Enhancing the Clinical Utility of Radiomics: Addressing the Challenges of Repeatability and Reproducibility in CT and MRI
Abstract
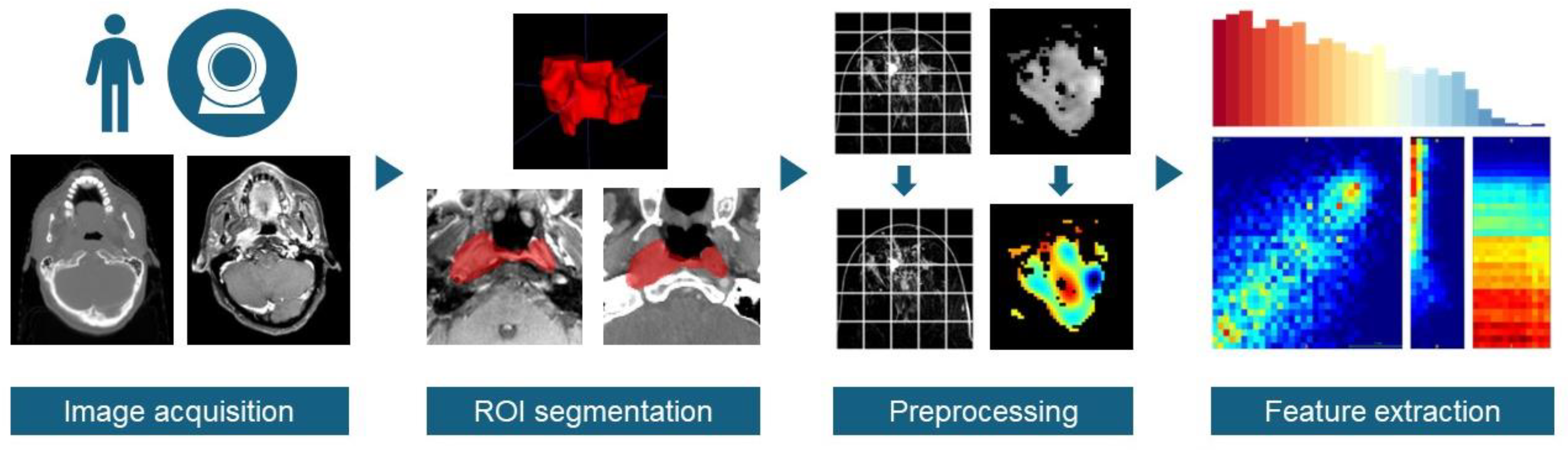
1. Introduction
2. Materials and Methods
2.1. Eligibility Criteria
2.2. Research Strategy
2.3. Data Extraction
3. Results
3.1. Overall Results
3.2. Random Effects Affecting CT-Based Radiomic Features
3.3. Bias Affecting CT-Based Radiomic Features
3.4. Random Effects Affecting MR-Based Radiomic Features
3.5. Bias Affecting MR-Based Radiomic Features
4. Discussion
4.1. Significance of Repeatability and Reproducibility in Radiomic Studies
4.2. Randomness: A Fundamental Source of Variation in Radiomic Studies
4.3. Bias: Inter-Observer and Inter-Scanner Variations—A Significant Hurdle to Generalizable Radiomic Signatures
4.4. Efforts to Mitigate Randomness for Repeatable Radiomic Signatures
4.5. Efforts to Address Bias for Generalizable Radiomic Signatures
4.6. Enhancing the Reporting of Repeatability and Reproducibility in Radiomic Feature Studies
- (1)
- Detailed Reporting of Variation Sources: Authors should meticulously document any sources of variation encountered across different measurement settings. These include, but are not limited to, changes in scanner types, imaging protocols, and segmentation processes. Such detailed reporting will provide valuable context for understanding the conditions under which the radiomic features were assessed.
- (2)
- Transparent Disclosure of Calculation Parameters: It is imperative to transparently disclose all parameters used in the calculation of radiomic features. This transparency ensures that other researchers can accurately replicate the feature extraction process, facilitating a more reliable comparison of results across different studies.
- (3)
- Careful Selection of a Suitable Reliability Index: Choosing an appropriate reliability index is critical for assessing the repeatability and reproducibility of radiomic features. Researchers should select indices that most accurately reflect the nature of the variations.
- (4)
- Comprehensive Reporting of Reliability Metrics: The reliability metrics for individual features should be thoroughly reported. This comprehensive reporting will allow other researchers to discern which features are most stable and reliable across different datasets and conditions, thereby informing the selection of features for their own radiomic signatures.
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- O’Connor, J.P.B.; Aboagye, E.O.; Adams, J.E.; Aerts, H.J.W.L.; Barrington, S.F.; Beer, A.J.; Boellaard, R.; Bohndiek, S.E.; Brady, M.; Brown, G.; et al. Imaging biomarker roadmap for cancer studies. Nat. Rev. Clin. Oncol. 2017, 14, 169–186. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Desideri, I.; Loi, M.; Francolini, G.; Becherini, C.; Livi, L.; Bonomo, P. Application of Radiomics for the Prediction of Radiation-Induced Toxicity in the IMRT Era: Current State-of-the-Art. Front. Oncol. 2020, 10, 1708. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Zhu, T.; Zhang, X.; Li, W.; Zheng, X.; Cheng, M.; Ji, F.; Zhang, L.; Yang, C.; Wu, Z.; et al. Longitudinal MRI-based fusion novel model predicts pathological complete response in breast cancer treated with neoadjuvant chemotherapy: A multicenter, retrospective study. eClinicalMedicine 2023, 58, 101899. [Google Scholar] [CrossRef]
- Teng, X.; Zhang, J.; Zhang, X.; Fan, X.; Zhou, T.; Huang, Y.; Wang, L.; Lee, E.Y.P.; Yang, R.; Cai, J. Noninvasive imaging signatures of HER2 and HR using ADC in invasive breast cancer: Repeatability, reproducibility, and association with pathological complete response to neoadjuvant chemotherapy. Breast Cancer Res. 2023, 25, 77. [Google Scholar] [CrossRef]
- Peng, H.; Dong, D.; Fang, M.-J.; Li, L.; Tang, L.-L.; Chen, L.; Li, W.-F.; Mao, Y.-P.; Fan, W.; Liu, L.-Z.; et al. Prognostic Value of Deep Learning PET/CT-Based Radiomics: Potential Role for Future Individual Induction Chemotherapy in Advanced Nasopharyngeal Carcinoma. Clin. Cancer Res. 2019, 25, 4271–4279. [Google Scholar] [CrossRef]
- Steyerberg, E.W. Clinical Prediction Models: A Practical Approach to Development, Validation, and Updating; Statistics for Biology and Health; Springer: Cham, Switzerland, 2019; ISBN 978-3-030-16398-3. [Google Scholar]
- Sullivan, D.C.; Obuchowski, N.A.; Kessler, L.G.; Raunig, D.L.; Gatsonis, C.; Huang, E.P.; Kondratovich, M.; McShane, L.M.; Reeves, A.P.; Barboriak, D.P.; et al. Metrology Standards for Quantitative Imaging Biomarkers. Radiology 2015, 277, 813–825. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef]
- Gourtsoyianni, S.; Doumou, G.; Prezzi, D.; Taylor, B.; Stirling, J.J.; Taylor, N.J.; Siddique, M.; Cook, G.J.R.; Glynne-Jones, R.; Goh, V. Primary Rectal Cancer: Repeatability of Global and Local-Regional MR Imaging Texture Features. Radiology 2017, 284, 552–561. [Google Scholar] [CrossRef]
- Van Timmeren, J.E.; Leijenaar, R.T.H.; Van Elmpt, W.; Wang, J.; Zhang, Z.; Dekker, A.; Lambin, P. Lambin, Test–Retest Data for Radiomics Feature Stability Analysis: Generalizable or Study-Specific? Tomography 2016, 2, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Fiset, S.; Welch, M.L.; Weiss, J.; Pintilie, M.; Conway, J.L.; Milosevic, M.; Fyles, A.; Traverso, A.; Jaffray, D.; Metser, U.; et al. Repeatability and reproducibility of MRI-based radiomic features in cervical cancer. Radiother. Oncol. 2019, 135, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Traverso, A.; Wee, L.; Dekker, A.; Gillies, R. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, 1143–1158. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Leger, S.; Agolli, L.; Pilz, K.; Troost, E.G.C.; Richter, C.; Löck, S. Assessing robustness of radiomic features by image perturbation. Sci. Rep. 2019, 9, 614. [Google Scholar] [CrossRef]
- Teng, X.; Zhang, J.; Zwanenburg, A.; Sun, J.; Huang, Y.; Lam, S.; Zhang, Y.; Li, B.; Zhou, T.; Xiao, H.; et al. Building reliable radiomic models using image perturbation. Sci. Rep. 2022, 12, 10035. [Google Scholar] [CrossRef]
- Teng, X.; Zhang, J.; Ma, Z.; Zhang, Y.; Lam, S.; Li, W.; Xiao, H.; Li, T.; Li, B.; Zhou, T.; et al. Improving radiomic model reliability using robust features from perturbations for head-and-neck carcinoma. Front. Oncol. 2022, 12, 974467. [Google Scholar] [CrossRef]
- Zhao, B. Understanding Sources of Variation to Improve the Reproducibility of Radiomics. Front. Oncol. 2021, 11, 633176. [Google Scholar] [CrossRef]
- Euler, A.; Laqua, F.C.; Cester, D.; Lohaus, N.; Sartoretti, T.; Pinto Dos Santos, D.; Alkadhi, H.; Baessler, B. Virtual Monoenergetic Images of Dual-Energy CT—Impact on Repeatability, Reproducibility, and Classification in Radiomics. Cancers 2021, 13, 4710. [Google Scholar] [CrossRef]
- Carbonell, G.; Kennedy, P.; Bane, O.; Kirmani, A.; El Homsi, M.; Stocker, D.; Said, D.; Mukherjee, P.; Gevaert, O.; Lewis, S.; et al. Precision of MRI radiomics features in the liver and hepatocellular carcinoma. Eur. Radiol. 2022, 32, 2030–2040. [Google Scholar] [CrossRef]
- Mitchell-Hay, R.N.; Ahearn, T.S.; Murray, A.D.; Waiter, G.D. Investigation of the Inter- and Intrascanner Reproducibility and Repeatability of Radiomics Features in T1 -Weighted Brain MRI. Magn. Reson. Imaging 2022, 56, 1559–1568. [Google Scholar] [CrossRef]
- Chen, Y.; Zhong, J.; Wang, L.; Shi, X.; Lu, W.; Li, J.; Feng, J.; Xia, Y.; Chang, R.; Fan, J.; et al. Robustness of CT radiomics features: Consistency within and between single-energy CT and dual-energy CT. Eur. Radiol. 2022, 32, 5480–5490. [Google Scholar] [CrossRef]
- Li, Y.; Tan, G.; Vangel, M.; Hall, J.; Cai, W. Influence of feature calculating parameters on the reproducibility of CT radiomic features: A thoracic phantom study. Quant. Imaging Med. Surg. 2020, 10, 1775–1785. [Google Scholar] [CrossRef]
- Lee, J.; Steinmann, A.; Ding, Y.; Lee, H.; Owens, C.; Wang, J.; Yang, J.; Followill, D.; Ger, R.; MacKin, D.; et al. Radiomics feature robustness as measured using an MRI phantom. Sci. Rep. 2021, 11, 3973. [Google Scholar] [CrossRef]
- Duron, L.; Balvay, D.; Vande Perre, S.; Bouchouicha, A.; Savatovsky, J.; Sadik, J.-C.; Thomassin-Naggara, I.; Fournier, L.; Lecler, A. Gray-level discretization impacts reproducible MRI radiomics texture features. PLoS ONE 2019, 14, e0213459. [Google Scholar] [CrossRef] [PubMed]
- Moradmand, H.; Aghamiri, S.M.R.; Ghaderi, R. Impact of image preprocessing methods on reproducibility of radiomic features in multimodal magnetic resonance imaging in glioblastoma. J. Appl. Clin. Med. Phys. 2020, 21, 179–190. [Google Scholar] [CrossRef]
- Chen, K.; Deng, L.; Li, Q.; Luo, L. Are computed-tomography-based hematoma radiomics features reproducible and predictive of intracerebral hemorrhage expansion? an in vitro experiment and clinical study. Br. J. Radiol. 2021, 94, 20200724. [Google Scholar] [CrossRef]
- Mahon, R.N.; Hugo, G.D.; Weiss, E. Repeatability of texture features derived from magnetic resonance and computed tomography imaging and use in predictive models for non-small cell lung cancer outcome. Phys. Med. Biol. 2019, 64, 145007. [Google Scholar] [CrossRef] [PubMed]
- Muenzfeld, H.; Nowak, C.; Riedlberger, S.; Hartenstein, A.; Hamm, B.; Jahnke, P.; Penzkofer, T. Intra-scanner repeatability of quantitative imaging features in a 3D printed semi-anthropomorphic CT phantom. Eur. J. Radiol. 2021, 141, 109818. [Google Scholar] [CrossRef] [PubMed]
- Prayer, F.; Hofmanninger, J.; Weber, M.; Kifjak, D.; Willenpart, A.; Pan, J.; Röhrich, S.; Langs, G.; Prosch, H. Variability of computed tomography radiomics features of fibrosing interstitial lung disease: A test-retest study. Methods 2021, 188, 98–104. [Google Scholar] [CrossRef]
- Duan, J.; Qiu, Q.; Zhu, J.; Shang, D.; Dou, X.; Sun, T.; Yin, Y.; Meng, X. Reproducibility for Hepatocellular Carcinoma CT Radiomic Features: Influence of Delineation Variability Based on 3D-CT, 4D-CT and Multiple-Parameter MR Images. Front. Oncol. 2022, 12, 881931. [Google Scholar] [CrossRef]
- Kocak, B.; Durmaz, E.S.; Kaya, O.K.; Ates, E.; Kilickesmez, O. Reliability of Single-Slice–Based 2D CT Texture Analysis of Renal Masses: Influence of Intra- and Interobserver Manual Segmentation Variability on Radiomic Feature Reproducibility. Am. J. Roentgenol. 2019, 213, 377–383. [Google Scholar] [CrossRef]
- Le, E.P.V.; Rundo, L.; Tarkin, J.M.; Evans, N.R.; Chowdhury, M.M.; Coughlin, P.A.; Pavey, H.; Wall, C.; Zaccagna, F.; Gallagher, F.A.; et al. Assessing robustness of carotid artery CT angiography radiomics in the identification of culprit lesions in cerebrovascular events. Sci. Rep. 2021, 11, 3499. [Google Scholar] [CrossRef]
- Müller-Franzes, G.; Nebelung, S.; Schock, J.; Haarburger, C.; Khader, F.; Pedersoli, F.; Schulze-Hagen, M.; Kuhl, C.; Truhn, D. Reliability as a Precondition for Trust—Segmentation Reliability Analysis of Radiomic Features Improves Survival Prediction. Diagnostics 2022, 12, 247. [Google Scholar] [CrossRef]
- Denzler, S.; Vuong, D.; Bogowicz, M.; Pavic, M.; Frauenfelder, T.; Thierstein, S.; Eboulet, E.I.; Maurer, B.; Schniering, J.; Gabryś, H.S.; et al. Impact of CT convolution kernel on robustness of radiomic features for different lung diseases and tissue types. Br. J. Radiol. 2021, 94, 20200947. [Google Scholar] [CrossRef] [PubMed]
- Gruzdev, I.S.; Zamyatina, K.A.; Tikhonova, V.S.; Kondratyev, E.V.; Glotov, A.V.; Karmazanovsky, G.G.; Revishvili, A.S. Reproducibility of CT texture features of pancreatic neuroendocrine neoplasms. Eur. J. Radiol. 2020, 133, 109371. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.; Widaatalla, Y.; Refaee, T.; Primakov, S.; Miclea, R.L.; Öcal, O.; Fabritius, M.P.; Ingrisch, M.; Ricke, J.; Hustinx, R.; et al. Reproducibility of CT-Based Hepatocellular Carcinoma Radiomic Features across Different Contrast Imaging Phases: A Proof of Concept on SORAMIC Trial Data. Cancers 2021, 13, 4638. [Google Scholar] [CrossRef] [PubMed]
- Lennartz, S.; O’Shea, A.; Parakh, A.; Persigehl, T.; Baessler, B.; Kambadakone, A. Robustness of dual-energy CT-derived radiomic features across three different scanner types. Eur. Radiol. 2022, 32, 1959–1970. [Google Scholar] [CrossRef]
- Meyer, M.; Ronald, J.; Vernuccio, F.; Nelson, R.C.; Ramirez-Giraldo, J.C.; Solomon, J.; Patel, B.N.; Samei, E.; Marin, D. Reproducibility of CT Radiomic Features within the Same Patient: Influence of Radiation Dose and CT Reconstruction Settings. Radiology 2019, 293, 583–591. [Google Scholar] [CrossRef]
- Perrin, T.; Midya, A.; Yamashita, R.; Chakraborty, J.; Saidon, T.; Jarnagin, W.R.; Gonen, M.; Simpson, A.L.; Do, R.K.G. Short-term reproducibility of radiomic features in liver parenchyma and liver malignancies on contrast-enhanced CT imaging. Abdom. Radiol. 2018, 43, 3271–3278. [Google Scholar] [CrossRef] [PubMed]
- Refaee, T.; Salahuddin, Z.; Widaatalla, Y.; Primakov, S.; Woodruff, H.C.; Hustinx, R.; Mottaghy, F.M.; Ibrahim, A.; Lambin, P. CT Reconstruction Kernels and the Effect of Pre- and Post-Processing on the Reproducibility of Handcrafted Radiomic Features. J. Pers. Med. 2022, 12, 553. [Google Scholar] [CrossRef]
- Rinaldi, L.; De Angelis, S.P.; Raimondi, S.; Rizzo, S.; Fanciullo, C.; Rampinelli, C.; Mariani, M.; Lascialfari, A.; Cremonesi, M.; Orecchia, R.; et al. Reproducibility of radiomic features in CT images of NSCLC patients: An integrative analysis on the impact of acquisition and reconstruction parameters. Eur. Radiol. Exp. 2022, 6, 2. [Google Scholar] [CrossRef]
- Bianconi, F.; Fravolini, M.L.; Palumbo, I.; Pascoletti, G.; Nuvoli, S.; Rondini, M.; Spanu, A.; Palumbo, B. Impact of Lesion Delineation and Intensity Quantisation on the Stability of Texture Features from Lung Nodules on CT: A Reproducible Study. Diagnostics 2021, 11, 1224. [Google Scholar] [CrossRef] [PubMed]
- Haarburger, C.; Müller-Franzes, G.; Weninger, L.; Kuhl, C.; Truhn, D.; Merhof, D. Radiomics feature reproducibility under inter-rater variability in segmentations of CT images. Sci. Rep. 2020, 10, 12688. [Google Scholar] [CrossRef] [PubMed]
- Könik, A.; Miskin, N.; Guo, Y.; Shinagare, A.B.; Qin, L. Robustness and performance of radiomic features in diagnosing cystic renal masses. Abdom. Radiol. 2021, 46, 5260–5267. [Google Scholar] [CrossRef]
- Pandey, U.; Saini, J.; Kumar, M.; Gupta, R.; Ingalhalikar, M. Normative Baseline for Radiomics in Brain MRI: Evaluating the Robustness, Regional Variations, and Reproducibility on FLAIR Images. Magn. Reson. Imaging 2021, 53, 394–407. [Google Scholar] [CrossRef]
- Dewi, D.E.O.; Sunoqrot, M.R.S.; Nketiah, G.A.; Sandsmark, E.; Giskeødegård, G.F.; Langørgen, S.; Bertilsson, H.; Elschot, M.; Bathen, T.F. The impact of pre-processing and disease characteristics on reproducibility of T2-weighted MRI radiomics features. Magn. Reson. Mater. Phy. 2023, 36, 945–956. [Google Scholar] [CrossRef] [PubMed]
- Raisi-Estabragh, Z.; Gkontra, P.; Jaggi, A.; Cooper, J.; Augusto, J.; Bhuva, A.N.; Davies, R.H.; Manisty, C.H.; Moon, J.C.; Munroe, P.B.; et al. Repeatability of Cardiac Magnetic Resonance Radiomics: A Multi-Centre Multi-Vendor Test-Retest Study. Front. Cardiovasc. Med. 2020, 7, 586236. [Google Scholar] [CrossRef] [PubMed]
- Duron, L.; Savatovsky, J.; Fournier, L.; Lecler, A. Can we use radiomics in ultrasound imaging? Impact of preprocessing on feature repeatability. Diagn. Interv. Imaging 2021, 102, 659–667. [Google Scholar] [CrossRef]
- Scalco, E.; Belfatto, A.; Mastropietro, A.; Rancati, T.; Avuzzi, B.; Messina, A.; Valdagni, R.; Rizzo, G. T2w-MRI signal normalization affects radiomics features reproducibility. Med. Phys. 2020, 47, 1680–1691. [Google Scholar] [CrossRef]
- Haniff, N.S.M.; Abdul Karim, M.K.; Osman, N.H.; Saripan, M.I.; Che Isa, I.N.; Ibahim, M.J. Stability and Reproducibility of Radiomic Features Based Various Segmentation Technique on MR Images of Hepatocellular Carcinoma (HCC). Diagnostics 2021, 11, 1573. [Google Scholar] [CrossRef]
- Zhang, J.; Lam, S.-K.; Teng, X.; Ma, Z.; Han, X.; Zhang, Y.; Cheung, A.L.-Y.; Chau, T.-C.; Ng, S.C.-Y.; Lee, F.K.-H.; et al. Radiomic feature repeatability and its impact on prognostic model generalizability: A multi-institutional study on nasopharyngeal carcinoma patients. Radiother. Oncol. 2023, 183, 109578. [Google Scholar] [CrossRef]
- Kocak, B.; Baessler, B.; Bakas, S.; Cuocolo, R.; Fedorov, A.; Maier-Hein, L.; Mercaldo, N.; Müller, H.; Orlhac, F.; Pinto Dos Santos, D.; et al. CheckList for EvaluAtion of Radiomics research (CLEAR): A step-by-step reporting guideline for authors and reviewers endorsed by ESR and EuSoMII. Insights Imaging 2023, 14, 75. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; De Jong, E.E.C.; Van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Hatt, M.; Krizsan, A.K.; Rahmim, A.; Bradshaw, T.J.; Costa, P.F.; Forgacs, A.; Seifert, R.; Zwanenburg, A.; El Naqa, I.; Kinahan, P.E.; et al. Joint EANM/SNMMI guideline on radiomics in nuclear medicine: Jointly supported by the EANM Physics Committee and the SNMMI Physics, Instrumentation and Data Sciences Council. Eur. J. Nucl. Med. Mol. Imaging 2023, 50, 352–375. [Google Scholar] [CrossRef] [PubMed]
- Koo, T.K.; Li, M.Y. A Guideline of Selecting and Reporting Intraclass Correlation Coefficients for Reliability Research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Teng, X.; Zhang, X.; Lam, S.-K.; Lin, Z.; Liang, Y.; Yu, H.; Siu, S.W.K.; Chang, A.T.Y.; Zhang, H.; et al. Comparing effectiveness of image perturbation and test retest imaging in improving radiomic model reliability. Sci. Rep. 2023, 13, 18263. [Google Scholar] [CrossRef]
- Hoebel, K.V.; Patel, J.B.; Beers, A.L.; Chang, K.; Singh, P.; Brown, J.M.; Pinho, M.C.; Batchelor, T.T.; Gerstner, E.R.; Rosen, B.R.; et al. Radiomics Repeatability Pitfalls in a Scan-Rescan MRI Study of Glioblastoma. Radiol. Artif. Intell. 2021, 3, e190199. [Google Scholar] [CrossRef]
- Pfaehler, E.; Zhovannik, I.; Wei, L.; Boellaard, R.; Dekker, A.; Monshouwer, R.; El Naqa, I.; Bussink, J.; Gillies, R.; Wee, L.; et al. A systematic review and quality of reporting checklist for repeatability and reproducibility of radiomic features. Phys. Imaging Radiat. Oncol. 2021, 20, 69–75. [Google Scholar] [CrossRef]

| Author | Modality | Sources of Variation | Criteria for High Repeatablity/Reproduciblity | Site | Highly Repeatable/Reproducible Features | Availability of Reliability Index |
|---|---|---|---|---|---|---|
| Chen et al. (2021) [27] | CT | Intra-scanner test–retest | CV < 10% | Phantom and hematoma | Phantom: 79.05% to 81.43% Hematoma: 42.54% to 45.4% | No |
| Chen et al. (2022) [22] | DECT SECT | Intra-scanner test–retest | Bland–Altman analysis > 0.90 | Phantom | DECT: 87.02 ± 5.79% SECT: 92.91 ± 1.89% | Yes |
| Euler et al. (2021) [19] | CT | Intra-scanner test–retest | CCC and DR ≥ 0.9 | Liver | 74% to 86% repeatable features under acquisition settings | No |
| Mahon et al. (2019) [28] | CT | Intra-scanner test–retest | CCC > 0.9 | Lung | Tumor: 54.4% Normal tissue: 78.5% | Yes |
| Muenzfeld et al. (2021) [29] | CT | Intra-scanner test–retest | CCC > 0.85 | Phantom | 19/86 (22%) | Yes |
| Prayer et al. (2020) [30] | CT | Intra-scanner test–retest | ICC > 0.9 | Lung | 65/86 (75.58%) | Yes |
| Duan et al. (2022) [31] | MIP | Intra-observer variability | ICC > 0.75 | Liver | 77/107 (71.96%) | Yes |
| CECT | Intra-observer variability | 84/107 (78.50%) | ||||
| Kocak et al. (2019) [32] | CECT | Intra-observer variability | ICC > 0.75 | Kidney | Texture features: 693/744 (93.1%) | No |
| CT | Intra-observer variability | Texture features: 686/744 (92.2%) | ||||
| EPV Le et al. (2021) [33] | CT | Perturbations | ICC > 0.9 | Heart | 14/93 (15.1%) | No |
| Müller-Franzes et al. (2022) [34] | CT | Autogenerated segmentations | ICC > 0.99 | Multi-site | Lung: 269/439 (61.28%) Liver: 292/439 (66.51%) Kidney: 377/439 (85.88%) | Yes |
| Author | Modality | Sources of Variation | Criteria for High Repeatablity/ Reproduciblity | Site | Highly Repeatable/Reproducible Features | Availability of Reliability Index |
|---|---|---|---|---|---|---|
| Chen et al. (2021) [27] | CT | Acquisition parameters | CV < 10% | Phantom and hematoma | Phantom: 48.89% to 53.97% Hematoma: 43% to 42.38% | No |
| Chen et al. (2022) [22] | CT | Acquisition parameters | ICC/CCC > 0.90 | Phantom | DECT: 10.76 ± 2.05% SECT: 10.28 ± 2.05% | Yes |
| Inter-scanner variability | CV/QCD < 10% | DECT: 15.16 ± 3.26%, 32.78 ± 5.62% SECT: 17.09 ± 2.60%, 27.73 ± 4.07% | ||||
| Denzler et al. (2021) [35] | CT | Acquisition parameters | ICC > 0.9 | Lung | 360/1386 (26%) | Yes |
| Euler et al. (2021) [19] | CT | Acquisition parameters | CCC and DR ≥ 0.9 | Liver | 32.7% to 99.2% reproducible features across different energies | No |
| Gruzdev et al. (2020) [36] | CECT | Inter-observer variability | Kendall’s concordance coefficient > 0.7 | Pancreas | 52/52 (100%) features for all phases | No |
| Inter-scanner variability | 74% reproducible texture features | |||||
| Inter-scanner and inter-observer variability | 67% reproducible texture features | |||||
| Ibrahim et al. (2021) [37] | CT | Contrast-enhanced phases | CCC > 0.9 | Liver | 42/167 (25.15%) | No |
| Lennartz et al. (2022) [38] | DECT | Inter-scanner variability | CCC > 0.9 | Phantom and multi-sites | Phantom: None Patients: 2.5% to 16.1% of features | No |
| Meyer et al. (2019) [39]) | CT | Acquisition parameters | R2 ≥ 0.95 | Liver | 12/106 (11%) | Yes |
| Muenzfeld et al. (2021) [29] | CT | Acquisition parameters | CCC > 0.85 | Phantom | 11/86 (12.8%) | Yes |
| Perrin et al. (2018) [40] | CECT | Contrast-agent injection rate | CCC > 0.9 | Liver | Liver parenchyma: 63/254 (24.8%) and 0/254 (0%) Liver malignancies: 68/254 (26.77%) and 50/254 (19.69%) | Yes |
| Acquisition parameters | Liver parenchyma: 20/254 (7.87%), 0/254 (0%); Liver malignancies: 34/254 (13.39%) | |||||
| Prayer et al. (2020) [30] | CT | Inter-scanner variability | ICC > 0.9 | Lung | ICC ranges from 0.471 to 0.927 | Yes |
| Refaee et al. (2022) [41] | CT | Acquisition parameters | CCC > 0.9 | Phantom | 6/91 (6.59%) to 78/91 (85.71%) | No |
| Rinaldi et al. (2022) [42] | CT | Acquisition parameters | OCCC ≥ 0.85 | Lung | 1260/1414 (89.11%) | Yes |
| Bianconi et al. (2021) [43] | CT | Inter-observer variability | Average symmetric mean absolute percentage error < 10% | Lung | 30/88 (34.09%) | Yes |
| Duan et al. (2022) [31] | MIP | Inter-observer variability | ICC > 0.75 | Liver | 71/107 (66.36%) | Yes |
| CECT | Inter-observer variability | 74/107 (69.16%) | ||||
| Haarburger et al. (2020) [44] | CT | Inter-observer variability and automatic segmentation | ICC > 0.9 | Multi-site | Lung: 71/105 (67.62%) Kidney: 61/105 (58.10%) Liver: 75/105 (71.43%) | Yes |
| Kocak et al. (2019) [32] | CECT | Inter-observer variability | ICC > 0.75 | Kidney | Texture features: 632/744 (84.9%) | No |
| CT | Inter-observer variability | Texture features: 571/744 (76.7%) | ||||
| Konik et al. (2021) [45] | CT | Inter-observer variability | ICC > 0.85 | Kidney | 78/169 (46.15%) | Yes |
| Li et al. (2020) [23] | CT | Preprocessing parameters | ICC > 0.8 and CV < 20% | Phantom | 44/88 (50%) | No |
| Le et al. (2021) [33] | CT | Preprocessing parameters | ICC > 0.9 | Heart | 52/93 (55.9%) | No |
| Bianconi et al. (2021) [43] | CT | Preprocessing parameters | Averaging symmetric mean absolute percentage error < 10% | Lung | 28/88 (31.82%) | Yes |
| Author | Modality | Sources of Variation | Criteria for High Repeatablity/Reproduciblity | Site | Highly Repeatable/Reproducible Features | Availability of Reliability Index |
|---|---|---|---|---|---|---|
| Carbonell et al. (2022) [20] | MRI | Intra-scanner test–retest | ICC > 0.9 and CV < 20% | Liver | HCC-T1WIpre: 45/108 (41.67%), T1WIpvp: 47/108 (43.52%), T2WI: 39/108 (36.11%), ADC: 21/108 (19.44%) Liver-T1WIpre: 32/92 (34.78%), T1WIpvp: 16/92 (17.39%), T2WI: 12/92 (13.04%), ADC: 2/92 (2.17%) | Yes |
| Fiset et al. (2019) [13] | MRI (T2WI) | Intra-scanner test–retest | ICC ≥ 0.75 | Cervical | Cervical: 917/1761 (52.1%) | Yes |
| Mahon et al. (2019) [28] | MRI | Intra-scanner test–retest | CCC ≥ 0.9 | Lung | Lung (TRUFISP): 64.4% (tumor), 67.8% (normal tissue) Lung (VIBE): 54.4% (tumor), 72.9% (normal tissue) | Yes |
| Mitchell-Hay et al. (2022) [21] | MRI (T1WI) | Intra-scanner test–retest | CCC/DR > 0.9 | Brain | 8/1596 (0.50%) features were repeatable in all centers | Yes |
| Pandey et al. (2021) [46] | MRI (T2WI) | Intra-scanner test–retest | ICC > 0.5 | Brain | ICC: 0.73 for right grey matter, 0.78 for left grey matter ICC: 0.65 for right white matter, 0.67 for left white matter | Yes |
| Dewi et al. (2023) [47] | MRI (T2WI) | Intra-scanner test–retest | ICC > 0.75 | Prostate | 25/107 (23.36%) at fixed bin count discretization of 64 | Yes |
| Duan et al. (2022) [31] | MRI | Intra-observer variability | ICC > 0.75 | Liver | 98/107 (91.6%) | Yes |
| Müller-Franzes et al. (2022) [34] | MRI | Automatic segmentations | ICC > 0.99 | Brain | 77/439 (17.54%) | Yes |
| Author | Modality | Sources of Variation | Criteria for High Repeatablity/Reproduciblity | Site | Highly Repeatable/Reproducible Features | Availability of Reliability Index |
|---|---|---|---|---|---|---|
| Carbonell et al. (2022) [20] | MRI | Inter-observer variability | CCC > 0.75 and CV < 20% | Liver | HCC-T1WIpre: 95/108 (87.96%), T1WIpvp: 102/108 (94.44%), T2WI: 61/108 (56.48%), ADC: 91/108 (84.26%) Liver-T1WIpre: 25/92 (27.17%), T1WIpvp: 37/92 (40.22%), T2WI: 8/92 (8.70%), ADC: 49/92 (53.26%) | Yes |
| Inter-scanner variability | CCC > 0.75, CV < 20% | HCC-T1WIpre: 23/108 (21.30%), T1WIpvp: 25/108 (23.15%), T2WI: 11/108 (10.19%), ADC: 7/108 (6.48%) Liver-T1WIpre: 0/92 (0%), T1WIpvp: 0/92 (0%), T2WI: 0/92 (0%), ADC: 0/92 (0%) | ||||
| Fiset et al. (2019) [13] | MRI (T2W) | Inter-observer variability | ICC > 0.9 | Cervix | 1301/1761 (73.88%) | Yes |
| Inter-scanner variability | ICC ≥ 0.75 | 248/1761 (14.1%) | Yes | |||
| Lee et al. (2021) [24] | MRI | Acquisition parameters | ICC > 0.9, CV < 20% | Phantom and brain (healthy volunteers) | Phantom: average ICC, 0.963 (T1WI) and 0.959 (T2WI) Brain: average ICC, 0.856 (T1WI) and 0.859 (T2WI) | Yes |
| Mitchell-Hay et al. (2022) [21] | MRI (T1WI) | Inter-scanner variability | ICC > 0.9 | Brain | 40/1595 (2.51%) features were excellent in terms of reproducibility | Yes |
| Pandey et al. (2020) [46] | MRI (T2WI) | Spatial variability | ICC > 0.5 | Brain | 29.04% of gray matter and 38.7% of white matter features demonstrated an ICC > 0.5 | Yes |
| Inter-scanner variability | 18% of gray matter and 21.5% of white matter features demonstrated an ICC > 0.5 | |||||
| Raisi-Estabragh et al. (2020) [48] | MRI | Inter-scanner variability | ICC > 0.9 | Cardiac | LV myocardium: 4/16 (25%) for repeatable shape features, (28/38, 74%) for repeatable first order features, (125/146, 86%) for repeatable texture features | Yes |
| Duron et al. (2019) [49] | MRI | Preprocessing parameters | ICC > 0.8 and CCC > 0.9 | Lacrimal gland | 54/69 (78.26%) for T2WI, 37/69 (53.62%) for T1WI, and 31/69 (44.93%) for ADC | No |
| Breast | 32/69 (46.38%) for DISCO sequence | |||||
| Moradmand (2019) [26] | MRI | Preprocessing parameters | CCC/DR > 0.9 | Brain (glioblastoma) | 703/4066 (17.3%) | No |
| Scalco et al. (2020) [50] | T2w-MRI | Preprocessing parameters | ICC > 0.9 | Prostate | Prostate: 14% Obturators: 12% Bulb: 13/91 (14%) | Yes |
| Duan et al. (2022) [31] | MRI | Inter-observer variability | ICC > 0.75 | Liver | 85/107 (79.4%) | Yes |
| Fiset et al. (2019) [13] | MRI (T2W) | Inter-observer variability | ICC > 0.9 | Cervix | 1301/1761 (73.88%) | Yes |
| Haniff (2021) [51] | MRI | Semi-automatic segmentation | ICC ≥ 0.8 | Liver | 640/662 (96.7%) | Yes (partial) |
| Inter-observer variability | 517/662 (78.1%) | |||||
| Müller-Franzes et al. (2022) [34] | MRI | Automatic segmentations | ICC > 0.99 | Brain | 77/439 (17.54%) | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Teng, X.; Wang, Y.; Nicol, A.J.; Ching, J.C.F.; Wong, E.K.Y.; Lam, K.T.C.; Zhang, J.; Lee, S.W.-Y.; Cai, J. Enhancing the Clinical Utility of Radiomics: Addressing the Challenges of Repeatability and Reproducibility in CT and MRI. Diagnostics 2024, 14, 1835. https://doi.org/10.3390/diagnostics14161835
Teng X, Wang Y, Nicol AJ, Ching JCF, Wong EKY, Lam KTC, Zhang J, Lee SW-Y, Cai J. Enhancing the Clinical Utility of Radiomics: Addressing the Challenges of Repeatability and Reproducibility in CT and MRI. Diagnostics. 2024; 14(16):1835. https://doi.org/10.3390/diagnostics14161835
Chicago/Turabian StyleTeng, Xinzhi, Yongqiang Wang, Alexander James Nicol, Jerry Chi Fung Ching, Edwin Ka Yiu Wong, Kenneth Tsz Chun Lam, Jiang Zhang, Shara Wee-Yee Lee, and Jing Cai. 2024. "Enhancing the Clinical Utility of Radiomics: Addressing the Challenges of Repeatability and Reproducibility in CT and MRI" Diagnostics 14, no. 16: 1835. https://doi.org/10.3390/diagnostics14161835
APA StyleTeng, X., Wang, Y., Nicol, A. J., Ching, J. C. F., Wong, E. K. Y., Lam, K. T. C., Zhang, J., Lee, S. W.-Y., & Cai, J. (2024). Enhancing the Clinical Utility of Radiomics: Addressing the Challenges of Repeatability and Reproducibility in CT and MRI. Diagnostics, 14(16), 1835. https://doi.org/10.3390/diagnostics14161835








