Evaluation of Open-Source Ciliary Analysis Software in Primary Ciliary Dyskinesia: A Comparative Assessment
Abstract
1. Introduction
2. Materials and Methods
2.1. Subjects
2.2. Sample Collection and Preparation
2.3. Highspeed Video Microscopy Analysis (HSVA)
2.4. Manual Method for CBF Count
2.5. Software Analysis
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hannah, W.B.; Seifert, B.A.; Truty, R.; Zariwala, M.A.; Ameel, K.; Zhao, Y.; Nykamp, K.; Gaston, B. The global prevalence and ethnic heterogeneity of primary ciliary dyskinesia gene variants: A genetic database analysis. Lancet Respir. Med. 2022, 10, 459–468. [Google Scholar] [CrossRef]
- Knowles, M.R.; Daniels, L.A.; Davis, S.D.; Zariwala, M.A.; Leigh, M.W. Primary ciliary dyskinesia. Recent advances in diagnostics, genetics, and characterization of clinical disease. Am. J. Respir. Crit. Care Med. 2013, 188, 913–922. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, M.G.; Mosquera, R.; Metjian, H.; Marmor, M.; Olivier, K.N.; Shapiro, A.J. Primary Ciliary Dyskinesia. CHEST Pulm. 2023, 1, 100004. [Google Scholar] [CrossRef]
- Nawroth, J.C.; Van Der Does, A.M.; Ryan, A.; Kanso, E. Multiscale mechanics of mucociliary clearance in the lung. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020, 375, 20190160. [Google Scholar] [CrossRef]
- Shapiro, A.J.; Zariwala, M.A.; Ferkol, T.; Davis, S.D.; Sagel, S.D.; Dell, S.D.; Rosenfeld, M.; Olivier, K.N.; Milla, C.; Daniel, S.J.; et al. Diagnosis, monitoring, and treatment of primary ciliary dyskinesia: PCD foundation consensus recommendations based on state of the art review. Pediatr. Pulmonol. 2016, 51, 115–132. [Google Scholar] [CrossRef]
- Kuehni, C.E.; Frischer, T.; Strippoli, M.F.; Maurer, E.; Bush, A.; Nielsen, K.G.; Escribano, A.; Lucas, J.S.; Yiallouros, P.; Omran, H.; et al. Factors influencing age at diagnosis of primary ciliary dyskinesia in European children. Eur. Respir. J. 2010, 36, 1248–1258. [Google Scholar] [CrossRef] [PubMed]
- Bricmont, N.; Alexandru, M.; Louis, B.; Papon, J.F.; Kempeneers, C. Ciliary Videomicroscopy: A Long Beat from the European Respiratory Society Guidelines to the Recognition as a Confirmatory Test for Primary Ciliary Dyskinesia. Diagnostics 2021, 11, 1700. [Google Scholar] [CrossRef]
- Lucas, J.S. Diagnosis of primary ciliary dyskinesia: Searching for a gold standard. Eur. Respir. J. 2014, 44, 1418–1422. [Google Scholar] [CrossRef]
- Shoemark, A.; Dell, S.; Shapiro, A.; Lucas, J.S. ERS and ATS diagnostic guidelines for primary ciliary dyskinesia: Similarities and differences in approach to diagnosis. Eur. Respir. J. 2019, 54, 1901066. [Google Scholar] [CrossRef]
- Kouis, P.; Yiallouros, P.K.; Middleton, N.; Evans, J.S.; Kyriacou, K.; Papatheodorou, S.I. Prevalence of primary ciliary dyskinesia in consecutive referrals of suspect cases and the transmission electron microscopy detection rate: A systematic review and meta-analysis. Pediatr. Res. 2017, 81, 398–405. [Google Scholar] [CrossRef]
- Zariwala, M.A.; Knowles, M.R.; Leigh, M.W. Primary Ciliary Dyskinesia. GeneReviews®. 2019. Available online: http://europepmc.org/books/NBK1122 (accessed on 18 August 2023).
- Rumman, N.; Jackson, C.; Collins, S.; Goggin, P.; Coles, J.; Lucas, J.S. Diagnosis of primary ciliary dyskinesia: Potential options for resource-limited countries. Eur. Respir. Rev. 2017, 26, 160058. [Google Scholar] [CrossRef] [PubMed]
- Lucas, J.S.; Barbato, A.; Collins, S.A.; Goutaki, M.; Behan, L.; Caudri, D.; Dell, S.; Eber, E.; Escudier, E.; Hirst, R.A.; et al. European Respiratory Society guidelines for the diagnosis of primary ciliary dyskinesia. Eur. Respir. J. 2017, 49, 1601090. [Google Scholar] [CrossRef]
- Shapiro, A.J.; Davis, S.D.; Polineni, D.; Manion, M.; Rosenfeld, M.; Dell, S.D.; Chilvers, M.A.; Ferkol, T.W.; Zariwala, M.A.; Dagel, S.D.; et al. Diagnosis of primary ciliary dyskinesia: An official American thoracic society clinical practice guideline. Am. J. Respir. Crit. Care Med. 2018, 197, e24–e39. [Google Scholar] [CrossRef] [PubMed]
- Lucas, J.S.; Paff, T.; Goggin, P.; Haarman, E. Diagnostic Methods in Primary Ciliary Dyskinesia. Paediatr. Respir. Rev. 2016, 18, 8–17. [Google Scholar] [CrossRef]
- Bush, A.; Chodhari, R.; Collins, N.; Copeland, F.; Hall, P.; Harcourt, J.; Hariri, M.; Hogg, C.; Lucas, J.; Mitchison, H.M.; et al. Primary ciliary dyskinesia: Current state of the art. Arch. Dis. Child. 2007, 92, 1136–1140. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.M.; Djakow, J.; Free, R.C.; Djakow, P.; Lonnen, R.; Williams, G.; Pohunek, P.; Hirst, R.A.; Easton, A.J.; Andrew, P.W.; et al. ciliaFA: A research tool for automated, high-throughput measurement of ciliary beat frequency using freely available software. Cilia 2012, 1, 1–7. [Google Scholar] [CrossRef]
- Sisson, J.H.; Stoner, J.A.; Ammons, B.A.; Wyatt, T.A. All-digital image capture and whole-field analysis of ciliary beat frequency. J. Microsc. 2003, 211, 103–111. [Google Scholar] [CrossRef]
- Armengot, M.; Bonet, M.; Carda, C.; Gómez, M.J.; Milara, J.; Mata, M.; Cortijo, J. Development and validation of a method of cilia motility analysis for the early diagnosis of primary ciliary dyskinesia. Acta Otorrinolaringol. Esp. 2012, 63, 1–8. [Google Scholar] [CrossRef]
- Schneiter, M.; Tschanz, S.A.; Müller, L.; Frenz, M. The Cilialyzer—A freely available open-source software for a standardised identification of impaired mucociliary activity facilitating the diagnostic testing for PCD. bioRxiv 2022, 506514. [Google Scholar] [CrossRef]
- Sampaio, P.; da Silva, M.F.; Vale, I.; Roxo-Rosa, M.; Pinto, A.; Constant, C.; Pereira, L.; Quintão, C.M.; Lopes, S.S. Ciliarmove: New software for evaluating ciliary beat frequency helps find novel mutations by a portuguese multidisciplinary team on primary ciliary dyskinesia. ERJ Open Res. 2021, 7, 1–12. [Google Scholar] [CrossRef]
- De Jesús-Rojas, W.; Meléndez-Montañez, J.; Muñiz-Hernández, J.; Marra-Nazario, A.; Alvarado-Huerta, F.; Santos-López, A.; Ramos-Benitez, M.J.; Mosquera, R.A. The RSPH4A Gene in Primary Ciliary Dyskinesia. Int. J. Mol. Sci. 2023, 24, 1936. [Google Scholar] [CrossRef]
- Müller, L.; Savas, S.T.; Tschanz, S.A.; Stokes, A.; Escher, A.; Nussbaumer, M.; Bullo, M.; Kuehni, C.E.; Blanchon, S.; Jung, A.; et al. A Comprehensive Approach for the Diagnosis of Primary Ciliary Dyskinesia-Experiences from the First 100 Patients of the PCD-UNIBE Diagnostic Center. Diagnostics 2021, 11, 1540. [Google Scholar] [CrossRef]
- Jackson, C.L.; Bottier, M. Methods for the assessment of human airway ciliary function. Eur. Respir. J. 2022, 60, 2102300. [Google Scholar] [CrossRef] [PubMed]
- Thévenaz, P.; Ruttimann, U.E.; Unser, M. A pyramid approach to subpixel registration based on intensity. IEEE Trans. Image Process. 1998, 7, 27–41. [Google Scholar] [CrossRef]
- Puybareau, E.; Talbot, H.; Bequignon, E.; Louis, B.; Pelle, G.; Papon, J.F.; Coste, A.; Najman, L. Automating the measurement of physiological parameters: A case study in the image analysis of cilia motion. In Proceedings of the 2016 IEEE International Conference on Image Processing (ICIP), IEEE, Phoenix, AZ, USA, 25–28 September 2016; pp. 1240–1244. [Google Scholar] [CrossRef]
- Puybareau, E.; Talbot, H.; Pelle, G.; Louis, B.; Papon, J.F.; Coste, A.; Najman, L. A regionalized automated measurement of ciliary beating frequency. In Proceedings of the 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), IEEE, New York, NY, USA, 16–19 April 2015; pp. 528–531. [Google Scholar] [CrossRef]
- Alton, E.W.; Armstrong, D.K.; Ashby, D.; Bayfield, K.J.; Bilton, D.; Bloomfield, E.V.; Boyd, A.C.; Brand, J.; Buchan, R.; Calcedo, R.; et al. A randomised, double-blind, placebo-controlled trial of repeated nebulisation of non-viral cystic fibrosis transmembrane conductance regulator (CFTR) gene therapy in patients with cystic fibrosis. Effic. Mech. Eval. 2016, 3, 1–210. [Google Scholar] [CrossRef] [PubMed]
- Jorissen, M.; Willems, T.; Van Der Schueren, B. Nasal ciliary beat frequency is age independent. Laryngoscope 1998, 108, 1042–1047. [Google Scholar] [CrossRef]
- Ho, J.C.; Chan, K.N.; Hu, W.H.; Lam, W.K.; Zheng, L.; Tipoe, G.L.; Sun, J.; Leung, R.; Tsang, K.W. The effect of aging on nasal mucociliary clearance, beat frequency, and ultrastructure of respiratory cilia. Am. J. Respir. Crit. Care Med. 2001, 163, 983–988. [Google Scholar] [CrossRef]
- Bailey, K.L.; Bonasera, S.J.; Wilderdyke, M.; Hanisch, B.W.; Pavlik, J.A.; DeVasure, J.; Robinson, J.E.; Sisson, J.H.; Wyatt, T.A. Aging causes a slowing in ciliary beat frequency, mediated by PKCε. Am. J. Physiol. Lung Cell. Mol. Physiol. 2014, 306, L584–L589. [Google Scholar] [CrossRef]
- Svartengren, M.; Falk, R.; Philipson, K. Long-term clearance from small airways decreases with age. Eur. Respir. J. 2005, 26, 609–615. [Google Scholar] [CrossRef]
- Rubbo, B.; Shoemark, A.; Jackson, C.L.; Hirst, R.; Thompson, J.; Hayes, J.; Frost, E.; Copeland, F.; Hogg, C.; O’Callaghan, C.; et al. Accuracy of High-Speed Video Analysis to Diagnose Primary Ciliary Dyskinesia. Chest 2019, 155, 1008–1017. [Google Scholar] [CrossRef]
- Stannard, W.A.; Chilvers, M.A.; Rutman, A.R.; Williams, C.D.; O’Callaghan, C. Diagnostic testing of patients suspected of primary ciliary dyskinesia. Am. J. Respir. Crit. Care Med. 2010, 181, 307–314. [Google Scholar] [CrossRef] [PubMed]
| Cilialyzer [20] | CiliarMove [21] | |
|---|---|---|
| Authors | Schneiter M, et al. | Sampaio P, et al. |
| Year of Publication | 2022 | 2021 |
| Purpose | Standardized identification of impaired mucociliary activity, facilitating diagnostic testing for PCD. | Evaluating CBF to aid in the correct diagnosis of PCD. |
| Innovation | Provides new quantitative information for PCD diagnostics. | Provides an open-source, fast, and intuitive tool for CBF evaluation. |
| Main Features | Semi-automated analysis of mucociliary activity captured by HSVA. | Semi-automated evaluation of CBF using HSVA. |
| Analyzes ciliated epithelial cell cluster samples, as well as intact mucociliary epithelium culture. | Analyzes CBF from respiratory epithelial cell clusters. | |
| Capable of analyzing CBF from HSVA at a rate of 500 frames per second as recommended by the European Respiratory Society (ERS). | ||
| Compute CBF within a sequence of images in greyscale by calculating the fast Fourier transform method for each pixel of a given region of interest (ROI). | ||
| Pre-processing features Sample-type application: Brushed human nasal epithelial cells immersed in medium | Manual rotation of image | Manual selection of ROI Video visualization panelIncludes play/pause feature, adjustable frame speed, and displays size of selected ROI measured in pixels. |
| Manual selection of ROI | ||
| Image stabilization | ||
| Motion extraction | ||
| Post-processing features Sample-type application: Brushed human nasal epithelial cells immersed in medium | Power spectrum Displays CBF as the peak bandwidth of | Heat map of CBF Color gradient indicates faster vs. slower frequency |
| Visual assessment Display panel includes adjustable frame speed, play/pause feature, forward/backward playback option of image sequence, zoom function, and image contrast settings. | Fast Fourier transform plot For any selected pixel | |
| Histogram of CBF distribution From the selected ROI | ||
| Table display of CBF percentages From the selected ROI | ||
| Pre/post-processing features Sample-type application: Intact cultures of mucociliary epithelium | “Pre-processing” features here include allmethods as described above. | Software not programmed to analyze intact mucociliary epithelium. |
| Power spectrum | ||
| Activity map of spatial CBF distribution | ||
| Frequency correlation length | ||
| Particle extraction | ||
| Particle tracking | ||
| Mucociliary transport speed | ||
| Validation/testing | The computational analysis methods of Cilialyzer are demonstrated using simulated and representative sample data from clinical practice. | Correlation coefficient R2 = 0.9895 when comparing CiliarMove to the manual observation method. CBF values were obtained from a healthy control group, a PCD-excluded group, and a PCD-confirmed group. |
| Availability | Freely available under the terms of the Massachusetts Institute of Technology (MIT) license. | Open-source program, but the specific license is not mentioned. |
| User-friendliness | The software stands out due to its ease of customization and extension. It requires no license or virtual machine. It provides many pre-processing and replaying options, making it easy to use. | The user-friendly interface allows less-trained users to perform the evaluation of the CBF. |
| Cost-effectiveness | The cost-effectiveness of using the Cilialyzer for PCD diagnosis has not been explicitly discussed. | Provides cost benefits by adapting to any high-speed image setup, enabling low-budget HSVA in resource-limited settings. |
| Manual Count | CiliarMove | Cilialyzer | ||||
|---|---|---|---|---|---|---|
| PCD (RSPH4A) | HC | PCD (RSPH4A) | HC | PCD (RSPH4A) | HC | |
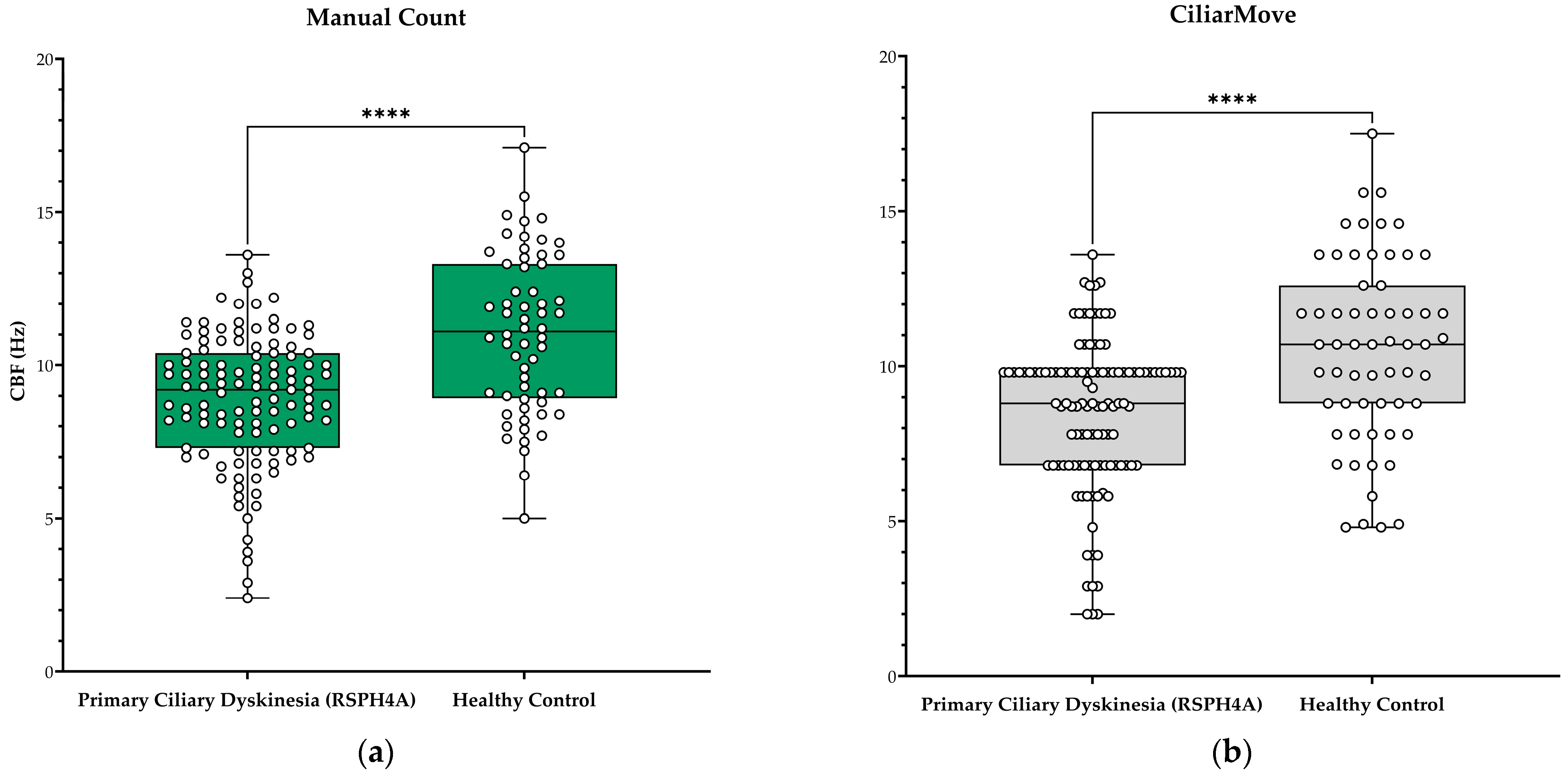
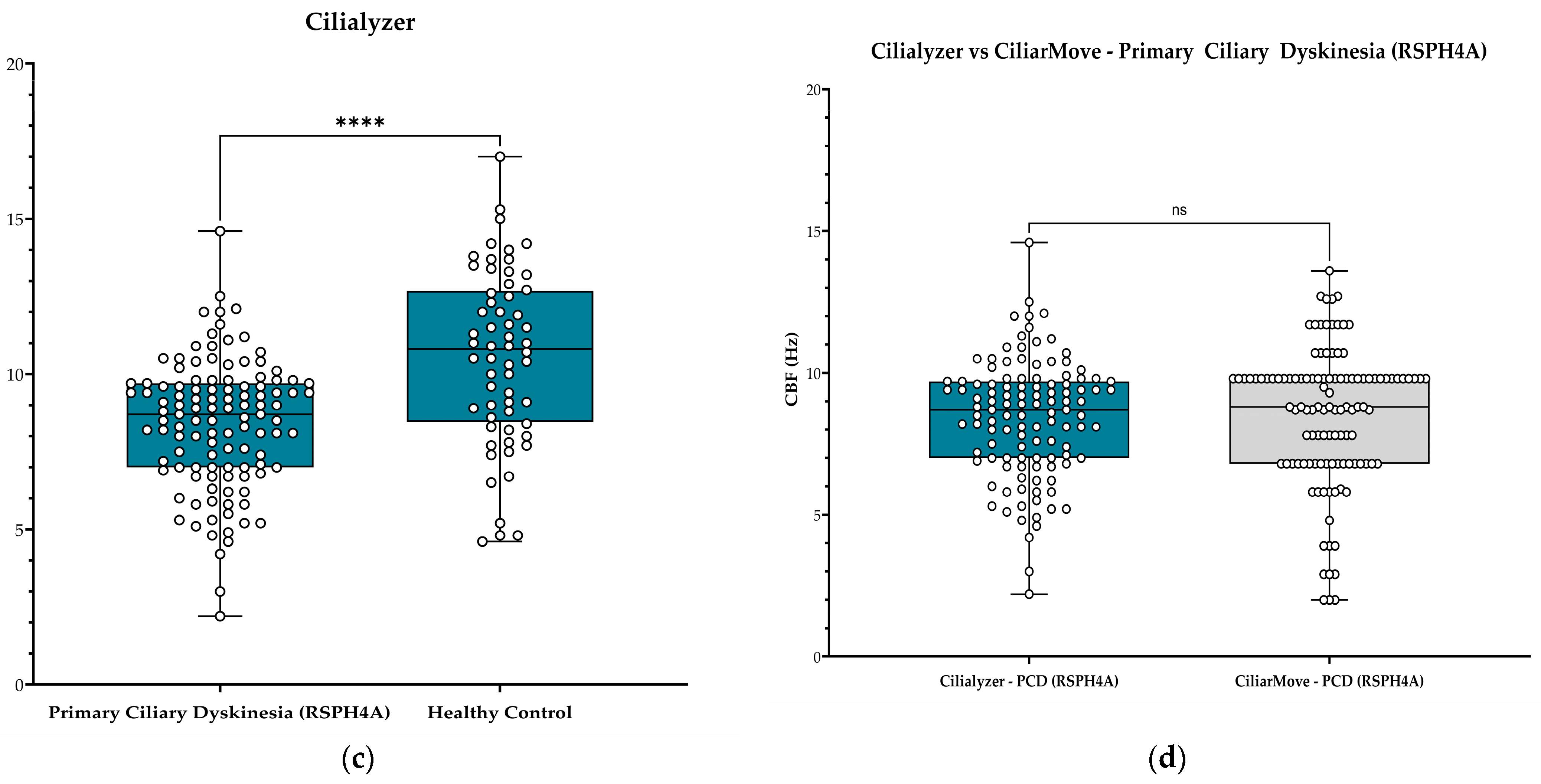
| Median CBF (Hz) | 9.2 | 11.1 | 8.8 | 10.7 | 8.7 | 10.8 |
| Actual median difference | 1.9 | 1.9 | 2.1 | |||
| p-value | ** <0.0001 | * <0.0001 | ** <0.0001 | |||
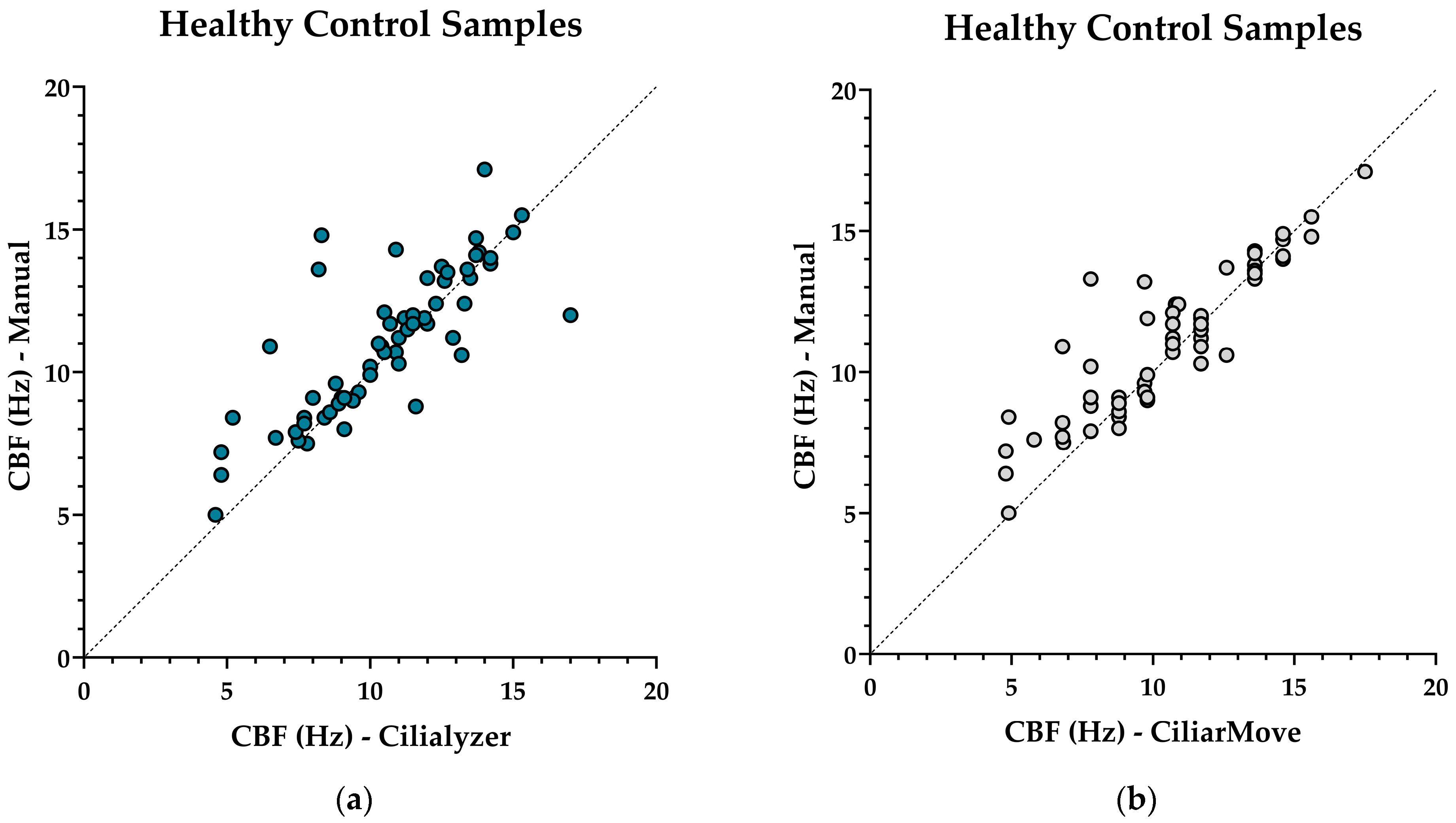
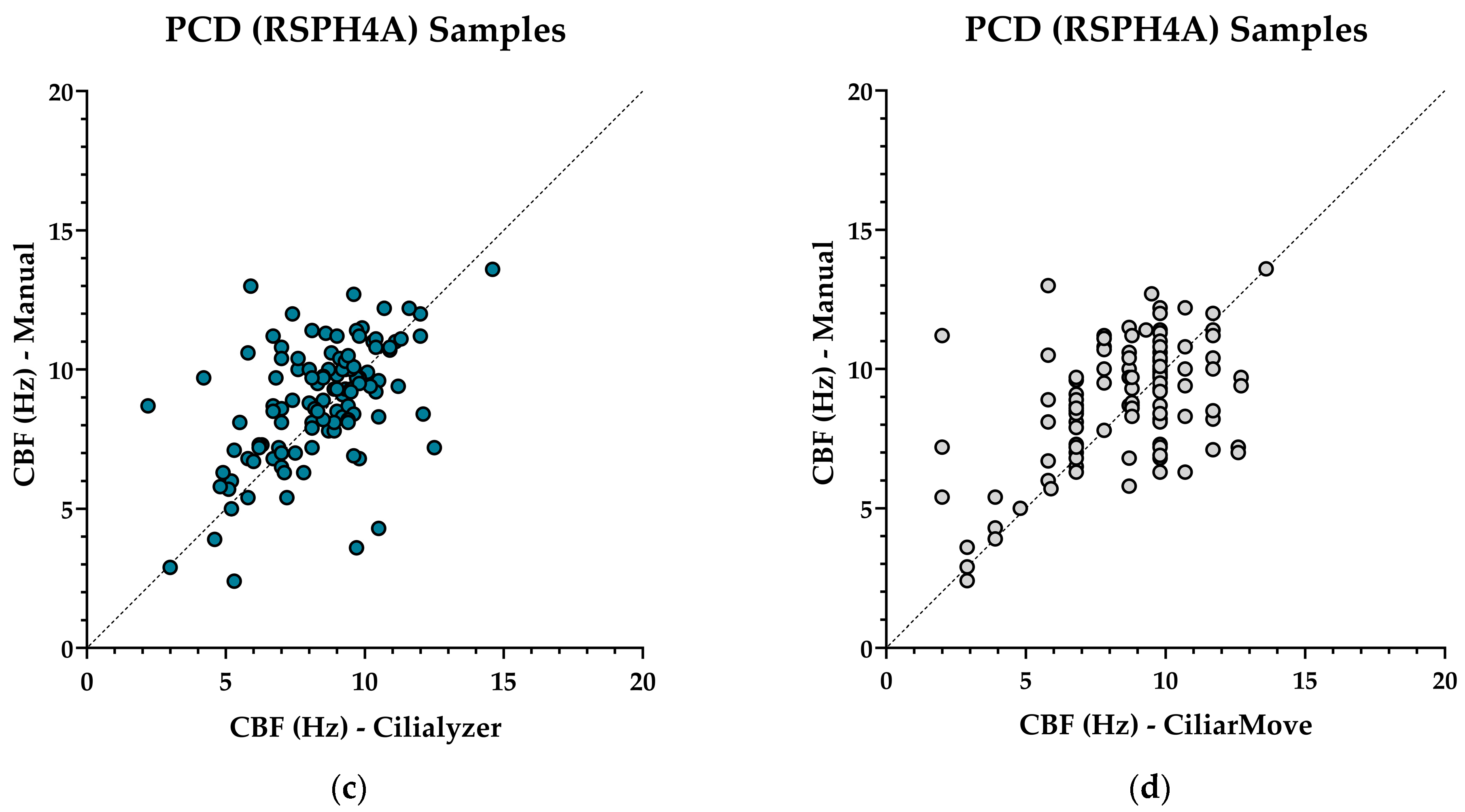
| Correlation coefficient r-value | 0.3830 | 0.8947 | 0.5206 | 0.7964 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Demetriou, Z.J.; Muñiz-Hernández, J.; Rosario-Ortiz, G.; Quiñones, F.M.; Gonzalez-Diaz, G.; Ramos-Benitez, M.J.; Mosquera, R.A.; De Jesús-Rojas, W. Evaluation of Open-Source Ciliary Analysis Software in Primary Ciliary Dyskinesia: A Comparative Assessment. Diagnostics 2024, 14, 1814. https://doi.org/10.3390/diagnostics14161814
Demetriou ZJ, Muñiz-Hernández J, Rosario-Ortiz G, Quiñones FM, Gonzalez-Diaz G, Ramos-Benitez MJ, Mosquera RA, De Jesús-Rojas W. Evaluation of Open-Source Ciliary Analysis Software in Primary Ciliary Dyskinesia: A Comparative Assessment. Diagnostics. 2024; 14(16):1814. https://doi.org/10.3390/diagnostics14161814
Chicago/Turabian StyleDemetriou, Zachary J., José Muñiz-Hernández, Gabriel Rosario-Ortiz, Frances M. Quiñones, Gabriel Gonzalez-Diaz, Marcos J. Ramos-Benitez, Ricardo A. Mosquera, and Wilfredo De Jesús-Rojas. 2024. "Evaluation of Open-Source Ciliary Analysis Software in Primary Ciliary Dyskinesia: A Comparative Assessment" Diagnostics 14, no. 16: 1814. https://doi.org/10.3390/diagnostics14161814
APA StyleDemetriou, Z. J., Muñiz-Hernández, J., Rosario-Ortiz, G., Quiñones, F. M., Gonzalez-Diaz, G., Ramos-Benitez, M. J., Mosquera, R. A., & De Jesús-Rojas, W. (2024). Evaluation of Open-Source Ciliary Analysis Software in Primary Ciliary Dyskinesia: A Comparative Assessment. Diagnostics, 14(16), 1814. https://doi.org/10.3390/diagnostics14161814








