Artificial Intelligence Models for the Detection of Microsatellite Instability from Whole-Slide Imaging of Colorectal Cancer
Abstract
1. Introduction
2. The Ongoing Road to Digitalization of Histopathology
2.1. Pathologists’ Role in Identifying Histological Predicts of Microsatellite Instability in CRC
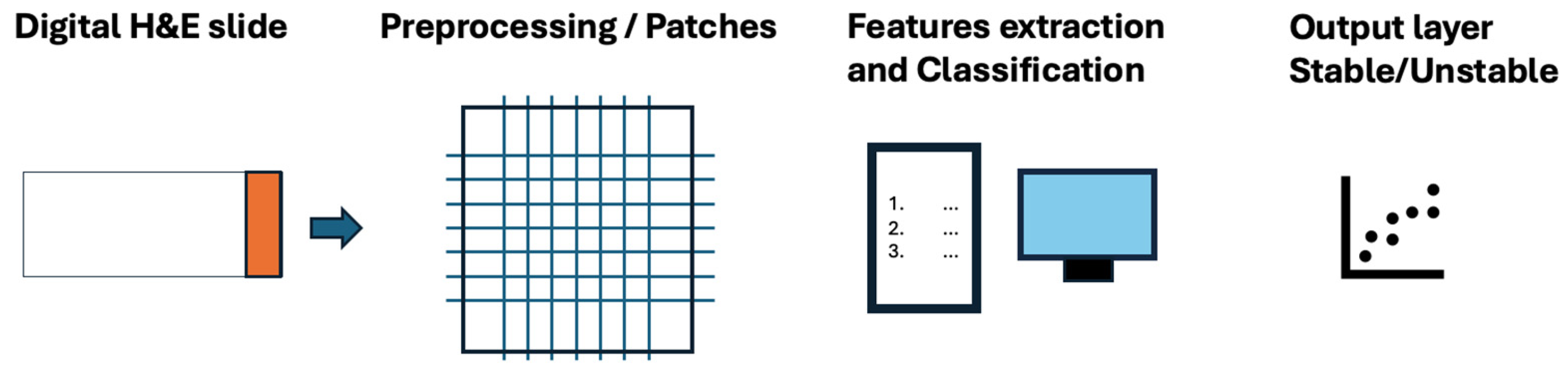
2.2. The Role of Deep Learning Models for the Identification of the MSI Status
2.3. Algorithm Sharing: New Approaches to Decentralize Artificial Intelligence in Histopathology
2.4. Bias and Limitations of Deep Learning Approach to Histopathology
3. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Faa, G.; Castagnola, M.; Didaci, L.; Coghe, F.; Scartozzi, M.; Saba, L.; Fraschini, M. The Quest for the Application of Artificial Intelligence to Whole Slide Imaging: Unique Prospective from New Advanced Tools. Algorithms 2024, 17, 254. [Google Scholar] [CrossRef]
- Kumar, N.; Gupta, R.; Gupta, S. Whole Slide Imaging (WSI) in Pathology: Current Perspectives and Future Directions. J. Digit. Imaging 2020, 33, 1034–1040. [Google Scholar] [CrossRef] [PubMed]
- Fell, C.; Mohammadi, M.; Morrison, D.; Arandjelovic, O.; Caie, P.; Harris-Birtill, D. Reproducibility of Deep Learning in Digital Pathology Whole Slide Image Analysis. PLoS Digit. Health 2022, 1, e0000145. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.C.; Bodmer, W.F. Genomic Landscape of Colorectal Carcinogenesis. J. Cancer Res. Clin. Oncol. 2022, 148, 533–545. [Google Scholar] [CrossRef]
- Kawakami, H.; Zaanan, A.; Sinicrope, F.A. Microsatellite Instability Testing and Its Role in the Management of Colorectal Cancer. Curr. Treat. Options Oncol. 2015, 16, 30. [Google Scholar] [CrossRef] [PubMed]
- Le Dung, T.; Uram Jennifer, N.; Hao, W.; Bartlett Bjarne, R.; Holly, K.; Eyring Aleksandra, D.; Skora Andrew, D.; Luber Brandon, S.; Azad Nilofer, S.; Dan, L.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Overman, M.J.; McDermott, R.; Leach, J.L.; Lonardi, S.; Lenz, H.-J.; Morse, M.A.; Desai, J.; Hill, A.; Axelson, M.; Moss, R.A.; et al. Nivolumab in Patients with Metastatic DNA Mismatch Repair-Deficient or Microsatellite Instability-High Colorectal Cancer (CheckMate 142): An Open-Label, Multicentre, Phase 2 Study. Lancet Oncol. 2017, 18, 1182–1191. [Google Scholar] [CrossRef] [PubMed]
- Rubenstein, J.H.; Enns, R.; Heidelbaugh, J.; Barkun, A.; Clinical Guidelines Committee. American Gastroenterological Association Institute Guideline on the Diagnosis and Management of Lynch Syndrome. Gastroenterology 2015, 149, 777–782, quiz e16–e17. [Google Scholar] [CrossRef]
- Bonadona, V.; Bonaïti, B.; Olschwang, S.; Grandjouan, S.; Huiart, L.; Longy, M.; Guimbaud, R.; Buecher, B.; Bignon, Y.-J.; Caron, O.; et al. Cancer Risks Associated with Germline Mutations in MLH1, MSH2, and MSH6 Genes in Lynch Syndrome. JAMA 2011, 305, 2304–2310. [Google Scholar] [CrossRef]
- Saldanha, O.L.; Quirke, P.; West, N.P.; James, J.A.; Loughrey, M.B.; Grabsch, H.I.; Salto-Tellez, M.; Alwers, E.; Cifci, D.; Ghaffari Laleh, N.; et al. Swarm Learning for Decentralized Artificial Intelligence in Cancer Histopathology. Nat. Med. 2022, 28, 1232–1239. [Google Scholar] [CrossRef]
- Eriksson, J.; Amonkar, M.; Al-Jassar, G.; Lambert, J.; Malmenäs, M.; Chase, M.; Sun, L.; Kollmar, L.; Vichnin, M. Mismatch Repair/Microsatellite Instability Testing Practices among US Physicians Treating Patients with Advanced/Metastatic Colorectal Cancer. J. Clin. Med. 2019, 8, 558. [Google Scholar] [CrossRef] [PubMed]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Werneck Krauss Silva, V.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-Grade Computational Pathology Using Weakly Supervised Deep Learning on Whole Slide Images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Greenson, J.K.; Bonner, J.D.; Ben-Yzhak, O.; Cohen, H.I.; Miselevich, I.; Resnick, M.B.; Trougouboff, P.; Tomsho, L.D.; Kim, E.; Low, M.; et al. Phenotype of Microsatellite Unstable Colorectal Carcinomas: Well-Differentiated and Focally Mucinous Tumors and the Absence of Dirty Necrosis Correlate with Microsatellite Instability. Am. J. Surg. Pathol. 2003, 27, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Mitrovic, B.; Schaeffer, D.F.; Riddell, R.H.; Kirsch, R. Tumor Budding in Colorectal Carcinoma: Time to Take Notice. Mod. Pathol. 2012, 25, 1315–1325. [Google Scholar] [CrossRef] [PubMed]
- Smyrk, T.C.; Watson, P.; Kaul, K.; Lynch, H.T. Tumor-Infiltrating Lymphocytes Are a Marker for Microsatellite Instability in Colorectal Carcinoma. Cancer 2001, 91, 2417–2422. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.-F.; Arbman, G.; Wadhra, T.I.; Zhang, H.; Sun, X.-F. Relationships of Tumor Inflammatory Infiltration and Necrosis with Microsatellite Instability in Colorectal Cancers. World J. Gastroenterol. 2005, 11, 2179–2183. [Google Scholar] [CrossRef] [PubMed]
- Gologan, A.; Krasinskas, A.; Hunt, J.; Thull, D.L.; Farkas, L.; Sepulveda, A.R. Performance of the Revised Bethesda Guidelines for Identification of Colorectal Carcinomas with a High Level of Microsatellite Instability. Arch. Pathol. Lab. Med. 2005, 129, 1390–1397. [Google Scholar] [CrossRef] [PubMed]
- Halvarsson, B.; Anderson, H.; Domanska, K.; Lindmark, G.; Nilbert, M. Clinicopathologic Factors Identify Sporadic Mismatch Repair-Defective Colon Cancers. Am. J. Clin. Pathol. 2008, 129, 238–244. [Google Scholar] [CrossRef]
- Greenson, J.K.; Huang, S.-C.; Herron, C.; Moreno, V.; Bonner, J.D.; Tomsho, L.P.; Ben-Izhak, O.; Cohen, H.I.; Trougouboff, P.; Bejhar, J.; et al. Pathologic Predictors of Microsatellite Instability in Colorectal Cancer. Am. J. Surg. Pathol. 2009, 33, 126–133. [Google Scholar] [CrossRef]
- Brazowski, E.; Rozen, P.; Pel, S.; Samuel, Z.; Solar, I.; Rosner, G. Can a Gastrointestinal Pathologist Identify Microsatellite Instability in Colorectal Cancer with Reproducibility and a High Degree of Specificity? Fam. Cancer 2012, 11, 249–257. [Google Scholar] [CrossRef]
- Shia, J.; Schultz, N.; Kuk, D.; Vakiani, E.; Middha, S.; Segal, N.H.; Hechtman, J.F.; Berger, M.F.; Stadler, Z.K.; Weiser, M.R.; et al. Morphological Characterization of Colorectal Cancers in The Cancer Genome Atlas Reveals Distinct Morphology-Molecular Associations: Clinical and Biological Implications. Mod. Pathol. 2017, 30, 599–609. [Google Scholar] [CrossRef]
- Yamashita, R.; Long, J.; Longacre, T.; Peng, L.; Berry, G.; Martin, B.; Higgins, J.; Rubin, D.L.; Shen, J. Deep Learning Model for the Prediction of Microsatellite Instability in Colorectal Cancer: A Diagnostic Study. Lancet Oncol. 2021, 22, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Hildebrand, L.A.; Pierce, C.J.; Dennis, M.; Paracha, M.; Maoz, A. Artificial Intelligence for Histology-Based Detection of Microsatellite Instability and Prediction of Response to Immunotherapy in Colorectal Cancer. Cancers 2021, 13, 391. [Google Scholar] [CrossRef]
- Kuntz, S.; Krieghoff-Henning, E.; Kather, J.N.; Jutzi, T.; Höhn, J.; Kiehl, L.; Hekler, A.; Alwers, E.; von Kalle, C.; Fröhling, S.; et al. Gastrointestinal Cancer Classification and Prognostication from Histology Using Deep Learning: Systematic Review. Eur. J. Cancer 2021, 155, 200–215. [Google Scholar] [CrossRef] [PubMed]
- Guitton, T.; Allaume, P.; Rabilloud, N.; Rioux-Leclercq, N.; Henno, S.; Turlin, B.; Galibert-Anne, M.-D.; Lièvre, A.; Lespagnol, A.; Pécot, T.; et al. Artificial Intelligence in Predicting Microsatellite Instability and KRAS, BRAF Mutations from Whole-Slide Images in Colorectal Cancer: A Systematic Review. Diagnostics 2024, 14, 99. [Google Scholar] [CrossRef]
- Kather, J.N.; Pearson, A.T.; Halama, N.; Jäger, D.; Krause, J.; Loosen, S.H.; Marx, A.; Boor, P.; Tacke, F.; Neumann, U.P.; et al. Deep Learning Can Predict Microsatellite Instability Directly from Histology in Gastrointestinal Cancer. Nat. Med. 2019, 25, 1054–1056. [Google Scholar] [CrossRef] [PubMed]
- Echle, A.; Grabsch, H.I.; Quirke, P.; van den Brandt, P.A.; West, N.P.; Hutchins, G.G.A.; Heij, L.R.; Tan, X.; Richman, S.D.; Krause, J.; et al. Clinical-Grade Detection of Microsatellite Instability in Colorectal Tumors by Deep Learning. Gastroenterology 2020, 159, 1406–1416.e11. [Google Scholar] [CrossRef]
- Echle, A.; Ghaffari Laleh, N.; Quirke, P.; Grabsch, H.I.; Muti, H.S.; Saldanha, O.L.; Brockmoeller, S.F.; van den Brandt, P.A.; Hutchins, G.G.A.; Richman, S.D.; et al. Artificial Intelligence for Detection of Microsatellite Instability in Colorectal Cancer-a Multicentric Analysis of a Pre-Screening Tool for Clinical Application. ESMO Open 2022, 7, 100400. [Google Scholar] [CrossRef] [PubMed]
- Bilal, M.; Raza, S.E.A.; Azam, A.; Graham, S.; Ilyas, M.; Cree, I.A.; Snead, D.; Minhas, F.; Rajpoot, N.M. Development and Validation of a Weakly Supervised Deep Learning Framework to Predict the Status of Molecular Pathways and Key Mutations in Colorectal Cancer from Routine Histology Images: A Retrospective Study. Lancet Digit. Health 2021, 3, e763–e772. [Google Scholar] [CrossRef]
- Schirris, Y.; Gavves, E.; Nederlof, I.; Horlings, H.M.; Teuwen, J. DeepSMILE: Contrastive Self-Supervised Pre-Training Benefits MSI and HRD Classification Directly from H&E Whole-Slide Images in Colorectal and Breast Cancer. Med. Image Anal. 2022, 79, 102464. [Google Scholar] [CrossRef]
- Niehues, J.M.; Quirke, P.; West, N.P.; Grabsch, H.I.; van Treeck, M.; Schirris, Y.; Veldhuizen, G.P.; Hutchins, G.G.A.; Richman, S.D.; Foersch, S.; et al. Generalizable Biomarker Prediction from Cancer Pathology Slides with Self-Supervised Deep Learning: A Retrospective Multi-Centric Study. Cell Rep. Med. 2023, 4, 100980. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Mei, W.-J.; Xu, S.-Y.; Ling, Y.-H.; Li, W.-R.; Kuang, J.-B.; Li, H.-S.; Hui, H.; Li, J.-B.; Cai, M.-Y.; et al. Clinical Actionability of Triaging DNA Mismatch Repair Deficient Colorectal Cancer from Biopsy Samples Using Deep Learning. eBioMedicine 2022, 81, 104120. [Google Scholar] [CrossRef] [PubMed]
- Saillard, C.; Dubois, R.; Tchita, O.; Loiseau, N.; Garcia, T.; Adriansen, A.; Carpentier, S.; Reyre, J.; Enea, D.; von Loga, K.; et al. Validation of MSIntuit as an AI-Based Pre-Screening Tool for MSI Detection from Colorectal Cancer Histology Slides. Nat. Commun. 2023, 14, 6695. [Google Scholar] [CrossRef] [PubMed]
- Gerwert, K.; Schörner, S.; Großerueschkamp, F.; Kraeft, A.; Schuhmacher, D.; Sternemann, C.; Feder, I.S.; Wisser, S.; Lugnier, C.; Arnold, D.; et al. Fast and Label-Free Automated Detection of Microsatellite Status in Early Colon Cancer Using Artificial Intelligence Integrated Infrared Imaging. Eur. J. Cancer 2023, 182, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Macenko, M.; Niethammer, M.; Marron, J.S.; Borland, D.; Woosley, J.T.; Guan, X.; Schmitt, C.; Thomas, N.E. A Method for Normalizing Histology Slides for Quantitative Analysis. In Proceedings of the 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Boston, MA, USA, 28 June–1 July 2009; pp. 1107–1110. [Google Scholar]
- Muti, H.S.; Heij, L.R.; Keller, G.; Kohlruss, M.; Langer, R.; Dislich, B.; Cheong, J.-H.; Kim, Y.-W.; Kim, H.; Kook, M.-C.; et al. Development and Validation of Deep Learning Classifiers to Detect Epstein-Barr Virus and Microsatellite Instability Status in Gastric Cancer: A Retrospective Multicentre Cohort Study. Lancet Digit. Health 2021, 3, e654–e664. [Google Scholar] [CrossRef]
- Lu, M.Y.; Chen, R.J.; Kong, D.; Lipkova, J.; Singh, R.; Williamson, D.F.K.; Chen, T.Y.; Mahmood, F. Federated Learning for Computational Pathology on Gigapixel Whole Slide Images. Med. Image Anal. 2022, 76, 102298. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, C.; Liu, N.; Huang, H.; Zheng, Z.; Yan, Q. A Blockchain-Based Decentralized Federated Learning Framework with Committee Consensus. IEEE Netw. 2021, 35, 234–241. [Google Scholar] [CrossRef]
- Warnat-Herresthal, S.; Schultze, H.; Shastry, K.L.; Manamohan, S.; Mukherjee, S.; Garg, V.; Sarveswara, R.; Händler, K.; Pickkers, P.; Aziz, N.A.; et al. Swarm Learning for Decentralized and Confidential Clinical Machine Learning. Nature 2021, 594, 265–270. [Google Scholar] [CrossRef]
- Hägele, M.; Seegerer, P.; Lapuschkin, S.; Bockmayr, M.; Samek, W.; Klauschen, F.; Müller, K.-R.; Binder, A. Resolving Challenges in Deep Learning-Based Analyses of Histopathological Images Using Explanation Methods. Sci. Rep. 2020, 10, 6423. [Google Scholar] [CrossRef]
- Ciga, O.; Xu, T.; Nofech-Mozes, S.; Noy, S.; Lu, F.-I.; Martel, A.L. Overcoming the Limitations of Patch-Based Learning to Detect Cancer in Whole Slide Images. Sci. Rep. 2021, 11, 8894. [Google Scholar] [CrossRef]
- Chen, C.-L.; Chen, C.-C.; Yu, W.-H.; Chen, S.-H.; Chang, Y.-C.; Hsu, T.-I.; Hsiao, M.; Yeh, C.-Y.; Chen, C.-Y. An Annotation-Free Whole-Slide Training Approach to Pathological Classification of Lung Cancer Types Using Deep Learning. Nat. Commun. 2021, 12, 1193. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.A.; Abouzid, M.; Kaczmarek, E. Deep Learning Approaches in Histopathology. Cancers 2022, 14, 5264. [Google Scholar] [CrossRef]
- Ching, T.; Himmelstein, D.S.; Beaulieu-Jones, B.K.; Kalinin, A.A.; Do, B.T.; Way, G.P.; Ferrero, E.; Agapow, P.-M.; Zietz, M.; Hoffman, M.M.; et al. Opportunities and Obstacles for Deep Learning in Biology and Medicine. J. R. Soc. Interface 2018, 15, 20170387. [Google Scholar] [CrossRef] [PubMed]
- Hekler, A.; Utikal, J.S.; Enk, A.H.; Solass, W.; Schmitt, M.; Klode, J.; Schadendorf, D.; Sondermann, W.; Franklin, C.; Bestvater, F.; et al. Deep Learning Outperformed 11 Pathologists in the Classification of Histopathological Melanoma Images. Eur. J. Cancer 2019, 118, 91–96. [Google Scholar] [CrossRef]
- Park, J.H.; Kim, E.Y.; Luchini, C.; Eccher, A.; Tizaoui, K.; Shin, J.I.; Lim, B.J. Artificial Intelligence for Predicting Microsatellite Instability Based on Tumor Histomorphology: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 2462. [Google Scholar] [CrossRef] [PubMed]
| Authors | Year | AI Model |
|---|---|---|
| Kather et al. [26] | 2019 | CNN with deep residual learning (resnet18) |
| Yamashita et al. [22] | 2021 | MSINet: a modified MobileNetV2 architecture |
| Echle et al. [27] | 2020 | A modified ShuffleNet deep learning system |
| Echle et al. [28] | 2022 | ResNet18 neural network model |
| Bilal et al. [29] | 2021 | Model 1 (ResNet18) and model 2 (adapted ResNet34) |
| Schirris et al. [30] | 2022 | Self-supervised pre-training and feature variability-aware deep multiple instance learning |
| Niehues et al. [31] | 2023 | Compared six different DL architectures to predict biomarkers from pathology slides |
| Jiang et al. [32] | 2022 | Densenet121 integrated with focal loss |
| Saillard et al. [33] | 2023 | A variant of the Chowder model |
| Gerwert et al. [34] | 2022 | CompSegNet and VGG 11 Neural Network |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Faa, G.; Coghe, F.; Pretta, A.; Castagnola, M.; Van Eyken, P.; Saba, L.; Scartozzi, M.; Fraschini, M. Artificial Intelligence Models for the Detection of Microsatellite Instability from Whole-Slide Imaging of Colorectal Cancer. Diagnostics 2024, 14, 1605. https://doi.org/10.3390/diagnostics14151605
Faa G, Coghe F, Pretta A, Castagnola M, Van Eyken P, Saba L, Scartozzi M, Fraschini M. Artificial Intelligence Models for the Detection of Microsatellite Instability from Whole-Slide Imaging of Colorectal Cancer. Diagnostics. 2024; 14(15):1605. https://doi.org/10.3390/diagnostics14151605
Chicago/Turabian StyleFaa, Gavino, Ferdinando Coghe, Andrea Pretta, Massimo Castagnola, Peter Van Eyken, Luca Saba, Mario Scartozzi, and Matteo Fraschini. 2024. "Artificial Intelligence Models for the Detection of Microsatellite Instability from Whole-Slide Imaging of Colorectal Cancer" Diagnostics 14, no. 15: 1605. https://doi.org/10.3390/diagnostics14151605
APA StyleFaa, G., Coghe, F., Pretta, A., Castagnola, M., Van Eyken, P., Saba, L., Scartozzi, M., & Fraschini, M. (2024). Artificial Intelligence Models for the Detection of Microsatellite Instability from Whole-Slide Imaging of Colorectal Cancer. Diagnostics, 14(15), 1605. https://doi.org/10.3390/diagnostics14151605









