A Machine Learning-Based Clustering Using Radiomics of F-18 Fluorodeoxyglucose Positron Emission Tomography/Computed Tomography for the Prediction of Prognosis in Patients with Intrahepatic Cholangiocarcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients, Tissue Samples, and Clinical Analysis
2.2. F-18 FDG PET/CT Image Acquisition
2.3. Image Processing and Analysis
2.4. Identification of Differentially Expressed Genes and Activated Pathways
2.5. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Radiomics-Based Clustering of Patients
3.3. Survival Analysis
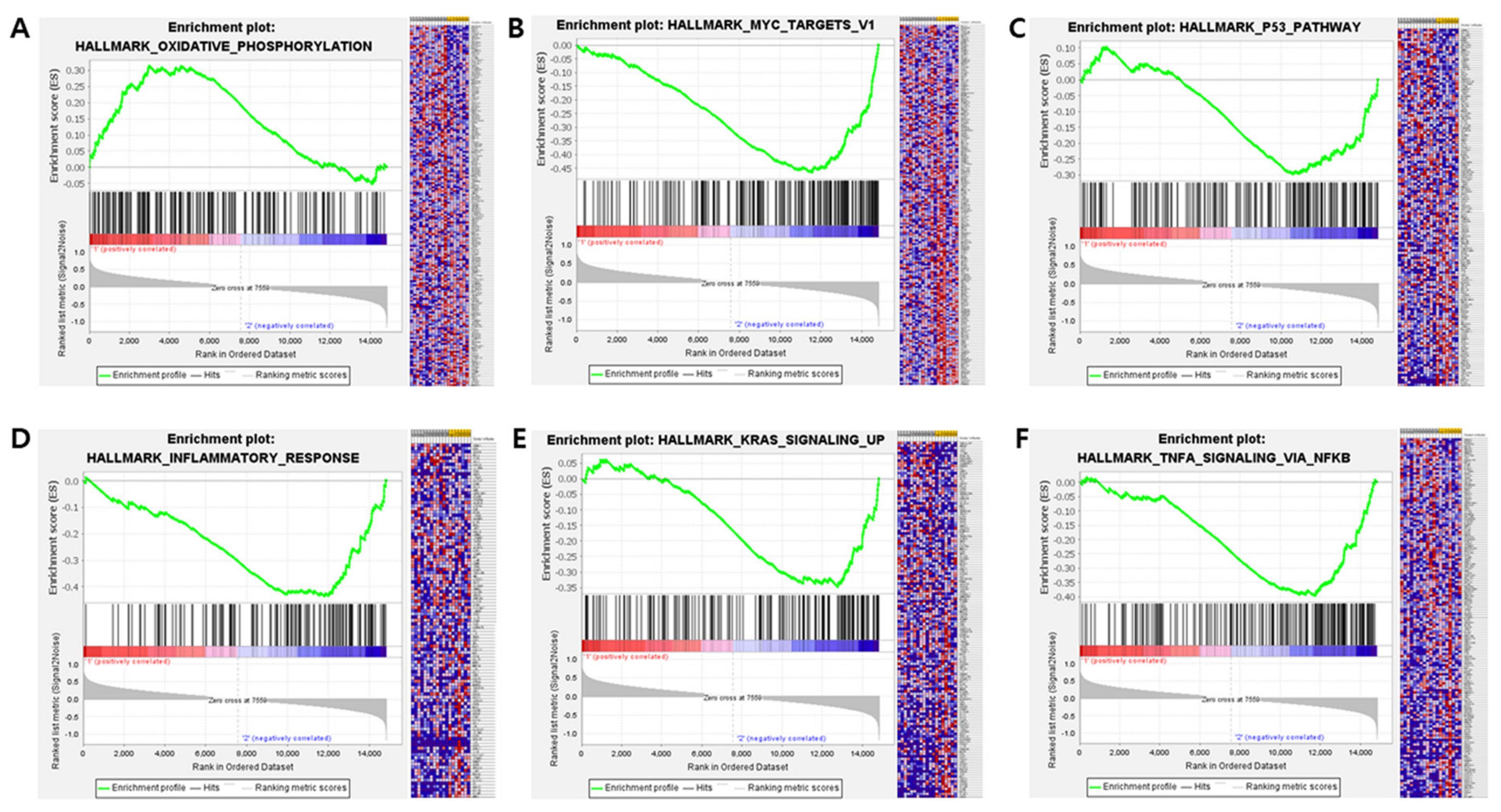
3.4. RNA Expression Profiling: Analysis of DEGs and Activated Pathways
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Buettner, S.; van Vugt, J.L.; IJzermans, J.N.; Groot Koerkamp, B. Intrahepatic Cholangiocarcinoma: Current Perspectives. Onco Targets Ther. 2017, 10, 1131–1142. [Google Scholar] [CrossRef] [PubMed]
- Bridgewater, J.; Galle, P.R.; Khan, S.A.; Llovet, J.M.; Park, J.-W.; Patel, T.; Pawlik, T.M.; Gores, G.J. Guidelines for the Diagnosis and Management of Intrahepatic Cholangiocarcinoma. J. Hepatol. 2014, 60, 1268–1289. [Google Scholar] [CrossRef] [PubMed]
- Bertuccio, P.; Malvezzi, M.; Carioli, G.; Hashim, D.; Boffetta, P.; El-Serag, H.B.; La Vecchia, C.; Negri, E. Global Trends in Mortality from Intrahepatic and Extrahepatic Cholangiocarcinoma. J. Hepatol. 2019, 71, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-F.; Beal, E.W.; Bagante, F.; Chakedis, J.; Weiss, M.; Popescu, I.; Marques, H.P.; Aldrighetti, L.; Maithel, S.K.; Pulitano, C.; et al. Early versus Late Recurrence of Intrahepatic Cholangiocarcinoma after Resection with Curative Intent. Br. J. Surg. 2018, 105, 848–856. [Google Scholar] [CrossRef] [PubMed]
- Mavros, M.N.; Economopoulos, K.P.; Alexiou, V.G.; Pawlik, T.M. Treatment and Prognosis for Patients with Intrahepatic Cholangiocarcinoma: Systematic Review and Meta-Analysis. JAMA Surg. 2014, 149, 565–574. [Google Scholar] [CrossRef]
- Chan, K.-M.; Tsai, C.-Y.; Yeh, C.-N.; Yeh, T.-S.; Lee, W.-C.; Jan, Y.-Y.; Chen, M.-F. Characterization of Intrahepatic Cholangiocarcinoma after Curative Resection: Outcome, Prognostic Factor, and Recurrence. BMC Gastroenterol. 2018, 18, 180. [Google Scholar] [CrossRef]
- Tanaka, M.; Tanaka, H.; Tsukuma, H.; Ioka, A.; Oshima, A.; Nakahara, T. Risk Factors for Intrahepatic Cholangiocarcinoma: A Possible Role of Hepatitis B Virus. J. Viral Hepat. 2010, 17, 742–748. [Google Scholar] [CrossRef]
- Labib, P.L.; Goodchild, G.; Pereira, S.P. Molecular Pathogenesis of Cholangiocarcinoma. BMC Cancer 2019, 19, 185. [Google Scholar] [CrossRef]
- Chaiteerakij, R.; Yang, J.D.; Harmsen, W.S.; Slettedahl, S.W.; Mettler, T.A.; Fredericksen, Z.S.; Kim, W.R.; Gores, G.J.; Roberts, R.O.; Olson, J.E.; et al. Risk Factors for Intrahepatic Cholangiocarcinoma: Association between Metformin Use and Reduced Cancer Risk. Hepatology 2013, 57, 648–655. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding Tumour Phenotype by Noninvasive Imaging Using a Quantitative Radiomics Approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Corvera, C.U.; Blumgart, L.H.; Akhurst, T.; DeMatteo, R.P.; D’Angelica, M.; Fong, Y.; Jarnagin, W.R. 18F-Fluorodeoxyglucose Positron Emission Tomography Influences Management Decisions in Patients with Biliary Cancer. J. Am. Coll. Surg. 2008, 206, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Onozato, Y.; Iwata, T.; Uematsu, Y.; Shimizu, D.; Yamamoto, T.; Matsui, Y.; Ogawa, K.; Kuyama, J.; Sakairi, Y.; Kawakami, E.; et al. Predicting Pathological Highly Invasive Lung Cancer from Preoperative [18F]FDG PET/CT with Multiple Machine Learning Models. Eur. J. Nucl. Med. Mol. Imaging 2023, 50, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Monaco, L.; De Bernardi, E.; Bono, F.; Cortinovis, D.; Crivellaro, C.; Elisei, F.; L’Imperio, V.; Landoni, C.; Mathoux, G.; Musarra, M.; et al. The “Digital Biopsy” in Non-Small Cell Lung Cancer (NSCLC): A Pilot Study to Predict the PD-L1 Status from Radiomics Features of [18F]FDG PET/CT. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 3401–3411. [Google Scholar] [CrossRef] [PubMed]
- Song, B.-I. A Machine Learning-Based Radiomics Model for the Prediction of Axillary Lymph-Node Metastasis in Breast Cancer. Breast Cancer 2021, 28, 664–671. [Google Scholar] [CrossRef]
- Eertink, J.J.; Zwezerijnen, G.J.C.; Cysouw, M.C.F.; Wiegers, S.E.; Pfaehler, E.A.G.; Lugtenburg, P.J.; van der Holt, B.; Hoekstra, O.S.; de Vet, H.C.W.; Zijlstra, J.M.; et al. Comparing Lesion and Feature Selections to Predict Progression in Newly Diagnosed DLBCL Patients with FDG PET/CT Radiomics Features. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 4642–4651. [Google Scholar] [CrossRef]
- Fiz, F.; Masci, C.; Costa, G.; Sollini, M.; Chiti, A.; Ieva, F.; Torzilli, G.; Viganò, L. PET/CT-Based Radiomics of Mass-Forming Intrahepatic Cholangiocarcinoma Improves Prediction of Pathology Data and Survival. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 3387–3400. [Google Scholar] [CrossRef]
- Hyun, S.H.; Ahn, M.S.; Koh, Y.W.; Lee, S.J. A Machine-Learning Approach Using PET-Based Radiomics to Predict the Histological Subtypes of Lung Cancer. Clin. Nucl. Med. 2019, 44, 956–960. [Google Scholar] [CrossRef]
- Alongi, P.; Laudicella, R.; Stefano, A.; Caobelli, F.; Comelli, A.; Vento, A.; Sardina, D.; Ganduscio, G.; Toia, P.; Ceci, F.; et al. Choline PET/CT Features to Predict Survival Outcome in High-Risk Prostate Cancer Restaging: A Preliminary Machine-Learning Radiomics Study. Q. J. Nucl. Med. Mol. Imaging 2022, 66, 352–360. [Google Scholar] [CrossRef]
- Figueroa, R.L.; Zeng-Treitler, Q.; Kandula, S.; Ngo, L.H. Predicting Sample Size Required for Classification Performance. BMC Med. Inform. Decis. Mak. 2012, 12, 8. [Google Scholar] [CrossRef]
- Avanzo, M.; Wei, L.; Stancanello, J.; Vallières, M.; Rao, A.; Morin, O.; Mattonen, S.A.; El Naqa, I. Machine and Deep Learning Methods for Radiomics. Med. Phys. 2020, 47, e185–e202. [Google Scholar] [CrossRef]
- Schöder, H.; Erdi, Y.E.; Chao, K.; Gonen, M.; Larson, S.M.; Yeung, H.W.D. Clinical Implications of Different Image Reconstruction Parameters for Interpretation of Whole-Body PET Studies in Cancer Patients. J. Nucl. Med. 2004, 45, 559–566. [Google Scholar] [PubMed]
- He, Y.; Jie, L.; Dehong, Y.; Pu, W. An Improved Algorithm of the Maximum Entropy Image Segmentation. In Proceedings of the 2014 Fifth International Conference on Intelligent Systems Design and Engineering Applications, Hunan, China, 15–16 June 2014; pp. 157–160. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
- Haga, A.; Takahashi, W.; Aoki, S.; Nawa, K.; Yamashita, H.; Abe, O.; Nakagawa, K. Standardization of Imaging Features for Radiomics Analysis. J. Med. Investig. 2019, 66, 35–37. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Ramakrishnan, R.; Livny, M. BIRCH: A New Data Clustering Algorithm and Its Applications. Data Min. Knowl. Discov. 1997, 1, 141–182. [Google Scholar] [CrossRef]
- Song, B.-I.; Lee, J.; Jung, W.; Kim, B.S. Pure Uric Acid Stone Prediction Model Using the Variant Coefficient of Stone Density Measured by Thresholding 3D Segmentation-Based Methods: A Multicenter Study. Comput. Methods Programs Biomed. 2023, 240, 107691. [Google Scholar] [CrossRef]
- Ahn, K.S.; O’Brien, D.; Kang, Y.N.; Mounajjed, T.; Kim, Y.H.; Kim, T.-S.; Kocher, J.-P.A.; Allotey, L.K.; Borad, M.J.; Roberts, L.R.; et al. Prognostic Subclass of Intrahepatic Cholangiocarcinoma by Integrative Molecular-Clinical Analysis and Potential Targeted Approach. Hepatol. Int. 2019, 13, 490–500. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, B.; He, Y.; Pang, L.; Yu, H.; Shi, H. Correlation among Maximum Standardized 18F-FDG Uptake and Pathological Differentiation, Tumor Size, and Ki67 in Patients with Moderately and Poorly Differentiated Intrahepatic Cholangiocarcinoma. Hell. J. Nucl. Med. 2022, 25, 38–42. [Google Scholar] [CrossRef]
- Hyun, S.H.; Eo, J.S.; Song, B.-I.; Lee, J.W.; Na, S.J.; Hong, I.K.; Oh, J.K.; Chung, Y.A.; Kim, T.-S.; Yun, M. Preoperative Prediction of Microvascular Invasion of Hepatocellular Carcinoma Using 18F-FDG PET/CT: A Multicenter Retrospective Cohort Study. Eur. J. Nucl. Med. Mol. Imaging 2018, 45, 720–726. [Google Scholar] [CrossRef]
- Jiang, C.; Zhao, L.; Xin, B.; Ma, G.; Wang, X.; Song, S. 18F-FDG PET/CT Radiomic Analysis for Classifying and Predicting Microvascular Invasion in Hepatocellular Carcinoma and Intrahepatic Cholangiocarcinoma. Quant. Imaging Med. Surg. 2022, 12, 4135–4150. [Google Scholar] [CrossRef] [PubMed]
- Song, B.-I.; Kim, H.W.; Won, K.S. Predictive Value of 18F-FDG PET/CT for Axillary Lymph Node Metastasis in Invasive Ductal Breast Cancer. Ann. Surg. Oncol. 2017, 24, 2174–2181. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Song, B.-I.; Kim, B.W.; Kim, H.W.; Won, K.S.; Bae, S.U.; Jeong, W.K.; Baek, S.K. Predictive Value of [18F]FDG PET/CT for Lymph Node Metastasis in Rectal Cancer. Sci. Rep. 2019, 9, 4979. [Google Scholar] [CrossRef]
- Song, B.-I. Nomogram Using F-18 Fluorodeoxyglucose Positron Emission Tomography/Computed Tomography for Preoperative Prediction of Lymph Node Metastasis in Gastric Cancer. World J. Gastrointest. Oncol. 2020, 12, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Na, S.J.; Oh, J.K.; Hyun, S.H.; Lee, J.W.; Hong, I.K.; Song, B.-I.; Kim, T.-S.; Eo, J.S.; Lee, S.W.; Yoo, I.R.; et al. (18)F-FDG PET/CT Can Predict Survival of Advanced Hepatocellular Carcinoma Patients: A Multicenter Retrospective Cohort Study. J. Nucl. Med. 2017, 58, 730–736. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kim, S.H.; Song, B.I.; Kim, H.W.; Won, K.S.; Son, Y.G.; Ryu, S.W. Prognostic Value of Restaging F-18 Fluorodeoxyglucose Positron Emission Tomography/Computed Tomography to Predict 3-Year Post-Recurrence Survival in Patients with Recurrent Gastric Cancer after Curative Resection. Korean J. Radiol. 2020, 21, 829–837. [Google Scholar] [CrossRef]
- Kim, S.H.; Song, B.-I.; Kim, H.W.; Won, K.S.; Son, Y.-G.; Ryu, S.W.; Kang, Y.N. Prognostic Value of the Metabolic Score Obtained via [18F]FDG PET/CT and a New Prognostic Staging System for Gastric Cancer. Sci. Rep. 2022, 12, 20681. [Google Scholar] [CrossRef]
- Marei, H.E.; Althani, A.; Afifi, N.; Hasan, A.; Caceci, T.; Pozzoli, G.; Morrione, A.; Giordano, A.; Cenciarelli, C. P53 Signaling in Cancer Progression and Therapy. Cancer Cell Int. 2021, 21, 703. [Google Scholar] [CrossRef]
- Andersen, J.B.; Spee, B.; Blechacz, B.R.; Avital, I.; Komuta, M.; Barbour, A.; Conner, E.A.; Gillen, M.C.; Roskams, T.; Roberts, L.R.; et al. Genomic and Genetic Characterization of Cholangiocarcinoma Identifies Therapeutic Targets for Tyrosine Kinase Inhibitors. Gastroenterology 2012, 142, 1021–1031. [Google Scholar] [CrossRef]
- Ahn, K.S.; O’Brien, D.R.; Kim, Y.H.; Kim, T.-S.; Yamada, H.; Park, J.-W.; Park, S.-J.; Kim, S.H.; Zhang, C.; Li, H.; et al. Associations of Serum Tumor Biomarkers with Integrated Genomic and Clinical Characteristics of Hepatocellular Carcinoma. Liver Cancer 2021, 10, 593–605. [Google Scholar] [CrossRef]
- Zhang, G.-F.; Qiu, L.; Yang, S.-L.; Wu, J.-C.; Liu, T.-J. Wnt/β-Catenin Signaling as an Emerging Potential Key Pharmacological Target in Cholangiocarcinoma. Biosci. Rep. 2020, 40, BSR20193353. [Google Scholar] [CrossRef] [PubMed]
- Cook, G.J.R.; Siddique, M.; Taylor, B.P.; Yip, C.; Chicklore, S.; Goh, V. Radiomics in PET: Principles and Applications. Clin. Transl. Imaging 2014, 2, 269–276. [Google Scholar] [CrossRef]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images Are More than Pictures, They Are Data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Fang, S.; Li, M.; Liu, Y.; Liang, X.; Li, Z.; Chen, G.; Peng, L.; Chen, N.; Liu, L.; et al. Genetic Alterations of KRAS and TP53 in Intrahepatic Cholangiocarcinoma Associated with Poor Prognosis. Open Life Sci. 2023, 18, 20220652. [Google Scholar] [CrossRef] [PubMed]
- Lozano, E.; Sanchon-Sanchez, P.; Morente-Carrasco, A.; Chinchilla-Tábora, L.M.; Mauriz, J.L.; Fernández-Palanca, P.; Marin, J.J.G.; Macias, R.I.R. Impact of Aberrant β-Catenin Pathway on Cholangiocarcinoma Heterogeneity. Cells 2023, 12, 1141. [Google Scholar] [CrossRef]
- Kim, G.R.; Ku, Y.J.; Kim, J.H.; Kim, E.-K. Correlation between MR Image-Based Radiomics Features and Risk Scores Associated with Gene Expression Profiles in Breast Cancer. Taehan Yongsang Uihakhoe Chi 2020, 81, 632–643. [Google Scholar] [CrossRef]
- He, X.; Zhao, J.; He, D.; Ren, J.; Zhao, L.; Huang, G. Radiogenomics Study to Predict the Nuclear Grade of Renal Clear Cell Carcinoma. Eur. J. Radiol. Open 2023, 10, 100476. [Google Scholar] [CrossRef]
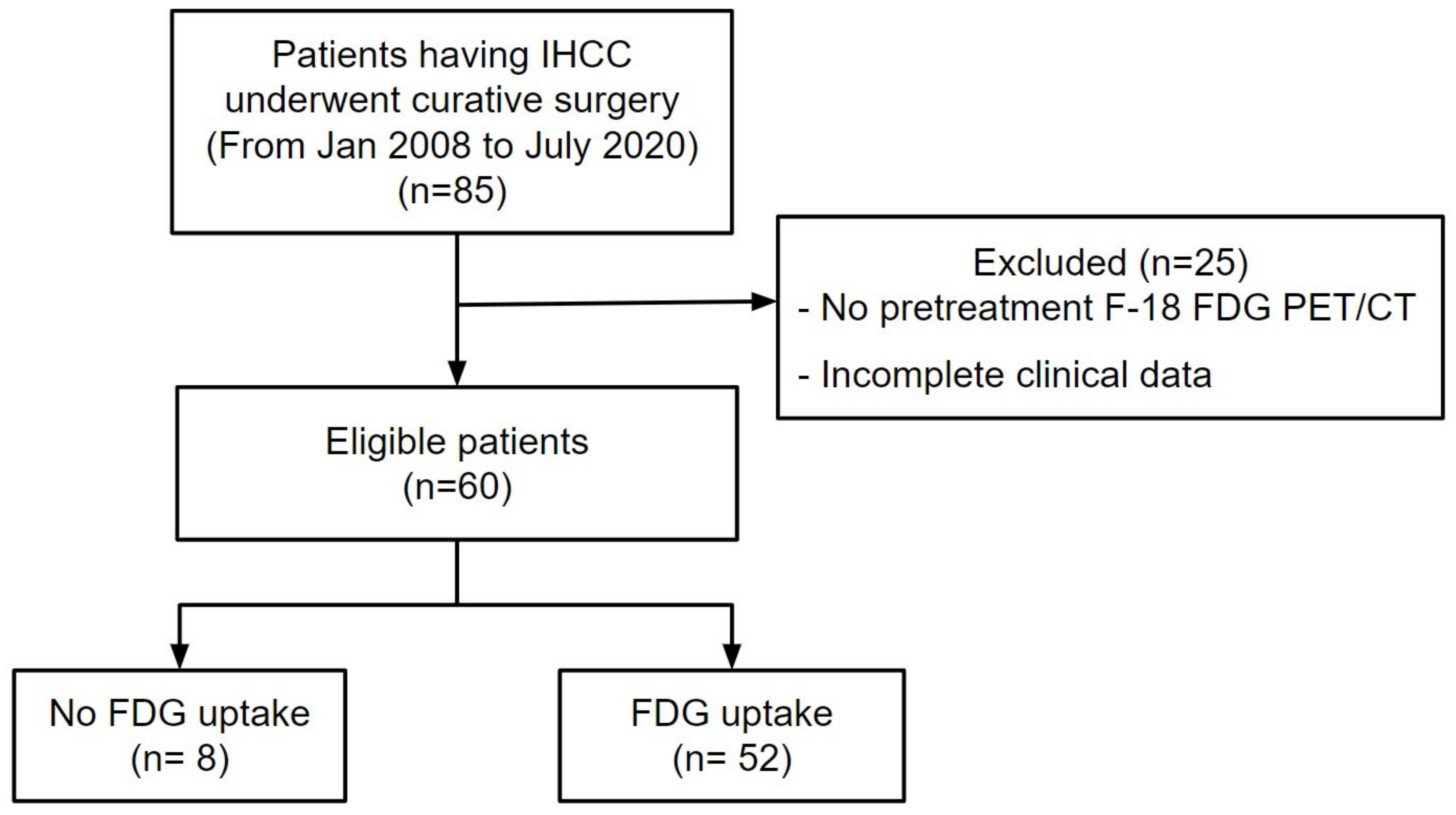

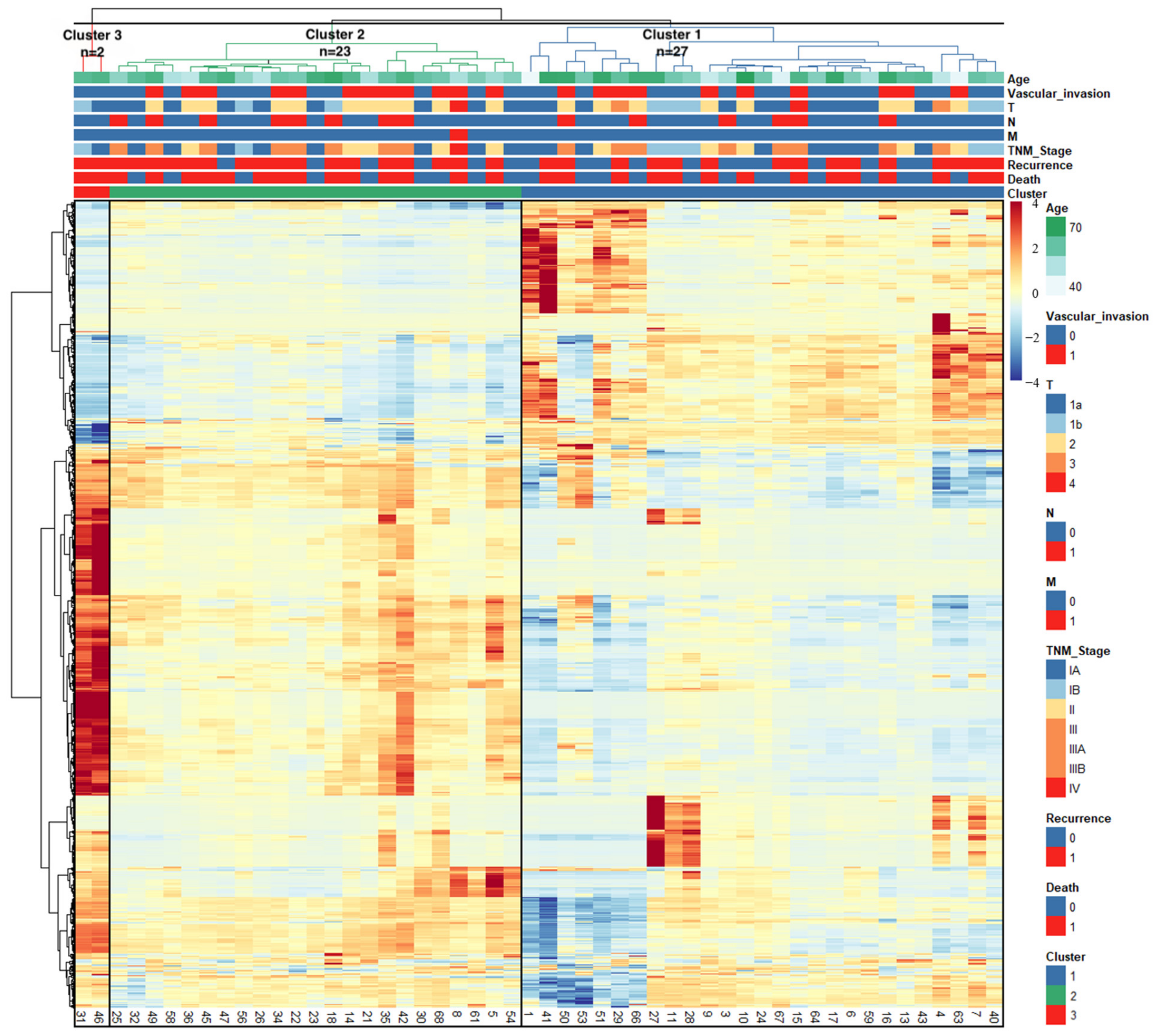
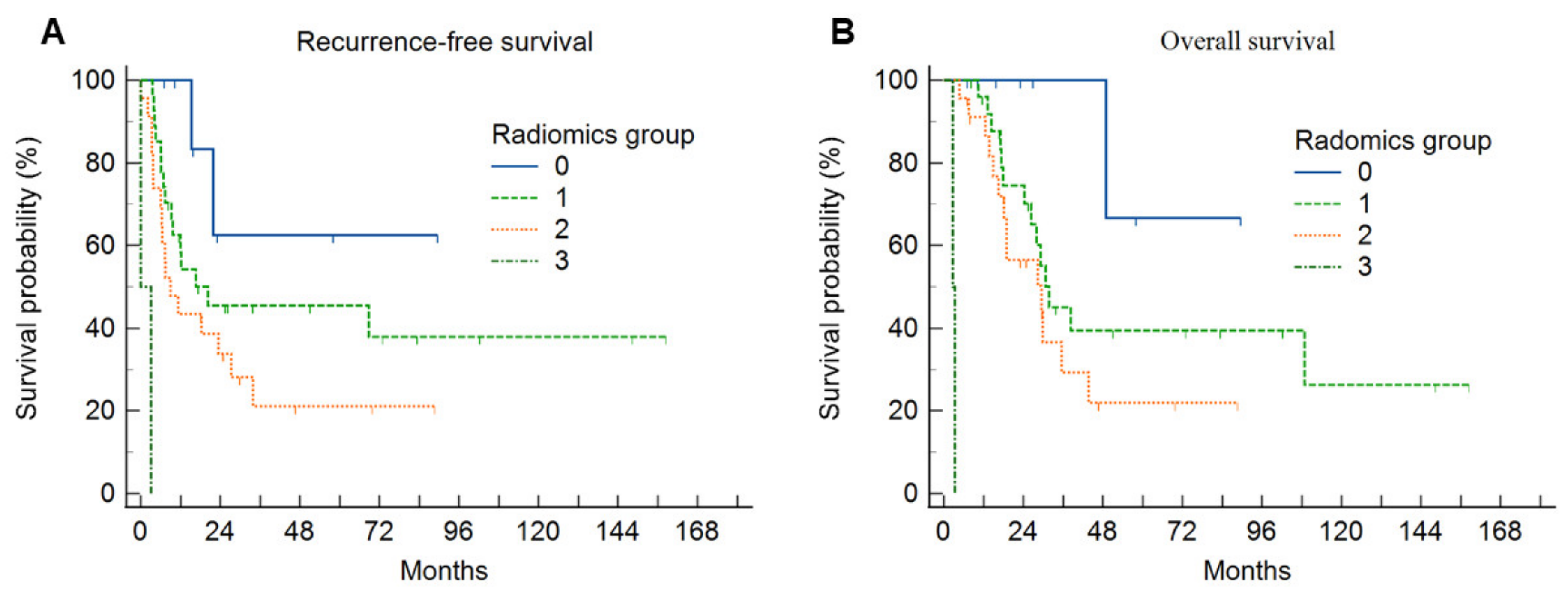
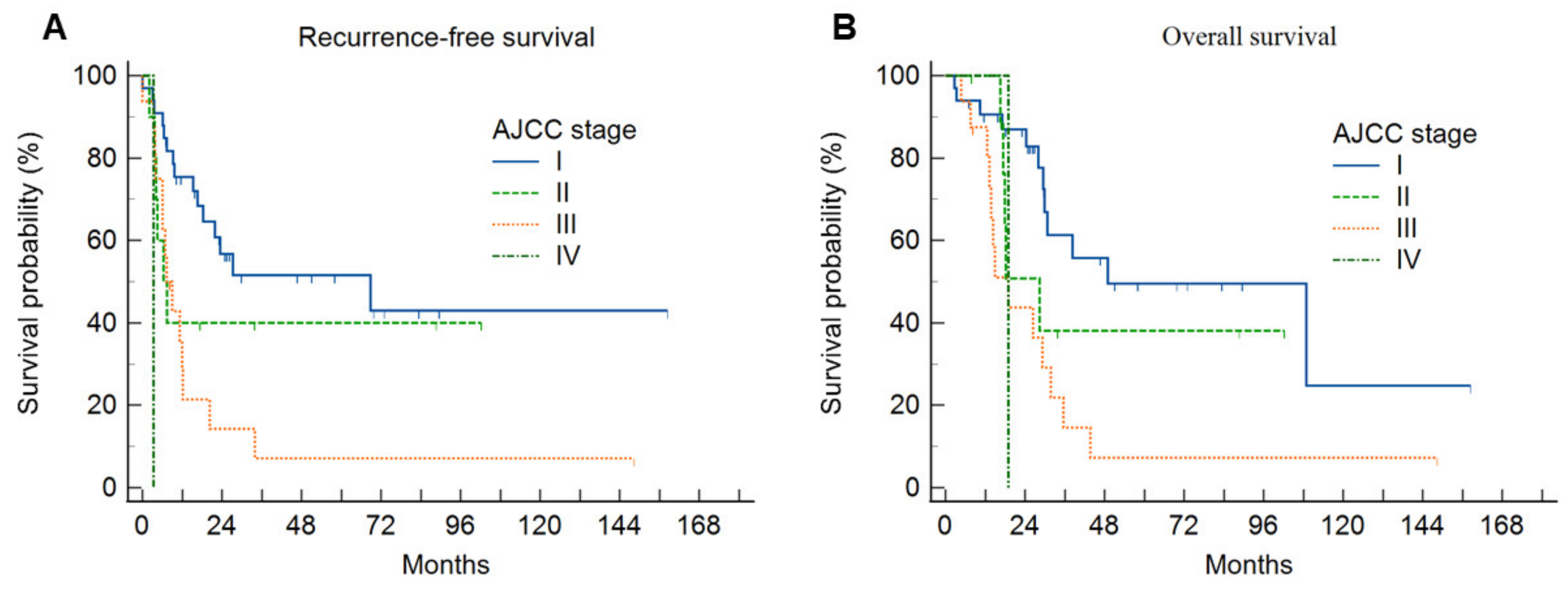
| Characteristics | Group 0 (n = 8) | Group 1 (n = 27) | Group 2 (n = 23) | Group 3 (n = 2) | p |
|---|---|---|---|---|---|
| Age | 67.0 ± 8.8 | 62.7 ± 11.9 | 62.4 ± 7.8 | 69.0 ± 1.4 | 0.576 |
| Sex | 0.175 | ||||
| Female | 2 (25.0%) | 10 (37.0%) | 6 (26.1%) | 2 (100.0%) | |
| Male | 6 (75.0%) | 17 (63.0%) | 17 (73.9%) | 0 (0.0%) | |
| CEA | 3.6 ± 2.2 | 2.3 ± 2.8 | 3.5 ± 1.6 | 2.4 ± 2.7 | 0.494 |
| CA19-9 | 295.8 ± 258.7 | 417.1 ± 1286.4 | 554.1 ± 1727.4 | 42.4 ± 18.4 | 0.955 |
| Differentiation | 0.667 | ||||
| Well | 0 (0.0%) | 1 (3.7%) | 0 (0.0%) | 0 (0.0%) | |
| Moderately | 7 (87.5%) | 15 (55.6%) | 16 (69.6%) | 1 (50.0%) | |
| Poorly | 1 (12.5%) | 11 (40.7%) | 7 (30.4%) | 1 (50.0%) | |
| Vascular invasion | 0.043 | ||||
| Negative | 8 (100.0%) | 17 (63.0%) | 11 (47.8%) | 2 (100.0%) | |
| Positive | 0 (0.0%) | 10 (37.0%) | 12 (52.2%) | 0 (0.0%) | |
| Gross appearance | 0.388 | ||||
| Mass forming | 6 (75.0%) | 25 (92.6%) | 18 (78.3%) | 2 (100.0%) | |
| Periductal infiltrating or mixed | 2 (25.0%) | 2 (7.4%) | 5 (21.7%) | 0 (0.0%) | |
| Microscopic type * | 0.827 | ||||
| Small duct type | 4 (57.1%) | 14 (56.0%) | 10 (47.6%) | 0 (0.0%) | |
| Large duct type | 3 (42.9%) | 11 (44.0%) | 11 (52.4%) | 1 (100.0%) | |
| Tumor size (cm) | 2.2 ± 0.9 | 5.6 ± 2.9 | 4.6 ± 1.5 | 4.3 ± 3.1 | 0.006 |
| LN metastasis | 0.154 | ||||
| Negative | 8 (100.0%) | 22 (81.5%) | 15 (65.2%) | 2 (100.0%) | |
| Positive | 0 (0.0%) | 5 (18.5%) | 8 (34.8%) | 0 (0.0%) | |
| Pathologic T stage | 0.146 | ||||
| 1a | 8 (100.0%) | 11 (40.7%) | 9 (39.1%) | 1 (50.0%) | |
| 1b | 0 (0.0%) | 5 (18.5%) | 2 (8.7%) | 1 (50.0%) | |
| 2 | 0 (0.0%) | 8 (29.6%) | 11 (47.8%) | 0 (0.0%) | |
| 3 | 0 (0.0%) | 2 (7.4%) | 0 (0.0%) | 0 (0.0%) | |
| 4 | 0 (0.0%) | 1 (3.7%) | 1 (4.3%) | 0 (0.0%) | |
| Pathologic N stage | 0.191 | ||||
| 0 | 8 (100.0%) | 21 (77.8%) | 15 (65.2%) | 2 (100.0%) | |
| 1 | 0 (0.0%) | 6 (22.2%) | 8 (34.8%) | 0 (0.0%) | |
| Pathologic M stage | 0.651 | ||||
| 0 | 8 (100.0%) | 27 (100.0%) | 22 (95.7%) | 2 (100.0%) | |
| 1 | 0 (0.0%) | 0 (0.0%) | 1 (4.3%) | 0 (0.0%) | |
| 8th AJCC TNM stage | 0.127 | ||||
| IA | 8 (100.0%) | 10 (37.0%) | 7 (30.4%) | 0 (0.0%) | |
| IB | 0 (0.0%) | 5 (18.5%) | 1 (4.3%) | 1 (50.0%) | |
| II | 0 (0.0%) | 5 (18.5%) | 6 (26.1%) | 1 (50.0%) | |
| III | 0 (0.0%) | 1 (3.7%) | 0 (0.0%) | 0 (0.0%) | |
| IIIA | 0 (0.0%) | 1 (3.7%) | 0 (0.0%) | 0 (0.0%) | |
| IIIB | 0 (0.0%) | 5 (18.5%) | 8 (34.8%) | 0 (0.0%) | |
| IV | 0 (0.0%) | 0 (0.0%) | 1 (4.3%) | 0 (0.0%) | |
| SUVmax | - | 6.8 ± 1.8 | 11.6 ± 2.8 | 28.8 ± 5.6 | 0 |
| MTV (mm3) | - | 76,508.0 ± 135,011.5 | 37,964.3 ± 37,340.1 | 99,738.0 ± 12,982.5 | 0.357 |
| Univariate Analysis | Multivariate Analysis | |||
|---|---|---|---|---|
| Variables | HR (95% CI) | p Value | HR (95% CI) | p Value |
| Age, years (<65 vs. ≥65) | 1.05 (0.55–2.02) | 0.886 | ||
| Sex (female vs. male) | 2.28 (1.03–5.04) | 0.043 | 0.69 (0.26–1.82) | 0.453 |
| CEA (<5 ng/mL vs. ≥5 ng/mL) | 0.92 (0.31–2.70) | 0.879 | ||
| CA 19-9 (<37 U/mL vs. ≥37 U/mL) | 2.57 (1.24–5.32) | 0.011 | 5.50 (2.12–14.25) | <0.001 |
| Tumor differentiation (well, moderately, poorly) | 1.77 (0.94–3.33) | 0.079 | ||
| Vascular invasion (No vs. Yes) | 2.54 (1.31–4.95) | 0.006 | 0.35 (0.10–1.27) | 0.111 |
| Tumor size (<5 cm vs. ≥5 cm) | 1.46 (0.75–2.85) | 0.262 | ||
| Pathologic T stage (T1, T2, T3, T4) | 1.79 (1.24–2.59) | 0.002 | 2.31 (1.16–4.61) | 0.018 |
| Pathologic N stage (N0, N1) | 2.45 (1.21–4.96) | 0.013 | 3.15 (1.25–7.93) | 0.015 |
| PET radiomics group (0, 1, 2, 3) | 2.19 (1.28–3.47) | 0.004 | 3.03 (1.55–5.95) | 0.001 |
| Univariate Analysis | Multivariate Analysis | |||
|---|---|---|---|---|
| Variables | HR (95% CI) | p Value | HR (95% CI) | p Value |
| Age, years (<65 vs. ≥65) | 1.54 (0.75–3.19) | 0.242 | ||
| Sex (female vs. male) | 1.89 (0.86–4.13) | 0.111 | ||
| CEA (<5 ng/mL vs. ≥5 ng/mL) | 0.61 (0.14–2.66) | 0.514 | ||
| CA 19-9 (<37 U/mL vs. ≥37 U/mL) | 2.72 (1.25–5.93) | 0.012 | 4.87 (1.76–13.44) | 0.002 |
| Tumor differentiation (well, moderately, poorly) | 1.72 (0.86–3.46) | 0.126 | ||
| Vascular invasion (no vs. yes) | 3.18 (1.53–6.62) | 0.002 | 1.01 (0.30–3.38) | 0.985 |
| Tumor size (<5 cm vs. ≥5 cm) | 1.26 (0.62–2.57) | 0.528 | ||
| Pathologic T stage (T1, T2, T3, T4) | 2.07 (1.39–3.08) | <0.001 | 1.55 (0.80–3.03) | 0.197 |
| Pathologic N stage (N0, N1) | 2.60 (1.24–5.47) | 0.012 | 4.09 (1.57–10.70) | 0.004 |
| PET radiomics group (0, 1, 2, 3) | 2.46 (1.32–4.56) | 0.004 | 2.39 (1.09–5.25) | 0.030 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwon, R.; Kim, H.; Ahn, K.S.; Song, B.-I.; Lee, J.; Kim, H.W.; Won, K.S.; Lee, H.W.; Kim, T.-S.; Kim, Y.; et al. A Machine Learning-Based Clustering Using Radiomics of F-18 Fluorodeoxyglucose Positron Emission Tomography/Computed Tomography for the Prediction of Prognosis in Patients with Intrahepatic Cholangiocarcinoma. Diagnostics 2024, 14, 2245. https://doi.org/10.3390/diagnostics14192245
Kwon R, Kim H, Ahn KS, Song B-I, Lee J, Kim HW, Won KS, Lee HW, Kim T-S, Kim Y, et al. A Machine Learning-Based Clustering Using Radiomics of F-18 Fluorodeoxyglucose Positron Emission Tomography/Computed Tomography for the Prediction of Prognosis in Patients with Intrahepatic Cholangiocarcinoma. Diagnostics. 2024; 14(19):2245. https://doi.org/10.3390/diagnostics14192245
Chicago/Turabian StyleKwon, Rosie, Hannah Kim, Keun Soo Ahn, Bong-Il Song, Jinny Lee, Hae Won Kim, Kyoung Sook Won, Hye Won Lee, Tae-Seok Kim, Yonghoon Kim, and et al. 2024. "A Machine Learning-Based Clustering Using Radiomics of F-18 Fluorodeoxyglucose Positron Emission Tomography/Computed Tomography for the Prediction of Prognosis in Patients with Intrahepatic Cholangiocarcinoma" Diagnostics 14, no. 19: 2245. https://doi.org/10.3390/diagnostics14192245
APA StyleKwon, R., Kim, H., Ahn, K. S., Song, B.-I., Lee, J., Kim, H. W., Won, K. S., Lee, H. W., Kim, T.-S., Kim, Y., & Kang, K. J. (2024). A Machine Learning-Based Clustering Using Radiomics of F-18 Fluorodeoxyglucose Positron Emission Tomography/Computed Tomography for the Prediction of Prognosis in Patients with Intrahepatic Cholangiocarcinoma. Diagnostics, 14(19), 2245. https://doi.org/10.3390/diagnostics14192245





