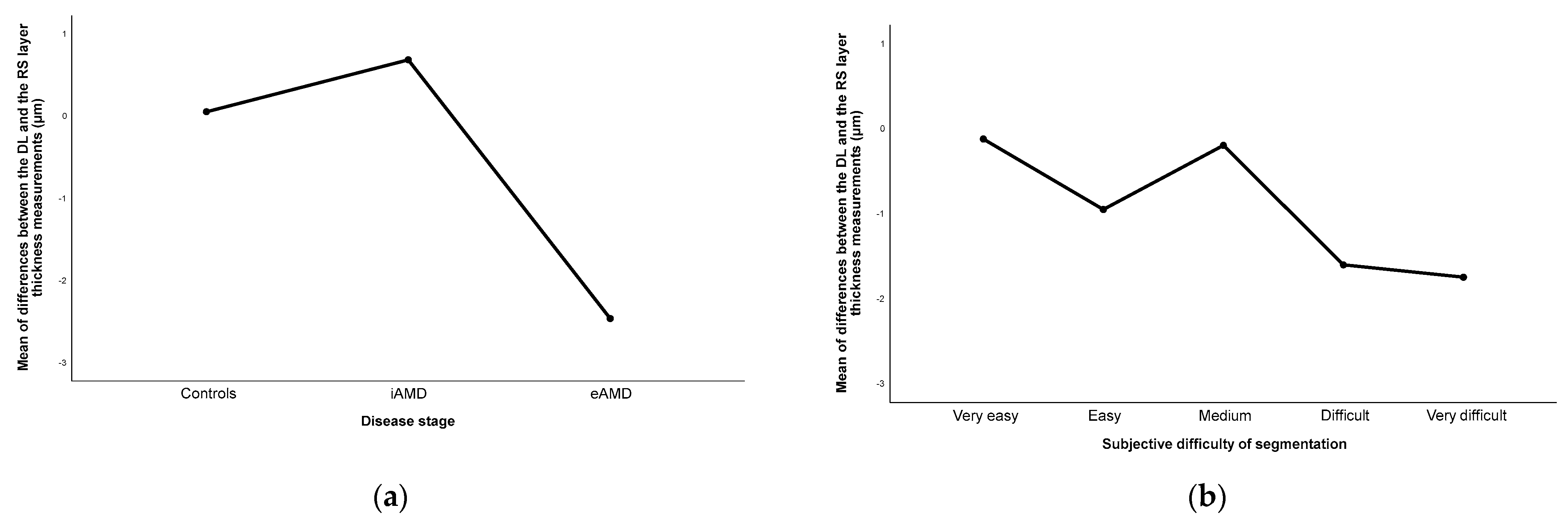
| Reference Standard (Mean ± SD) | DL Software (Mean ± SD) | Paired t-Test (p Value) | Pearson Correlation Coefficient (r) | Dice Coefficient | |
|---|---|---|---|---|---|
| Healthy Controls (n = 20) | |||||
| NFL | 20.10 ± 10.61 | 18.23 ± 11.24 | <0.001 | 0.924 | 0.989 |
| GCL–IPL | 71.43 ± 23.32 | 70.78 ± 24.40 | 0.101 | 0.956 | 0.995 |
| INL | 33.00 ± 10.21 | 32.00 ± 11.38 | <0.001 | 0.980 | 0.995 |
| OPL | 27.47 ± 9.55 | 26.21 ± 8.88 | <0.001 | 0.971 | 0.978 |
| ONL–ISM | 89.82 ± 20.20 | 96.83 ± 20.22 | <0.001 | 0.957 | 0.962 |
| ISE | 32.28 ± 5.29 | 20.91 ± 3.74 | <0.001 | 0.395 ⴕ | 0.783 |
| OS–RPE | 32.96 ± 3.78 | 42.43 ± 3.18 | <0.001 | 0.227 ⴕ | 0.928 |
| iAMD (n = 20) | |||||
| NFL | 21.96 ± 12.39 | 19.91 ± 12.61 | <0.001 | 0.382 ⴕ | 0.978 |
| GCL–IPL | 71.11 ± 23.28 | 72.03 ± 26.46 | 0.711 | 0.496 | 0.972 |
| INL | 33.00 ± 10.07 | 29.14 ± 11.25 | <0.001 | 0.952 | 0.951 |
| OPL | 27.24 ± 9.37 | 32.33 ± 11.34 | 0.002 | 0.695 | 0.923 |
| ONL–ISM | 89.49 ± 21.93 | 88.34 ± 22.96 | 0.274 | 0.930 | 0.989 |
| ISE | 28.48 ± 6.01 | 22.84 ± 6.83 | <0.001 | 0.148 ⴕ | 0.881 |
| OS–RPE | 45.46 ± 21.15 | 56.91 ± 23.94 | <0.001 | 0.477 | 0.927 |
| eAMD (n = 20) | |||||
| NFL | 22.08 ± 12.71 | 19.54 ± 12.69 | <0.001 | 0.814 | 0.938 |
| GCL–IPL | 69.96 ± 25.27 | 77.76 ± 29.75 | 0.019 | −0.061 ⴕ | 0.964 |
| INL | 34.64 ± 13.01 | 30.06 ± 12.44 | 0.002 | 0.388 ⴕ | 0.938 |
| OPL | 31.92 ± 12.33 | 28.29 ± 15.53 | 0.003 | 0.645 | 0.919 |
| ONL–ISM | 71.26 ± 20.54 | 80.82 ± 24.27 | <0.001 | 0.889 | 0.922 |
| ISE | 24.42 ± 7.61 | 20.36 ± 18.32 | 0.003 | 0.781 | 0.878 |
| OS–RPE | 87.08 ± 62.10 | 67.28 ± 47.26 | <0.001 | 0.733 | 0.992 |
| Fluid | 0.125 * | 0.127 * | 0.192 | 0.999 | 0.976 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miranda, M.; Santos-Oliveira, J.; Mendonça, A.M.; Sousa, V.; Melo, T.; Carneiro, Â. Human versus Artificial Intelligence: Validation of a Deep Learning Model for Retinal Layer and Fluid Segmentation in Optical Coherence Tomography Images from Patients with Age-Related Macular Degeneration. Diagnostics 2024, 14, 975. https://doi.org/10.3390/diagnostics14100975
Miranda M, Santos-Oliveira J, Mendonça AM, Sousa V, Melo T, Carneiro Â. Human versus Artificial Intelligence: Validation of a Deep Learning Model for Retinal Layer and Fluid Segmentation in Optical Coherence Tomography Images from Patients with Age-Related Macular Degeneration. Diagnostics. 2024; 14(10):975. https://doi.org/10.3390/diagnostics14100975
Chicago/Turabian StyleMiranda, Mariana, Joana Santos-Oliveira, Ana Maria Mendonça, Vânia Sousa, Tânia Melo, and Ângela Carneiro. 2024. "Human versus Artificial Intelligence: Validation of a Deep Learning Model for Retinal Layer and Fluid Segmentation in Optical Coherence Tomography Images from Patients with Age-Related Macular Degeneration" Diagnostics 14, no. 10: 975. https://doi.org/10.3390/diagnostics14100975
APA StyleMiranda, M., Santos-Oliveira, J., Mendonça, A. M., Sousa, V., Melo, T., & Carneiro, Â. (2024). Human versus Artificial Intelligence: Validation of a Deep Learning Model for Retinal Layer and Fluid Segmentation in Optical Coherence Tomography Images from Patients with Age-Related Macular Degeneration. Diagnostics, 14(10), 975. https://doi.org/10.3390/diagnostics14100975






