Demonstration of T-Cell Monotypia Using Anti-TCRbeta1/2 (TRBC1/2) Immunostaining as a Rapid and Cost-Effective Alternative to PCR-Based Clonality Studies for the Diagnosis of T-Cell Lymphoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Production of Recombinant Rabbit Antibodies
2.2. Cell Culture and Production of FFPE Cell Line Pellets
2.3. Western Blotting
2.4. FFPE Clinical Tissue Samples
2.5. Immunostaining
2.6. BaseScopeTM RNA In Situ Hybridization
2.7. Analysis of Histological Results
2.8. Quantitative Real-Time Reverse Transcription PCR
2.9. Bioinformatic Analysis of RNAseq Data
3. Results
3.1. Validating TCRbeta1 and TCRbeta2-Specific Antibodies Applicable to Formalin-Fixed Paraffin-Embedded (FFPE; Routine Clinical) Tissue
3.1.1. Western Blotting
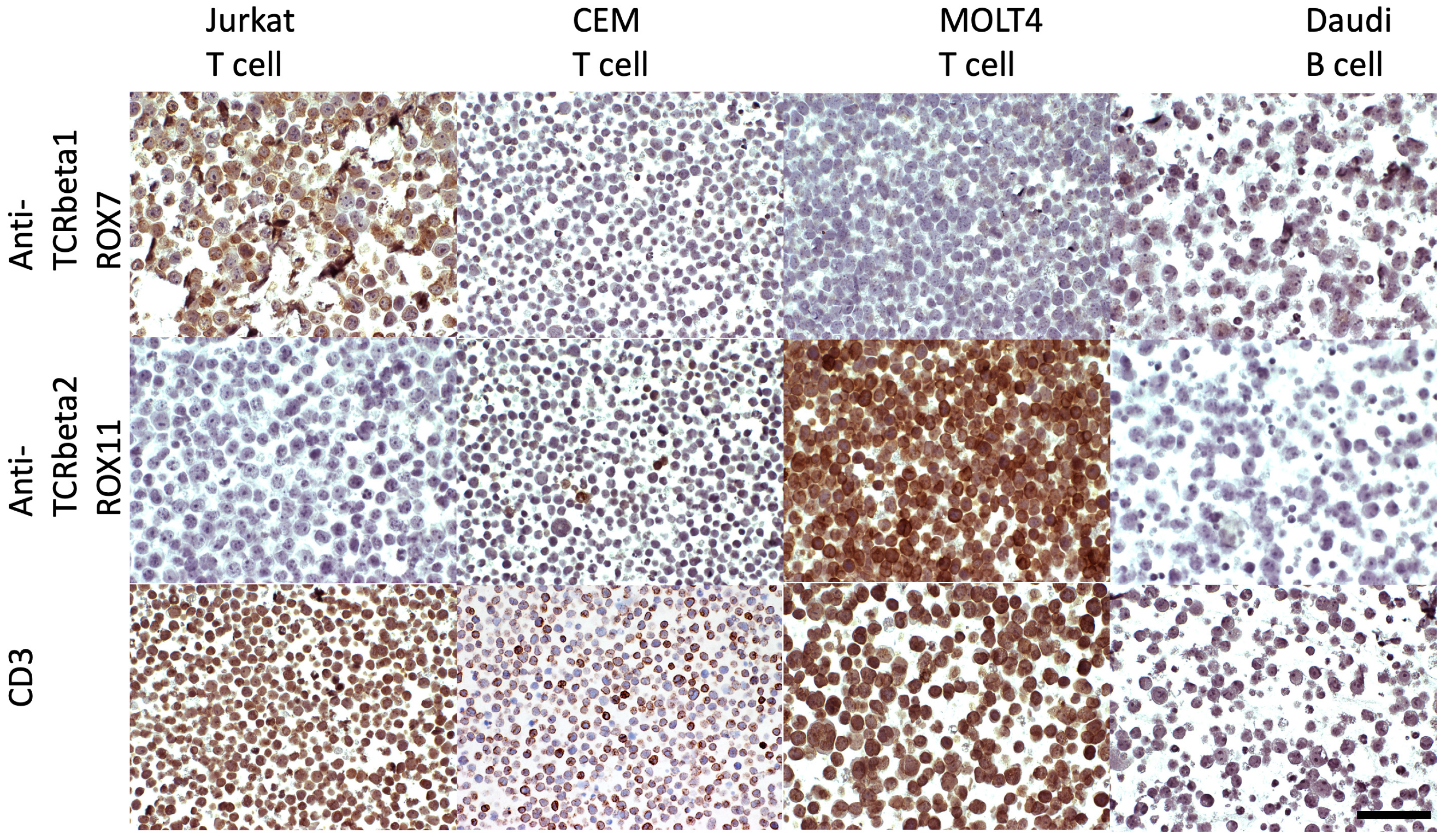
3.1.2. Immunostaining of FFPE Cell Line Pellets
3.2. Immunohistochemical Expression Pattern and TCRbeta2/TCRbeta1 Ratios in FFPE (Routine Clinical) Tissue Samples Containing Populations of Benign T-Cells
3.2.1. Corroboration by Quantitative Real-Time Reverse Transcription PCR (Q-PCR)
3.2.2. Analysis of TRBC2/TRBC1 Transcript Ratios in Publicly Available Datasets
3.2.3. Collation of TCRbeta2/TCRbeta1 Ratios in Published Data
3.2.4. Calculation of a Pragmatic Cut-Off for TCRbeta2/TCRbeta1 Ratios
3.3. Application of TCRbeta1 and TCRbeta2-Specific Antibodies to FFPE Tissue Samples of T-Cell Lymphoma
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Swerdlow, S.H.; World Health Organization; International Agency for Research on Cancer. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, revised 4th ed.; International Agency for Research on Cancer: Lyon, France, 2017; p. 585. [Google Scholar]
- Scarisbrick, J.J.; Quaglino, P.; Prince, H.M.; Papadavid, E.; Hodak, E.; Bagot, M.; Servitje, O.; Berti, E.; Ortiz-Romero, P.; Stadler, R.; et al. The PROCLIPI international registry of early-stage mycosis fungoides identifies substantial diagnostic delay in most patients. Br. J. Dermatol. 2019, 181, 350–357. [Google Scholar] [CrossRef] [PubMed]
- Spieth, K.; Grundmann-Kollmann, M.; Runne, U.; Staib, G.; Fellbaum, C.; Wolter, M.; Kaufmann, R.; Gille, J. Mycosis-fungoides-type cutaneous T cell lymphoma of the hands and soles: A variant causing delay in diagnosis and adequate treatment of patients with palmoplantar eczema. Dermatology 2002, 205, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Federico, M.; Rudiger, T.; Bellei, M.; Nathwani, B.N.; Luminari, S.; Coiffier, B.; Harris, N.L.; Jaffe, E.S.; Pileri, S.A.; Savage, K.J.; et al. Clinicopathologic characteristics of angioimmunoblastic T-cell lymphoma: Analysis of the international peripheral T-cell lymphoma project. J. Clin. Oncol. 2013, 31, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Lachenal, F.; Berger, F.; Ghesquieres, H.; Biron, P.; Hot, A.; Callet-Bauchu, E.; Chassagne, C.; Coiffier, B.; Durieu, I.; Rousset, H.; et al. Angioimmunoblastic T-cell lymphoma: Clinical and laboratory features at diagnosis in 77 patients. Medicine 2007, 86, 282–292. [Google Scholar] [CrossRef]
- Chandesris, M.O.; Malamut, G.; Verkarre, V.; Meresse, B.; Macintyre, E.; Delarue, R.; Rubio, M.T.; Suarez, F.; Deau-Fischer, B.; Cerf-Bensussan, N.; et al. Enteropathy-associated T-cell lymphoma: A review on clinical presentation, diagnosis, therapeutic strategies and perspectives. Gastroenterol. Clin. Biol. 2010, 34, 590–605. [Google Scholar] [CrossRef]
- Liu, H.; Brais, R.; Lavergne-Slove, A.; Jeng, Q.; Payne, K.; Ye, H.; Liu, Z.; Carreras, J.; Huang, Y.; Bacon, C.M.; et al. Continual monitoring of intraepithelial lymphocyte immunophenotype and clonality is more important than snapshot analysis in the surveillance of refractory coeliac disease. Gut 2010, 59, 452–460. [Google Scholar] [CrossRef]
- Davies, K.; Staniforth, J.; Xie, W.H.; Liu, H.; Salimi, M.; Ogg, G.; Soilleux, E. Advances in the assessment of T-cell clonality. Diagn. Histopathol. 2020, 26, 388–397. [Google Scholar] [CrossRef]
- Minca, E.C.; Wang, H.; Wang, Z.; Lanigan, C.; Billings, S.D.; Luo, Y.; Tubbs, R.R.; Ma, X.J. Detection of immunoglobulin light-chain restriction in cutaneous B-cell lymphomas by ultrasensitive bright-field mRNA in situ hybridization. J. Cutan. Pathol. 2015, 42, 82–89. [Google Scholar] [CrossRef]
- McNicol, A.M.; Farquharson, M.A.; Lee, F.D.; Foulis, A.K. Comparison of in situ hybridisation and polymerase chain reaction in the diagnosis of B cell lymphoma. J. Clin. Pathol. 1998, 51, 229–233. [Google Scholar] [CrossRef]
- Levy, R.; Warnke, R.; Dorfman, R.F.; Haimovich, J. The monoclonality of human B-cell lymphomas. J. Exp. Med. 1977, 145, 1014–1028. [Google Scholar] [CrossRef]
- van Dongen, J.J.; Langerak, A.W.; Bruggemann, M.; Evans, P.A.; Hummel, M.; Lavender, F.L.; Delabesse, E.; Davi, F.; Schuuring, E.; Garcia-Sanz, R.; et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: Report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 2003, 17, 2257–2317. [Google Scholar] [CrossRef] [PubMed]
- Laydon, D.J.; Bangham, C.R.; Asquith, B. Estimating T-cell repertoire diversity: Limitations of classical estimators and a new approach. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2015, 370, 20140291. [Google Scholar] [CrossRef]
- Lefranc, M.-P.; Lefranc, G. ScienceDirect (Online service). In The T Cell Receptor Factsbook; Factsbook series; Elsevier: Amsterdam, The Netherlands, 2001; Volume xiv, 397p. [Google Scholar]
- Devitt, K.A.; Kern, W.; Li, W.; Wang, X.; Wong, A.J.; Furtado, F.M.; Oak, J.S.; Illingworth, A. TRBC1 in flow cytometry: Assay development, validation, and reporting considerations. Cytom. B Clin. Cytom. 2024, 106, 192–202. [Google Scholar] [CrossRef]
- Horna, P.; Weybright, M.J.; Ferrari, M.; Jungherz, D.; Peng, Y.; Akbar, Z.; Tudor Ilca, F.; Otteson, G.E.; Seheult, J.N.; Ortmann, J.; et al. Dual T-cell constant beta chain (TRBC)1 and TRBC2 staining for the identification of T-cell neoplasms by flow cytometry. Blood Cancer J. 2024, 14, 34. [Google Scholar] [CrossRef] [PubMed]
- Maciocia, P.M.; Wawrzyniecka, P.A.; Philip, B.; Ricciardelli, I.; Akarca, A.U.; Onuoha, S.C.; Legut, M.; Cole, D.K.; Sewell, A.K.; Gritti, G.; et al. Targeting the T cell receptor beta-chain constant region for immunotherapy of T cell malignancies. Nat. Med. 2017, 23, 1416–1423. [Google Scholar] [CrossRef]
- Nocco, S.E.; Ewalt, M.D.; Moy, A.P.; Lewis, N.E.; Zhu, M.; Lezcano, C.; Busam, K.; Pulitzer, M. TRBC1 immunohistochemistry distinguishes cutaneous T-cell lymphoma from inflammatory dermatitis: A retrospective analysis of 39 cases. J. Am. Acad. Dermatol. 2024, 90, 839–841. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.; Cordell, J.L.; Beyers, A.D.; Tse, A.G.; Mason, D.Y. Detection of T and B cells in many animal species using cross-reactive anti-peptide antibodies. J. Immunol. 1993, 150, 5429–5435. [Google Scholar] [CrossRef]
- Drexler, H.G. The Leukemia-Lymphoma Cell Line Factsbook. In FactsBook; Academic Press: Cambridge, MA, USA, 2000; Volume 1, 751p. [Google Scholar]
- Anderson, C.M.; Zhang, B.; Miller, M.; Butko, E.; Wu, X.; Laver, T.; Kernag, C.; Kim, J.; Luo, Y.; Lamparski, H.; et al. Fully Automated RNAscope In Situ Hybridization Assays for Formalin-Fixed Paraffin-Embedded Cells and Tissues. J. Cell. Biochem. 2016, 117, 2201–2208. [Google Scholar] [CrossRef]
- Fowler, A.; FitzPatrick, M.; Shanmugarasa, A.; Ibrahim, A.S.F.; Kockelbergh, H.; Yang, H.C.; Williams-Walker, A.; Luu Hoang, K.N.; Evans, S.; Provine, N.; et al. An Interpretable Classification Model Using Gluten-Specific TCR Sequences Shows Diagnostic Potential in Coeliac Disease. Biomolecules 2023, 13, 1707. [Google Scholar] [CrossRef]
- Kasatskaya, S.A.; Ladell, K.; Egorov, E.S.; Miners, K.L.; Davydov, A.N.; Metsger, M.; Staroverov, D.B.; Matveyshina, E.K.; Shagina, I.A.; Mamedov, I.Z.; et al. Functionally specialized human CD4(+) T-cell subsets express physicochemically distinct TCRs. eLife 2020, 9, 57063. [Google Scholar] [CrossRef]
- Bolotin, D.A.; Poslavsky, S.; Mitrophanov, I.; Shugay, M.; Mamedov, I.Z.; Putintseva, E.V.; Chudakov, D.M. MiXCR: Software for comprehensive adaptive immunity profiling. Nat. Methods 2015, 12, 380–381. [Google Scholar] [CrossRef] [PubMed]
- Rossi, N.E.; Reine, J.; Pineda-Lezamit, M.; Pulgar, M.; Meza, N.W.; Swamy, M.; Risueno, R.; Schamel, W.W.; Bonay, P.; Fernandez-Malave, E.; et al. Differential antibody binding to the surface alphabetaTCR.CD3 complex of CD4+ and CD8+ T lymphocytes is conserved in mammals and associated with differential glycosylation. Int. Immunol. 2008, 20, 1247–1258. [Google Scholar] [CrossRef] [PubMed]
- Susac, L.; Vuong, M.T.; Thomas, C.; von Bulow, S.; O’Brien-Ball, C.; Santos, A.M.; Fernandes, R.A.; Hummer, G.; Tampe, R.; Davis, S.J. Structure of a fully assembled tumor-specific T cell receptor ligated by pMHC. Cell 2022, 185, 3201–3213.e19. [Google Scholar] [CrossRef] [PubMed]
- Zapata, D.A.; Pacheco-Castro, A.; Torres, P.S.; Ramiro, A.R.; San Jose, E.; Alarcon, B.; Alibaud, L.; Rubin, B.; Toribio, M.L.; Regueiro, J.R. Conformational and biochemical differences in the TCR.CD3 complex of CD8(+) versus CD4(+) mature lymphocytes revealed in the absence of CD3gamma. J. Biol. Chem. 1999, 274, 35119–35128. [Google Scholar] [CrossRef]
- Zapata, D.A.; Schamel, W.W.; Torres, P.S.; Alarcon, B.; Rossi, N.E.; Navarro, M.N.; Toribio, M.L.; Regueiro, J.R. Biochemical differences in the alphabeta T cell receptor.CD3 surface complex between CD8+ and CD4+ human mature T lymphocytes. J. Biol. Chem. 2004, 279, 24485–24492. [Google Scholar] [CrossRef]
- Van Leeuwen, J.E.; Kearse, K.P. Reglucosylation of N-linked glycans is critical for calnexin assembly with T cell receptor (TCR) alpha proteins but not TCRbeta proteins. J. Biol. Chem. 1997, 272, 4179–4186. [Google Scholar] [CrossRef] [PubMed]
- Horna, P.; Shi, M.; Jevremovic, D.; Craig, F.E.; Comfere, N.I.; Olteanu, H. Utility of TRBC1 Expression in the Diagnosis of Peripheral Blood Involvement by Cutaneous T-Cell Lymphoma. J. Invest. Dermatol. 2021, 141, 821–829.e2. [Google Scholar] [CrossRef]
- Ferrari, M.; Righi, M.; Baldan, V.; Wawrzyniecka, P.; Bulek, A.; Kinna, A.; Ma, B.; Bughda, R.; Akbar, Z.; Srivastava, S.; et al. Structure-guided engineering of immunotherapies targeting TRBC1 and TRBC2 in T cell malignancies. Nat. Commun. 2024, 15, 1583. [Google Scholar] [CrossRef]
- Berg, H.; Otteson, G.E.; Corley, H.; Shi, M.; Horna, P.; Jevremovic, D.; Olteanu, H. Flow cytometric evaluation of TRBC1 expression in tissue specimens and body fluids is a novel and specific method for assessment of T-cell clonality and diagnosis of T-cell neoplasms. Cytom. B Clin. Cytom. 2021, 100, 361–369. [Google Scholar] [CrossRef]
- Waldron, D.; O’Brien, D.; Smyth, L.; Quinn, F.; Vandenberghe, E. Reliable Detection of T-Cell Clonality by Flow Cytometry in Mature T-Cell Neoplasms Using TRBC1: Implementation as a Reflex Test and Comparison with PCR-Based Clonality Testing. Lab. Med. 2022, 53, 417–425. [Google Scholar] [CrossRef]
- Guo, L.; Wang, Z.; Anderson, C.M.; Doolittle, E.; Kernag, S.; Cotta, C.V.; Ondrejka, S.L.; Ma, X.J.; Cook, J.R. Ultrasensitive automated RNA in situ hybridization for kappa and lambda light chain mRNA detects B-cell clonality in tissue biopsies with performance comparable or superior to flow cytometry. Mod. Pathol. 2018, 31, 385–394. [Google Scholar] [CrossRef] [PubMed]
- Xu, D. Dual surface immunoglobulin light-chain expression in B-cell lymphoproliferative disorders. Arch. Pathol. Lab. Med. 2006, 130, 853–856. [Google Scholar] [CrossRef]
- Jiwani, S.; Bornhost, J.; Alapat, D. Biphenotypic plasma cell myeloma: Two cases of plasma cell neoplasm with a coexpression of kappa and lambda light chains. Int. J. Clin. Exp. Pathol. 2015, 8, 8536–8544. [Google Scholar]
- Schuldt, N.J.; Binstadt, B.A. Dual TCR T Cells: Identity Crisis or Multitaskers? J. Immunol. 2019, 202, 637–644. [Google Scholar] [CrossRef]
- Heath, W.R.; Carbone, F.R.; Bertolino, P.; Kelly, J.; Cose, S.; Miller, J.F. Expression of two T cell receptor alpha chains on the surface of normal murine T cells. Eur. J. Immunol. 1995, 25, 1617–1623. [Google Scholar] [CrossRef]
- Balomenos, D.; Balderas, R.S.; Mulvany, K.P.; Kaye, J.; Kono, D.H.; Theofilopoulos, A.N. Incomplete T cell receptor V beta allelic exclusion and dual V beta-expressing cells. J. Immunol. 1995, 155, 3308–3312. [Google Scholar] [CrossRef] [PubMed]
- Davodeau, F.; Peyrat, M.A.; Romagne, F.; Necker, A.; Hallet, M.M.; Vie, H.; Bonneville, M. Dual T cell receptor beta chain expression on human T lymphocytes. J. Exp. Med. 1995, 181, 1391–1398. [Google Scholar] [CrossRef] [PubMed]
- Peyrat, M.A.; Davodeau, F.; Houde, I.; Romagne, F.; Necker, A.; Leget, C.; Cervoni, J.P.; Cerf-Bensussan, N.; Vie, H.; Bonneville, M.; et al. Repertoire analysis of human peripheral blood lymphocytes using a human V delta 3 region-specific monoclonal antibody. Characterization of dual T cell receptor (TCR) delta-chain expressors and alpha beta T cells expressing V delta 3J alpha C alpha-encoded TCR chains. J. Immunol. 1995, 155, 3060–3067. [Google Scholar] [PubMed]
- Howie, B.; Sherwood, A.M.; Berkebile, A.D.; Berka, J.; Emerson, R.O.; Williamson, D.W.; Kirsch, I.; Vignali, M.; Rieder, M.J.; Carlson, C.S.; et al. High-throughput pairing of T cell receptor alpha and beta sequences. Sci. Transl. Med. 2015, 7, 301ra131. [Google Scholar] [CrossRef] [PubMed]
- Eltahla, A.A.; Rizzetto, S.; Pirozyan, M.R.; Betz-Stablein, B.D.; Venturi, V.; Kedzierska, K.; Lloyd, A.R.; Bull, R.A.; Luciani, F. Linking the T cell receptor to the single cell transcriptome in antigen-specific human T cells. Immunol. Cell Biol. 2016, 94, 604–611. [Google Scholar] [CrossRef]




| Case Number | Site | Diagnosis | IHC Cell TCRbeta2/TCRbeta1 | BaseScopeTM TRBC2:TRBC1 | Q-PCR TRBC2:TRBC1 |
|---|---|---|---|---|---|
| 1 | Tonsils | Benign, reactive | 1.5:1 | 1.5:1 | 2.82:1 |
| 2 | Lymph node | Benign, reactive | 1.5:1 | 2.3:1 | 1.6:1 |
| 3 | Lymph node | Benign, reactive | 1.5:1 | 1.5:1 | 0.83:1 |
| 4 | Lymph node | Benign, reactive | 1.5:1 | 1.5:1 | 1.92:1 |
| 5 | Skin (temple) | Lichenoid keratosis | 1:1 | 1:1 | 1.76:1 |
| 6 | Buccal mucosa | Lichen planus | 1:1 | 1.5:1 | 0.98:1 |
| 7 | Lymph node | T-cell/histiocyte-rich large B-cell lymphoma | 0.67:1 | 1:1 | 0.73:1 |
| 8 | Lymph node | Classic Hodgkin lymphoma | 1.5:1 | 1.5:1 | 1.88:1 |
| Sample | Clinical Characterization | TRBC2/TRBC1 Transcript Ratio (x:1) |
|---|---|---|
| TB15.0286 | Tonsillitis | 1.72:1 |
| TB14.1744 | Cholangiocarcinoma | 2.33:1 |
| TB15.0347 | Recurrent tonsillitis | 2.47:1 |
| TB15.2568 | Crohn’s disease | 2.62:1 |
| TB16.2261 | Ulcerative colitis | 3.41:1 |
| TB17.1570 | Inflamed skin | 3.80:1 |
| TB15.0750 | Crohn’s disease | 4.01:1 |
| Publication Reference for Dataset Analyzed | Anatomical Site/Pathological Status | T-Cell Subset | Estimated Mean TRBC2-Expressing/TRBC1-Expressing Cell Ratio (and Range) | Standard Deviation |
|---|---|---|---|---|
| Kasatskaya et al. [23] | Peripheral blood | CD4+ T follicular helper | 1.364 (range: 1.226 to 1.466) | 0.094 |
| CD4+ T helper 1 | 1.381 (range: 1.249 to 1.569) | 0.146 | ||
| CD4+ T helper 1-17 | 1.3 (range: 1.206 to 1.417) | 0.086 | ||
| CD4+ T helper 17 | 1.435 (range: 1.288 to 1.606) | 0.16 | ||
| CD4+ T helper 2 | 1.344 (range: 1.244 to 1.436) | 0.082 | ||
| CD4+ T helper 22 | 1.264 (range: 1.204 to 1.362) | 0.059 | ||
| CD4+ non-classical T helper 2 | 1.24 (range: 1.167 to 1.335) | 0.06 | ||
| CD4+ regulatory T | 1.333 (range: 1.225 to 1.52) | 0.093 | ||
| Fowler et al. [22] | Normal duodenum | CD4+ T-cells | 1.413 (range: 1.121 to 1.755) | 0.18 |
| CD8+ T-cells | 1.86 (range: 1.423 to 2.828) | 0.478 | ||
| Celiac disease duodenum | CD4+ T-cells | 1.605 (range: 1.368 to 1.938) | 0.196 | |
| CD8+ T-cells | 2.218 (range: 1.465 to 3.105) | 0.478 |
| Publication | Number and Diagnoses of Donors | Sample Type | Key Metrics Assessed | Results | Comments | Calculated TCRbeta2+/TCRbeta1+ Ratio (and Range) |
|---|---|---|---|---|---|---|
| Maciocia [17] | 27 healthy donors | Peripheral blood mononuclear cells | Assessed TCRbeta1 against pan-TCRbeta | 35% (range, 25–47%). MAITs and invariant natural killer T-cells contain a lower proportion of TCRbeta1+ cells. | 1.86:1 (1.13–3:1) | |
| Berg [32] | 97 benign samples | Lymph node, tonsils, spleen, and bodily fluids | Assessed TCRbeta1 in subsets of CD3+ population. | CD4+: 43.79% (median) or 43.34% (mean) of CD3+ cells were TCRbeta1+ (range: 28.03–53.70; 95th percentile: 35.82–51.14). CD8+: 37.98% (mean) or 37.89% (median) of CD3+ cells were TCRbeta1+. (range: 15.46–59.89; 95th percentile: 26.64–50.52). | CD4+ cells: 95th percentile of 35.8–51.1% TCRbeta1+: CD8+ cells: 95th percentile of 36.5–50.8%. The study chose conservative cutoffs, consistent with Horna et al. [16] | CD4+ cells: 1.31:1 (range: 0.86:1–2.57:1) CD8+ cells: 1.63:1 (range: 0.67:1–5.47:1) |
| Ferrari [31] | 4 healthy donors | Peripheral blood mononuclear cells | TCRbeta1 and TCRbeta2 dual staining | 38:62 TCRbeta1/TCRbeta2 | 1.63:1 | |
| Horna [30] | 24 healthy donors | Peripheral blood mononuclear cells | Assessed TCRbeta1 in subsets of CD3+ population. | Approximately 30–60% of CD4+ cells are TCRbeta1+ (JOVI1) across 4 subsets defined by CD7 and CD26 positivity/negativity | CD4: 0.67:1–2.33:1 | |
| Horna [16] | 104 benign patient samples and 39 healthy donor samples | Peripheral blood mononuclear cells | TCRbeta1 and TCRbeta2 dual staining | Not possible to extract exact results. | Monotypia defined as >85% TCRbeta1+ or TCRbeta2+ cells | Range: 0.18:1–5.7:1 |
| Waldron [33] | 46 healthy donors | Peripheral blood mononuclear cells | Assessed TCRbeta1 against CD3. | CD4+ T-cells: 41% (30–48%) TCRbeta1+; CD8+ T-cells: 33% (22–49%) TCRbeta1+ | TCRbeta monotypia defined as a TCRbeta1-negative population > 82% (CD4) or 88% (CD8), or a TCRbeta1-positive population > 68% (CD4) or 72% (CD8) | CD4: 1.44:1 (range: 1.08:1–2.33:1) CD8: 2.33:1 (range: 1.04:1–3.55:1) |
| Case Number | Site | Diagnosis | Biomed-2 Clonality Results | IHC Cell TCRbeta2/TCRbeta1 | BaseScopeTM TRBC2/TRBC1 | Q-PCR TRBC2/TRBC1 | Comments |
|---|---|---|---|---|---|---|---|
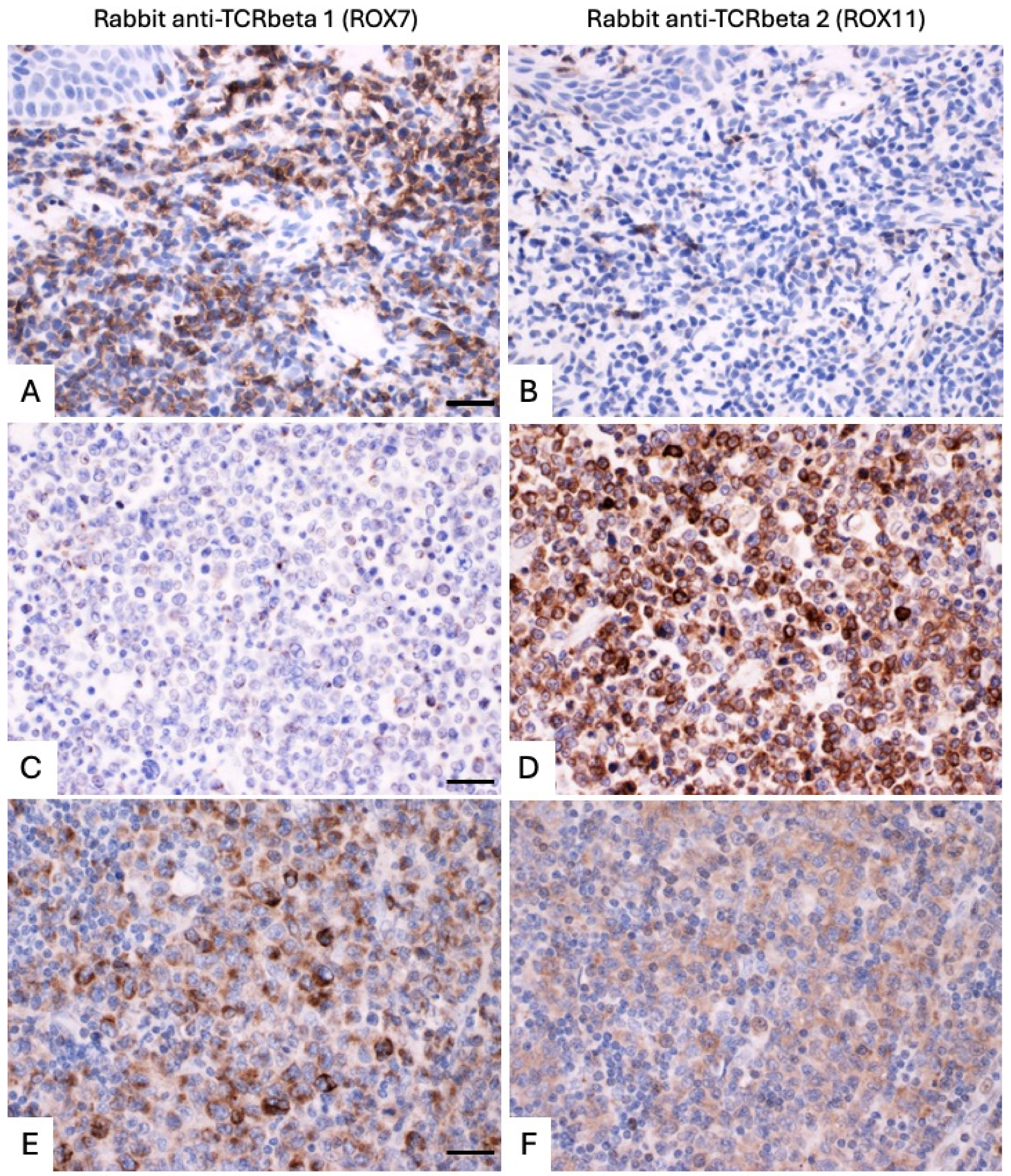
| 9 | Lymph node | Peripheral T-cell lymphoma, NOS | Clonal TRB and TRG | <0.01:1 (TCRbeta1/TCRbeta2 > 100:1 (when assessing strong staining)). Tumor cells are strongly TCRbeta1+ (membranous distribution) with weak cytoplasmic TCRbeta2 staining. | <0.01:1 (TRBC1/TRBC2 > 100:1 (when assessing strong staining)). Tumor cells contain approximately 10 times as many TRBC1 as TRBC2 transcripts. | 0.66:1 | Dual expression at the transcript and protein levels. Shown in Figure 5 and Supplementary Figure S5 panels E and F. |
| 10 | Lymph node | Peripheral T-cell lymphoma, NOS | Clonal TRB and TRG | 10:1 | 97:1 | 10.54:1 | |
| 11 | Lymph node | Peripheral T-cell lymphoma, NOS | Clonal TRB and TRG | >1000:1 | >1000:1 | 458.34:1 | Shown in Figure 5 and Supplementary Figure S5 panels C and D. |
| 12 | Skin (ear) | Indolent CD8+ T-cell lymphoma | Clonal TRB and TRG | >1000:1 | >1000:1 | 2.17:1 | Less TRBC2-skewed Q-PCR results than BaseScopeTM results are due to RNA extraction from peritumoral benign lymphocytes (no microdissection undertaken). |
| 13 | Skin (forehead) | CD4+ cutaneous T-cell lymphoma, unclassifiable | Clonal TRB and TRG | 0.125:1 (TCRbeta1/TCRbeta2 = 8:1) | <0.25:1 (TRBC1/TRBC2 8:1. | 0.74:1 | Shown in Figure 6 and Supplementary Figure S6 panels A and B. |
| 14 | Skin (back) | Sézary syndrome | Clonal TRB and TRG | 10:1 (but very low TCRbeta2 expression level) | 5:1 | 4.16:1 | |
| 15 | Skin (elbow) | Primary cutaneous anaplastic large cell lymphoma or transformed mycosis fungoides | Clonal TRB and TRG | 50:1 | >100:1 (when assessing strong staining). Each tumor cell contains approximately 5 times as many TRBC2 as TRBC1 transcripts. | 6.05:1 | Dual expression at the transcript, but not the protein level. |
| 16 | Skin (scrotum) | Transformed mycosis fungoides | Clonal TRB and TRG | 0.05:1 | 0.01:1 | 0.07:1 | Shown in Figure 5 and Supplementary Figure S5 panels A and B. |
| 17 | Skin (arm) | Transformed mycosis fungoides | Clonal TRB and TRG | >1000:1 | >1000:1 (for counting cytoplasmic transcripts only, but all tumor cells also have high levels of aberrantly distributed nuclear TRBC1 transcript.) | 2.77:1 | Shown in Figure 6 and Supplementary Figure S6 panels C and D. |
| 18 | Skin (back) | Mycosis fungoides | Clonal TRB and TRG | >100:1 | 99:1 | 2.81:1 | |
| 19 | Vulva | CD30+ cutaneous T-cell lymphoma, unclassifiable | Clonal TRB and TRG | <0.001:1 (TCRbeta1/TCRbeta2 > 1000:1) | <0.001:1 (TRBC1/TRBC2 > 1000:1), but all TRBC1+ tumor cells co-express TRBC2 transcripts with a TRBC1:TRBC2 transcript ratio in each cell at around 3:1. | 0.68:1 | |
| 20 | Skin (back) | Cutaneous T-cell lymphoma, unclassifiable | Clonal TRB and TRG | 6:1 | 10:1 | 1.35:1 | |
| 21 | Skin (buttock) | CD8 positive cutaneous T-cell lymphoma, unclassifiable | Clonal TRB and TRG | >1000:1 | >1000:1 | 10.63:1 | Shown in Figure 6 and Supplementary Figure S6 panels E and F. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soilleux, E.J.; Rodgers, D.T.; Situ, J.J.; Evans, S.C.; Konda, V.N.; Yang, H.-C.; Pang, J.; Gilbey Smith, I.; Rajesh, P.; Salimi, M.; et al. Demonstration of T-Cell Monotypia Using Anti-TCRbeta1/2 (TRBC1/2) Immunostaining as a Rapid and Cost-Effective Alternative to PCR-Based Clonality Studies for the Diagnosis of T-Cell Lymphoma. Diagnostics 2024, 14, 2479. https://doi.org/10.3390/diagnostics14222479
Soilleux EJ, Rodgers DT, Situ JJ, Evans SC, Konda VN, Yang H-C, Pang J, Gilbey Smith I, Rajesh P, Salimi M, et al. Demonstration of T-Cell Monotypia Using Anti-TCRbeta1/2 (TRBC1/2) Immunostaining as a Rapid and Cost-Effective Alternative to PCR-Based Clonality Studies for the Diagnosis of T-Cell Lymphoma. Diagnostics. 2024; 14(22):2479. https://doi.org/10.3390/diagnostics14222479
Chicago/Turabian StyleSoilleux, Elizabeth J., Daniel T. Rodgers, Jinlong J. Situ, Shelley C. Evans, Venkata N. Konda, Han-Chieh Yang, Jianxiong Pang, Isabella Gilbey Smith, Pete Rajesh, Maryam Salimi, and et al. 2024. "Demonstration of T-Cell Monotypia Using Anti-TCRbeta1/2 (TRBC1/2) Immunostaining as a Rapid and Cost-Effective Alternative to PCR-Based Clonality Studies for the Diagnosis of T-Cell Lymphoma" Diagnostics 14, no. 22: 2479. https://doi.org/10.3390/diagnostics14222479
APA StyleSoilleux, E. J., Rodgers, D. T., Situ, J. J., Evans, S. C., Konda, V. N., Yang, H.-C., Pang, J., Gilbey Smith, I., Rajesh, P., Salimi, M., Ng, S. W., Jones, J., Miller, J. L., Etherington, R., Ashton-Key, M., & Ogg, G. (2024). Demonstration of T-Cell Monotypia Using Anti-TCRbeta1/2 (TRBC1/2) Immunostaining as a Rapid and Cost-Effective Alternative to PCR-Based Clonality Studies for the Diagnosis of T-Cell Lymphoma. Diagnostics, 14(22), 2479. https://doi.org/10.3390/diagnostics14222479







