Ikaros Deletions among Bulgarian Patients with Acute Lymphoblastic Leukemia/Lymphoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients
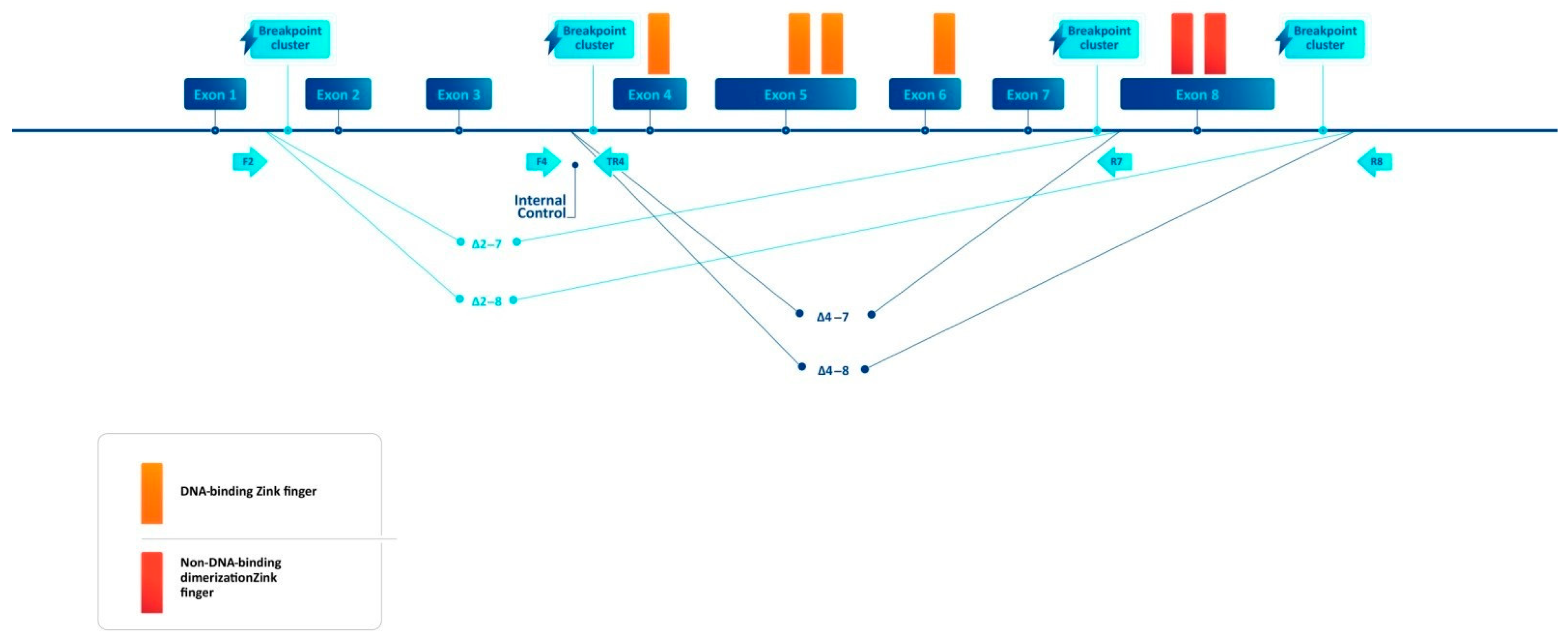
2.2. Breakpoint-Specific PCR Design
2.3. Statistical Considerations
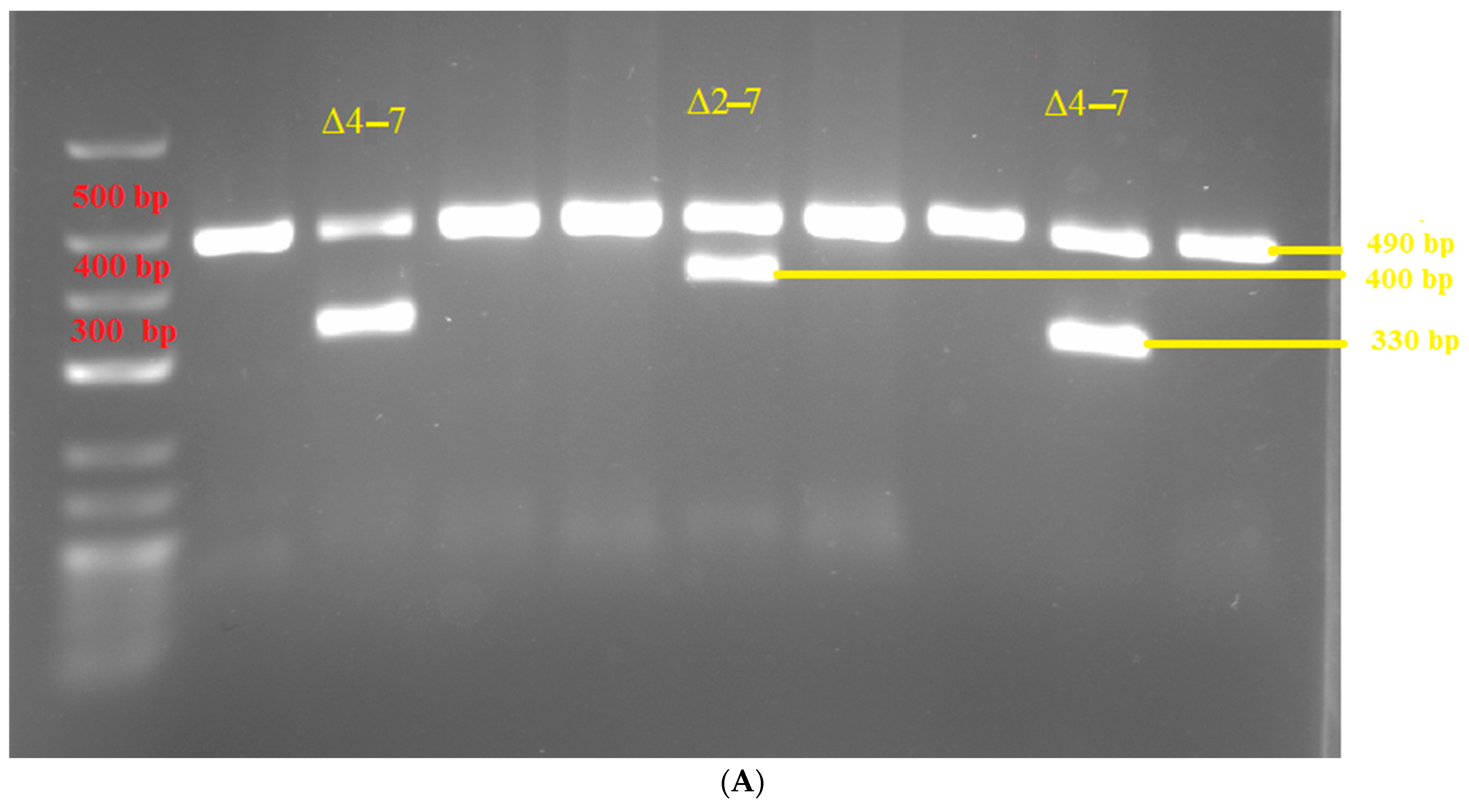
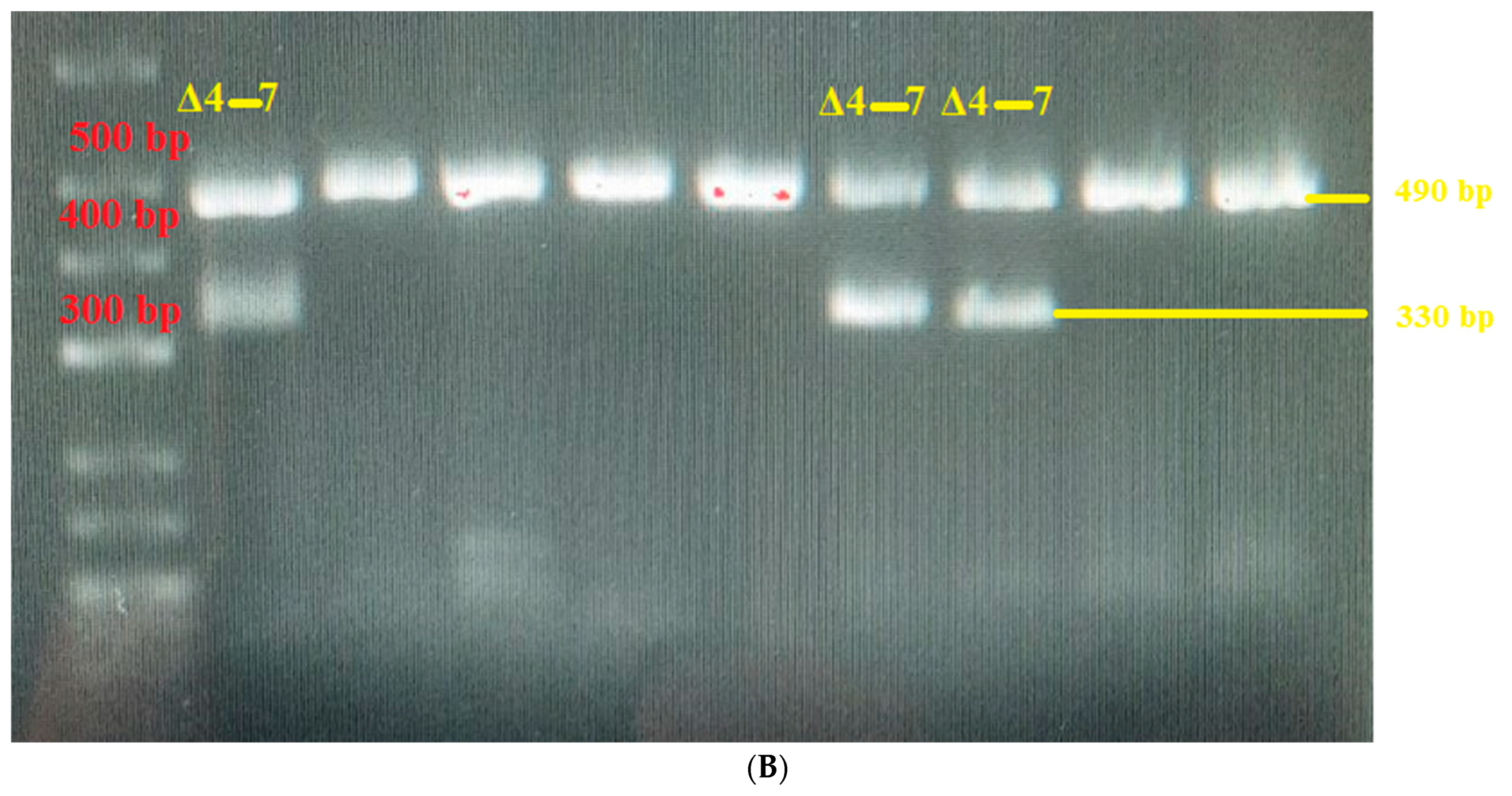
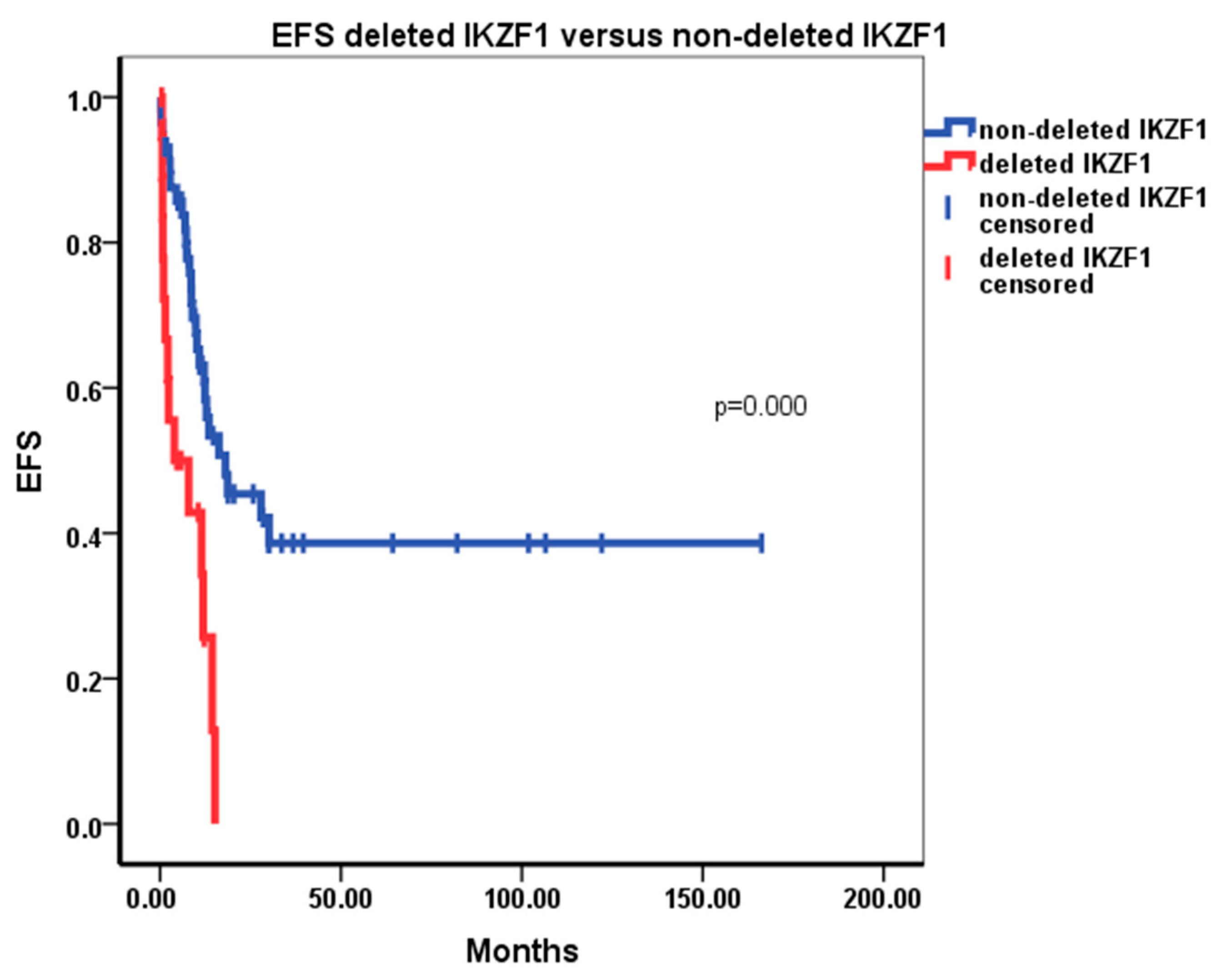
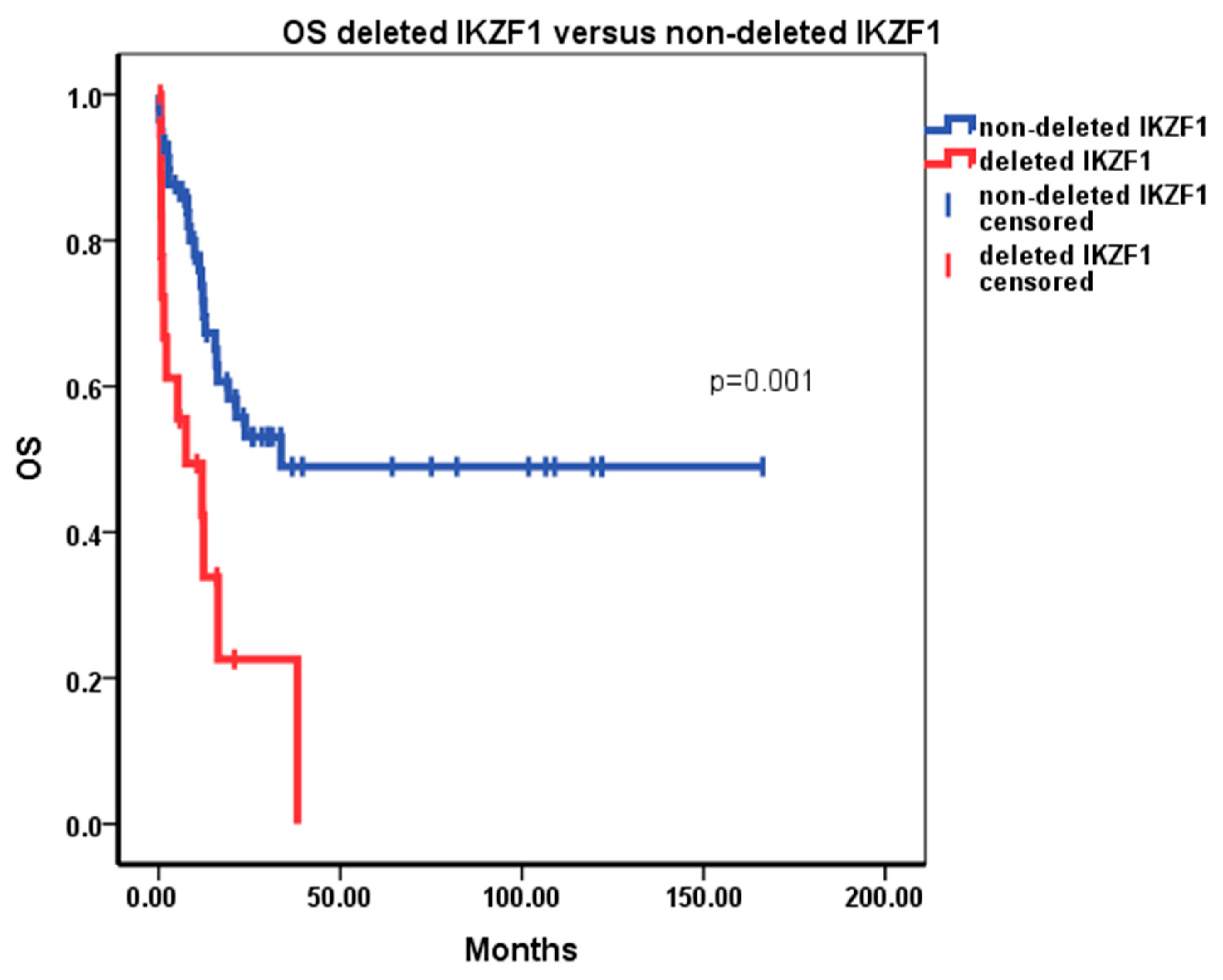
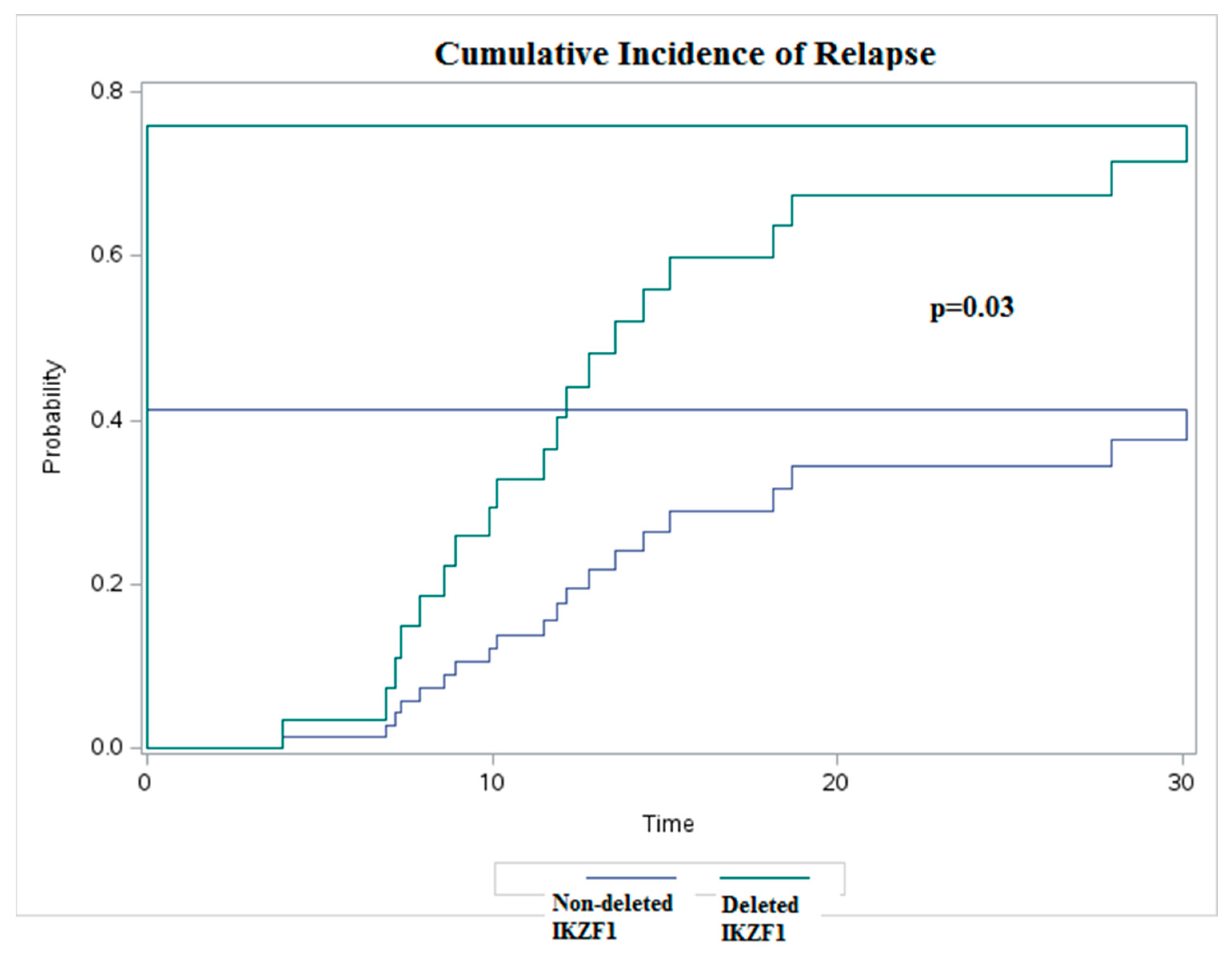
3. Results
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Georgopoulos, K.; Winandy, S.; Avitahl, N. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu. Rev. Immunol. 1997, 15, 155–176. [Google Scholar] [CrossRef]
- Sridharan, R.; Smale, S.T. Predominant interaction of both Ikaros and Helios with the NuRD complex in immature thymocytes. J. Biol. Chem. 2007, 282, 30227–30238. [Google Scholar] [CrossRef]
- Georgopoulos, K.; Moore, D.D.; Derfler, B. Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science 1992, 258, 808–812. [Google Scholar] [CrossRef] [PubMed]
- Payne, M.A. Zinc finger structure-function in Ikaros Marvin A Payne. World J. Biol. Chem. 2011, 2, 161–166. [Google Scholar] [CrossRef]
- Molnár, A.; Georgopoulos, K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol. Cell Biol. 1994, 14, 8292–8303. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.; Stadt, U.Z.; Escherich, G.; Hofmann, J.; Binato, R.; Barbosa, T.C.; Emerenciano, M.; Pombo-De-Oliveira, M.S.; Horstmann, M.; Marschalek, R. Refinement of IKZF1 recombination hotspots in pediatric BCP-ALL patients. Am. J. Blood Res. 2013, 3, 165–173. [Google Scholar] [PubMed]
- Bose, P.; Verstovsek, S. Prognosis of Primary Myelofibrosis in the Genomic Era. Clin. Lymphoma Myeloma Leuk. 2016, 16, S105–S113. [Google Scholar] [CrossRef]
- Kisiel, J.B.; Raimondo, M.; Taylor, W.R.; Yab, T.C.; Mahoney, D.W.; Sun, Z.; Middha, S.; Baheti, S.; Zou, H.; Smyrk, T.C.; et al. New DNA Methylation Markers for Pancreatic Cancer: Discovery, Tissue Validation, and Pilot Testing in Pancreatic Juice. Clin. Cancer Res. 2015, 21, 4473–4481. [Google Scholar] [CrossRef]
- Kuehn, H.S.; Nunes-Santos, C.J.; Rosenzweig, S.D. Germline IKZF1 mutations and their impact on immunity: IKAROS-associated diseases and pathophysiology. Expert Rev. Clin. Immunol. 2021, 17, 407–416. [Google Scholar] [CrossRef]
- Simonin, M.; Lhermitte, L.; Dourthe, M.-E.; Lengliné, E.; Graux, C.; Grardel, N.; Cayuela, J.-M.; Arnoux, I.; Gandemer, V.; Ifrah, N.; et al. IKZF1 alterations predict poor prognosis in adult and pediatric T-ALL. Blood 2021, 137, 1690–1694. [Google Scholar] [CrossRef]
- Mullighan, C.G.; Su, X.; Zhang, J.; Radtke, I.; Phillips, L.A.A.; Miller, C.B.; Ma, J.; Liu, W.; Cheng, C.; Schulman, B.A.; et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl. J. Med. 2009, 360, 470–480. [Google Scholar] [CrossRef] [PubMed]
- Kastner, P.; Chan, S. Role of Ikaros in T-cell acute lymphoblastic leukemia. World J. Biol. Chem. 2011, 2, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Kobitzsch, B.; Gökbuget, N.; Schwartz, S.; Reinhardt, R.; Brüggemann, M.; Viardot, A.; Wäsch, R.; Starck, M.; Thiel, E.; Hoelzer, D.; et al. Loss-of-function but not dominant-negative intragenic IKZF1 deletions are associated with an adverse prognosis in adult BCR-ABL-negative acute lymphoblastic leukemia. Haematologica 2017, 102, 1739–1747. [Google Scholar] [CrossRef]
- Martinelli, G.; Iacobucci, I.; Storlazzi, C.T.; Vignetti, M.; Paoloni, F.; Cilloni, D.; Soverini, S.; Vitale, A.; Chiaretti, S.; Cimino, G.; et al. IKZF1 (Ikaros) deletions in BCR-ABL1-positive acute lymphoblastic leukemia are associated with short disease-free survival and high rate of cumulative incidence of relapse: A GIMEMA AL WP report. J. Clin. Oncol. 2009, 27, 5202–5207. [Google Scholar] [CrossRef]
- Mitchell, R.J.; Kirkwood, A.A.; Barretta, E.; Clifton-Hadley, L.; Lawrie, E.; Lee, S.; Leongamornlert, D.; Marks, D.I.; McMillan, A.K.; Menne, T.F.; et al. IKZF1 alterations are not associated with outcome in 498 adults with B-precursor ALL enrolled in the UKALL14 trial. Blood Adv. 2021, 5, 3322–3332. [Google Scholar] [CrossRef] [PubMed]
- Iacobucci, I.; Lonetti, A.; Messa, F.; Cilloni, D.; Arruga, F.; Ottaviani, E.; Paolini, S.; Papayannidis, C.; Piccaluga, P.P.; Giannoulia, P.; et al. Expression of spliced oncogenic Ikaros isoforms in Philadelphia-positive acute lymphoblastic leukemia patients treated with tyrosine kinase inhibitors: Implications for a new mechanism of resistance. Blood 2008, 112, 3847–3855. [Google Scholar] [CrossRef] [PubMed]
- Van Der Veer, A.; Waanders, E.; Pieters, R.; Willemse, M.E.; Van Reijmersdal, S.V.; Russell, L.J.; Harrison, C.J.; Evans, W.E.; Van Der Velden, V.H.; Hoogerbrugge, P.M.; et al. Independent prognostic value of BCR-ABL1-like signature and IKZF1 deletion, but not high CRLF2 expression, in children with B-cell precursor ALL. Blood 2013, 122, 2622–2629. [Google Scholar] [CrossRef]
- Vrooman, L.M.; Blonquist, T.M.; Harris, M.H.; Stevenson, K.E.; Place, A.E.; Hunt, S.K.; O’brien, J.E.; Asselin, B.L.; Athale, U.H.; Clavell, L.A.; et al. Refining risk classification in childhood B acute lymphoblastic leukemia: Results of DFCI ALL Consortium Protocol 05-001. Blood Adv. 2018, 2, 1449–1458. [Google Scholar] [CrossRef]
- Kimura, H.; Onozawa, M.; Yoshida, S.; Miyashita, N.; Yokoyama, S.; Matsukawa, T.; Hirabayashi, S.; Goto, H.; Endo, T.; Oguri, S.; et al. Dominant-negative type of IKZF1 deletion showed a favorable prognosis in adult B-cell acute lymphoblastic leukemia. Ann. Hematol. 2023, 102, 3103–3113. [Google Scholar] [CrossRef]
- Kimura, H.; Onozawa, M.; Yoshida, S.; Miyashita, N.; Takahashi, S.; Yokoyama, S.; Matsukawa, T.; Hirabayashi, S.; Fujisawa, S.; Ota, S.; et al. Better Prognosis of Dominant-Negative Type of IKZF1 Deletion in BCR-ABL-positive Adult B-Cell Acute Lymphoblastic Leukemia. Blood 2022, 140, 9207–9208. [Google Scholar] [CrossRef]
- Marçais, A.; Jeannet, R.; Hernandez, L.; Soulier, J.; Sigaux, F.; Chan, S.; Kastner, P. Genetic inactivation of Ikaros is a rare event in human T-ALL. Leuk. Res. 2010, 34, 426–429. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Crotty, M.L.; Sensel, M.; Sather, H.; Navara, C.; Nachman, J.; Steinherz, P.G.; Gaynon, P.S.; Seibel, N.; Mao, C.; et al. Expression of dominant-negative Ikaros isoforms in T-cell acute lymphoblastic leukemia. Clin. Cancer Res. 1999, 5, 2112–2120. [Google Scholar]
- de Smith, A.J.; Wahlster, L.; Jeon, S.; Kachuri, L.; Black, S.; Langie, J.; Cato, L.D.; Nakatsuka, N.; Chan, T.-F.; Xia, G.; et al. A noncoding regulatory variant in IKZF1 increases acute lymphoblastic leukemia risk in Hispanic/Latino children. Cell Genom. 2024, 4, 100526. [Google Scholar] [CrossRef] [PubMed]
- Caye, A.; Beldjord, K.; Mass-Malo, K.; Drunat, S.; Soulier, J.; Gandemer, V.; Baruchel, A.; Bertrand, Y.; Cavé, H.; Clappier, E. Breakpoint-specific multiplex polymerase chain reaction allows the detection of IKZF1 intragenic deletions and minimal residual disease monitoring in B-cell precursor acute lymphoblastic leukemia. Haematologica 2013, 98, 597–601. [Google Scholar] [CrossRef]
- Reyes-León, A.; Juárez-Velázquez, R.; Medrano-Hernández, A.; Cuenca-Roldán, T.; Salas-Labadía, C.; Navarrete-Meneses, M.d.P.; Rivera-Luna, R.; López-Hernández, G.; Paredes-Aguilera, R.; Pérez-Vera, P. Expression of Ik6 and Ik8 Isoforms and Their Association with Relapse and Death in Mexican Children with Acute Lymphoblastic Leukemia. PLoS ONE 2015, 10, e0130756. [Google Scholar] [CrossRef]
- Rezayee, F.; Eisfeldt, J.; Skaftason, A.; Öfverholm, I.; Sayyab, S.; Syvänen, A.C.; Maqbool, K.; Lilljebjörn, H.; Johansson, B.; Olsson-Arvidsson, L.; et al. Feasibility to use whole-genome sequencing as a sole diagnostic method to detect genomic aberrations in pediatric B-cell acute lymphoblastic leukemia. Front. Oncol. 2023, 13, 1217712. [Google Scholar] [CrossRef]
- Brown, L.M.; Lonsdale, A.; Zhu, A.; Davidson, N.M.; Schmidt, B.; Hawkins, A.; Wallach, E.; Martin, M.; Mechinaud, F.M.; Khaw, S.L.; et al. The application of RNA sequencing for the diagnosis and genomic classification of pediatric acute lymphoblastic leukemia. Blood Adv. 2020, 4, 930–942. [Google Scholar] [CrossRef] [PubMed]
- Iacobucci, I.; Storlazzi, C.T.; Cilloni, D.; Lonetti, A.; Ottaviani, E.; Soverini, S.; Astolfi, A.; Chiaretti, S.; Vitale, A.; Messa, F.; et al. Identification and molecular characterization of recurrent genomic deletions on 7p12 in the IKZF1 gene in a large cohort of BCR-ABL1-positive acute lymphoblastic leukemia patients: On behalf of Gruppo Italiano Malattie Ematologiche dell’Adulto Acute Leukemia Working Party (GIMEMA AL WP). Blood 2009, 114, 2159–2167. [Google Scholar] [CrossRef]
- Lopes, B.A.; Meyer, C.; Bouzada, H.; Külp, M.; Maciel, A.L.T.; Larghero, P.; Barbosa, T.C.; Poubel, C.P.; Barbieri, C.; Venn, N.C.; et al. The recombinome of IKZF1 deletions in B-cell precursor ALL. Leukemia 2023, 37, 1727–1731. [Google Scholar] [CrossRef]
- Gökbuget, N.; Kneba, M.; Raff, T.; Trautmann, H.; Bartram, C.-R.; Arnold, R.; Fietkau, R.; Freund, M.; Ganser, A.; Ludwig, W.-D.; et al. Adult patients with acute lymphoblastic leukemia and molecular failure display a poor prognosis and are candidates for stem cell transplantation and targeted therapies. Blood 2012, 120, 1868–1876. [Google Scholar] [CrossRef]
- Kuehn, H.S.; Boast, B.; Rosenzweig, S.D. Inborn errors of human IKAROS: LOF and GOF variants associated with primary immunodeficiency. Clin. Exp. Immunol. 2023, 212, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Yeoh, A.E.J.; Lu, Y.; Ni Chin, W.H.; Chiew, E.K.H.; Lim, E.H.; Li, Z.; Kham, S.K.Y.; Chan, Y.H.; Abdullah, W.A.; Lin, H.P.; et al. Intensifying Treatment of Childhood B-Lymphoblastic Leukemia With IKZF1 Deletion Reduces Relapse and Improves Overall Survival: Results of Malaysia-Singapore ALL 2010 Study. J. Clin. Oncol. 2018, 36, 2726–2735. [Google Scholar] [CrossRef] [PubMed]
- Pieters, R.; de Groot-Kruseman, H.; Fiocco, M.; Verwer, F.; Van Overveld, M.; Sonneveld, E.; van der Velden, V.; Beverloo, H.B.; Bierings, M.; Dors, N.; et al. Improved Outcome for ALL by Prolonging Therapy for IKZF1 Deletion and Decreasing Therapy for Other Risk Groups. J. Clin. Oncol. 2023, 41, 4130–4142. [Google Scholar] [CrossRef]
- Tran, T.H.; Langlois, S.; Meloche, C.; Caron, M.; Saint-Onge, P.; Rouette, A.; Bataille, A.R.; Jimenez-Cortes, C.; Sontag, T.; Bittencourt, H.; et al. Whole-transcriptome analysis in acute lymphoblastic leukemia: A report from the DFCI ALL Consortium Protocol 16-001. Blood Adv. 2022, 6, 1329–1341. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | Patients with IKZF1 Deletion | Patients without IKZF1 Deletion | ||
|---|---|---|---|---|
| n = 19 | 24% | n = 59 | 76% | |
| Median age | 44 | 40 | ||
| Male | 12 | 63.2 | 34 | 57.6 |
| Female | 7 | 36.8 | 25 | 42.4 |
| B | 18 | 94.7 | 35 | 59.3 |
| T | 1 | 5.26 | 24 | 40.7 |
| Ph/+/ | 6 | 31.6 | 7 | 11.9 |
| KMT2A rearranged | 2 | 10.5 | 3 | 5.08 |
| t (1; 19) | 0 | 0 | 5 | 8.47 |
| B-ALL Hypoploidy | 1 | 1.69 | 1 | 1.69 |
| B-ALL Hyperploidy | 0 | 0 | 2 | 3.38 |
| Other cytogenetic abnormalities not listed above | 5 | 26.31 | 4 | 6.78 |
| Mean WBC count p = 0.703 | 48.5 | 42.7 | ||
| Mean bone marrow involvement p = 0.297 | 67.2 | 73.8 | ||
| Conventional risk groups as per the GMALL07/2003 initial risk classification | ||||
| VHR | 6 | 31.6 | 7 | 11.9 |
| HR | 9 | 47.4 | 29 | 49.2 |
| SR | 4 | 21.1 | 23 | 39 |
| CR rate | 12 | 63.2 | 46 | 78 |
| Allogeneic transplant rate | 4 | 21.1 | 21 | 35.6 |
| MRD negative after first consolidation p = 0.259 | 5 | 41.7 | 26 | 56.5 |
| Patient Number | Sex | Age | WHO Diagnosis | IKZF1del | Cytogenetics | Molecular Studies | CR | AlloHSCT |
|---|---|---|---|---|---|---|---|---|
| 3 | m | 45 | T-ALL/LBL | Δ4–8 | normal karyotype | not conducted | not achieved | No |
| 4 | f | 38 | B-ALL NOS | Δ4–7 | Trisomy 21 in 15% of the metaphasic plates | no abnormalities | achieved CR1 | No |
| 12 | m | 73 | B-ALL NOS | Δ4–7 and Δ4–8 | normal karyotype | no abnormalities | not achieved | No |
| 13 | m | 35 | B-ALL KMT2A rearranged | Δ4–7 | t (4; 11) | MLL-AF4 | achieved CR1 | Yes |
| 17 | m | 35 | B-ALL t (9; 22) | Δ4–7 | t (9; 22) | m BCR-ABL (p190) | not achieved | No |
| 20 | m | 44 | B-ALL NOS | Δ2–7 | 46, XX, t (3; 6) (q21; p23), −5, −7, add (12) (p13), +2 mar [9]/47, XYY [1]/46, XY [2] | no abnormalities | achieved CR1 | No |
| 23 | m | 41 | B-ALL t (9; 22) | Δ4–7 | t (9; 22) and near-tetraploid karyotype | m BCR-ABL (p190) | achieved CR1 | No |
| 31 | m | 45 | B-ALL NOS | Δ4–7 | 46, XY, del (3) (q22q26–27) [8]/46, XY [8] | no abnormalities | achieved CR1 | Yes |
| 47 | m | 30 | B-ALL t (9; 22) | Δ4–7 | t (9; 22) | m-BCR-ABL (p190) | achieved CR1 | Yes |
| 48 | f | 47 | B-ALL NOS | Δ4–7 | del 13q | no abnormalities | achieved CR1 | No |
| 49 | f | 23 | B-ALL NOS | Δ4–7 | normal karyotype | no abnormalities | not achieved | No |
| 55 | m | 16 | B-ALL NOS | Δ4–8 | no growth | no abnormalities | achieved CR1 | Yes |
| 75 | m | 50 | B-ALL t (9; 22) | Δ4–8 | t (9; 22) | M-BCR-ABL: p210 | achieved CR1 | No |
| 77 | f | 55 | B-ALL t (9; 22) | Δ4–7 | t (9; 22) | m-BCR-ABL (p190) | achieved CR1 | No |
| 84 | f | 37 | B-ALL NOS | Δ4–7 | 45, XX, −9 [7]/43–44, XX, −7 [1], −8 [5], −9, −13 [1], −22 [1], +mar [1] [cp 7]. | no abnormalities | not achieved | No |
| 85 | f | 71 | B-ALL NOS | Δ4–7 | normal karyotype | no abnormalities | achieved CR1 | No |
| 88 | f | 30 | B-ALL KMT2A rearranged | Δ4–7 and Δ4–8 | t (4; 11) (q21; q23) | MLL-AF4 | achieved CR1 | No |
| 89 | m | 45 | B-ALL t (9; 22) | Δ4–7 | no growth | M-BCR-ABL p210 | not achieved | No |
| 101 | m | 50 | B-ALL NOS | Δ4–7 | normal karyotype | no abnormalities | not achieved | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lozenov, S.; Tsoneva, Y.; Nikolaev, G.; Konakchieva, R. Ikaros Deletions among Bulgarian Patients with Acute Lymphoblastic Leukemia/Lymphoma. Diagnostics 2024, 14, 1953. https://doi.org/10.3390/diagnostics14171953
Lozenov S, Tsoneva Y, Nikolaev G, Konakchieva R. Ikaros Deletions among Bulgarian Patients with Acute Lymphoblastic Leukemia/Lymphoma. Diagnostics. 2024; 14(17):1953. https://doi.org/10.3390/diagnostics14171953
Chicago/Turabian StyleLozenov, Stefan, Yoanna Tsoneva, Georgi Nikolaev, and Rossitza Konakchieva. 2024. "Ikaros Deletions among Bulgarian Patients with Acute Lymphoblastic Leukemia/Lymphoma" Diagnostics 14, no. 17: 1953. https://doi.org/10.3390/diagnostics14171953






