Harnessing of Artificial Intelligence for the Diagnosis and Prevention of Hospital-Acquired Infections: A Systematic Review
Abstract
1. Introduction
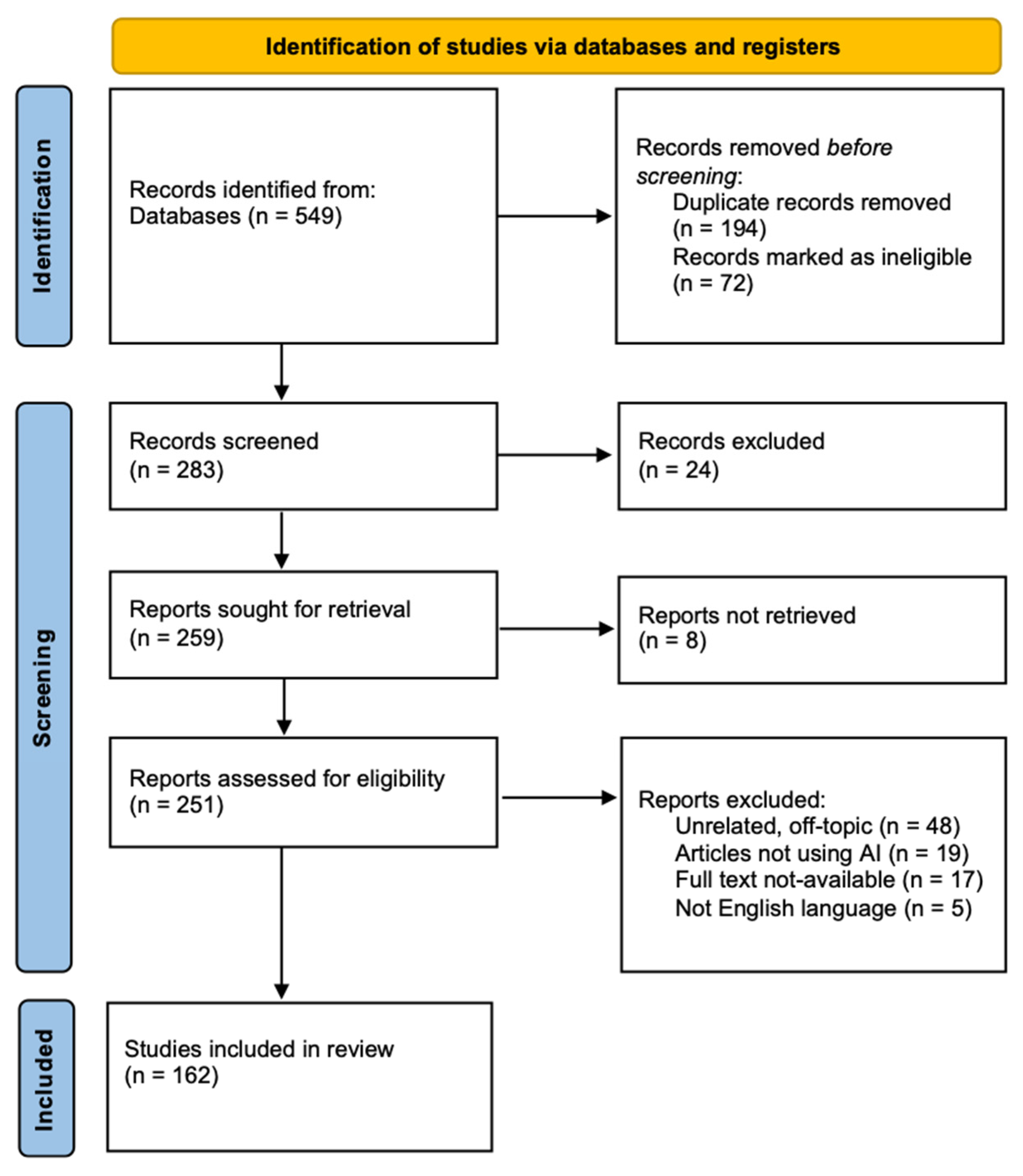
2. Materials and Methods
2.1. Search Strategy
2.2. Study Selection
2.3. Data Extraction and Assessment of Quality
3. Results
3.1. AI Applications in Microbiology
3.1.1. COVID-19
3.1.2. Image Analysis—Bacterial, Viral, Fungal, Parasitic
3.1.3. Automated Factor Analysis
3.1.4. Antimicrobial Resistance Analysis
3.1.5. Antimicrobial Discovery
3.1.6. Microbiome Analysis
3.2. AI and Hospital-Acquired Infections
3.2.1. Intensive Care Units (Predictions, Forecasting)
3.2.2. Ventilator-Associated Pneumonia (VAP)
3.2.3. Central Line-Associated Bloodstream Infections (CLABSIs)
3.2.4. Surgical Site Infections (SSIs)
3.2.5. Sepsis
3.2.6. Clostridium difficile Infection (CDI) and Complications
3.2.7. Multidrug-Resistant (MDR) Pathogens
3.2.8. Hand Hygiene
4. Future Perspectives
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| AFB | acid-fast bacilli |
| AFLP | amplified fragment length polymorphisms |
| AHHMS | automatic and electronically-assisted hand hygiene surveillance system |
| AI | artificial intelligence |
| ALP | alkaline phosphatase |
| ALT | alanine transaminase |
| AMCA | amplification and melting curve analysis |
| AMP | antimicrobial peptide |
| AMR | antimicrobial resistance |
| APAS | automated plate assessment system |
| ARDS | acute respiratory distress syndrome |
| AST | aspartate aminotransferase |
| AuNPs | gold nanoparticles |
| AUROC | area under the receiver operator curve |
| BiLSTM | bidirectional long-term short memory |
| CAUTI | catheter-associated urinary tract infection |
| CDC | Centers of Disease Control and Prevention |
| CDI | C. difficile infection |
| CFPNet-M | Channel-wise Feature Pyramid Network for Medicine |
| CFU | Colony-forming unit |
| CKD | chronic kidney disease |
| CLABSI | central line-associated bloodstream infection |
| CNN | convolutional neural network |
| COPD | chronic obstructive pulmonary disease |
| CPM | cross-point method |
| CR-GNB | carbapenem-resistant Gram-negative bacteria |
| CRP | C-reactive protein |
| CSF | cerebrospinal fluid |
| CT | computed tomography |
| CVC | central venous catheter |
| CVCBSI | central venous catheter bloodstream infection |
| CVL | central venous line |
| CX-R | chest X-ray |
| DASH | data analytics for safe healthcare |
| DNN | dense neural network |
| DT | decision tree |
| ET | extremely randomized trees |
| Faster R-CCN | Faster region-based CNN |
| FCN | fully convolutional neural network |
| GAN | generative adversarial network |
| GCS | Glasgow Coma Scale |
| GGT | gamma-glutamyl transferase |
| GPC | Gaussian process classifier |
| HAI | hospital-acquired infection |
| HDCP | high-definition care platform |
| HER | electronic health records |
| HH | hand hygiene |
| ICU | intensive care unit |
| IoT | Internet of Things |
| KNN | k-nearest neighbors |
| LC/MS-MS | liquid chromatography with tandem mass spectrometry |
| LCR | lymphocytes-to-blood CRP ratio |
| LDA | linear discriminant analysis |
| LDC | linear discriminant classification |
| LDC | linear discriminant classification |
| LDH | lactate dehydrogenase |
| LR | logistic regression |
| LR | logistic regression |
| LSP | laser-scribed graphene |
| LSTM | long short-term memory |
| MALDI-TOF | matrix-assisted laser desorption ionization–time of flight mass spectrometry |
| Mask R-CNN | mask regional-convolutional neural network |
| MAT | microscopic agglutination tests |
| MCHC | mean corpuscular hemoglobin concentration |
| MDR | multidrug resistant |
| MIC | minimum inhibitory concentration |
| MIMIC | Multiparameter Intelligent Monitoring in Intensive Care |
| ML | machine learning |
| MLP | multilayer perceptron |
| MLR | multiple logistic regression |
| MLST | multilocus sequence typing |
| MODE | multi-objective differential evolution |
| MPNN | message-passing neural network |
| MRSA | methicillin-resistant Staphylococcus aureus |
| NB | naïve Bayes |
| NLP | natural language processing |
| NLR | neutrophil-to-lymphocyte ratio |
| NN | neural network |
| NNC | nearest neighbor classification |
| NPA | negative percent agree |
| PaO2/FiO2 | partial pressure of the arterial oxygen/fraction of inspired |
| PCA | plate count agar |
| PFGE | pulse field gel electrophoresis |
| PPA | positive percent agree |
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| RAPD | random amplified polymorphic DNA |
| RCTA | random cover targets algorithm |
| RF | random forest |
| RFE | recursive feature elimination |
| RT–PCR | reverse transcriptase polymerase chain reaction |
| SARS-CoV-2 | severe acute respiratory syndrome coronavirus 2 |
| SERS | surface-enhanced Raman scattering |
| SGDC | stochastic gradient descent classifier |
| SHAP | SHapley Additive exPlanations |
| SOFA | sequential organ failure assessment |
| SpO2 | oxygen saturation |
| SR | soft-max regression |
| SSI | surgical site infection |
| suPAR | blood-soluble urokinase-type plasminogen activator receptor |
| SVC | support vector machine for classification |
| SVM-LK | support vector machine with linear kernel |
| SVM-RK | support vector machine with radial basis function kernel |
| SVM | support vector machine |
| TB | tuberculosis |
| VAP | ventilator-associated pneumonia |
| ViTs | Vision Transformers |
| WBC | white blood cell count |
| WGS | whole genome sequencing |
| XAI | explainable artificial intelligence |
| XGBoost | extreme gradient boosting |
References
- Murray, P.R. The Clinician and the Microbiology Laboratory. In Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2015; pp. 191–223. [Google Scholar]
- Naugler, C.; Church, D.L. Automation and artificial intelligence in the clinical laboratory. Crit. Rev. Clin. Lab. Sci. 2019, 56, 98–110. [Google Scholar] [CrossRef]
- Cassini, A.; Plachouras, D.; Eckmanns, T.; Abu Sin, M.; Blank, H.P.; Ducomble, T.; Haller, S.; Harder, T.; Klingeberg, A.; Sixtensson, M.; et al. Burden of Six Healthcare-Associated Infections on European Population Health: Estimating Incidence-Based Disability-Adjusted Life Years through a Population Prevalence-Based Modelling Study. PLoS Med. 2016, 13, e1002150. [Google Scholar] [CrossRef] [PubMed]
- Behnke, M.; Valik, J.K.; Gubbels, S.; Teixeira, D.; Kristensen, B.; Abbas, M.; van Rooden, S.M.; Gastmeier, P.; van Mourik, M.S.; Aspevall, O.; et al. Information technology aspects of large-scale implementation of automated surveillance of healthcare-associated infections. Clin. Microbiol. Infect. 2021, 27, S29–S39. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ. 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Naiyer, S.; Mansuri, S.; Soni, N.; Singh, V.; Bhat, K.H.; Singh, N.; Arora, G.; Mansuri, M.S. COVID-19 Diagnosis: A Comprehensive Review of the RT-qPCR Method for Detection of SARS-CoV-2. Diagnostics 2022, 12, 1503. [Google Scholar] [CrossRef] [PubMed]
- Alouani, D.J.; Rajapaksha, R.R.P.; Jani, M.; Rhoads, D.D.; Sadri, N. Specificity of SARS-CoV-2 Real-Time PCR Improved by Deep Learning Analysis. J. Clin. Microbiol. 2021, 59, e02959-20. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, Y.-S.; Lee, D.-I.; Jeong, S.; Kang, G.-H.; Jang, Y.S.; Kim, W.; Choi, H.Y.; Kim, J.G.; Choi, S.-H. The application of a deep learning system developed to reduce the time for RT-PCR in COVID-19 detection. Sci. Rep. 2022, 12, 1234. [Google Scholar] [CrossRef]
- Özbilge, E.; Sanlidag, T.; Ozbilge, E.; Baddal, B. Artificial Intelligence-Assisted RT-PCR Detection Model for Rapid and Reliable Diagnosis of COVID-19. Appl. Sci. 2022, 12, 9908. [Google Scholar] [CrossRef]
- Villarreal-González, R.; Acosta-Hoyos, A.J.; Garzon-Ochoa, J.A.; Galán-Freyle, N.J.; Amar-Sepúlveda, P.; Pacheco-Londoño, L.C. Anomaly Identification during Polymerase Chain Reaction for Detecting SARS-CoV-2 Using Artificial Intelligence Trained from Simulated Data. Molecules 2020, 26, 20. [Google Scholar] [CrossRef]
- Alvargonzález, C.J.; Janeiro, L.A.; Castro, P.S.; Torres, M.J.; Lamas, M.L.; Nuñez, D.C.; Del Campo-Pérez, V.; Luque, S.S.; García, R.B.; Fresco, P.J. Proof of concept of the potential of a machine learning algorithm to extract new information from conventional SARS-CoV-2 rRT-PCR results. Sci Rep. 2023, 13, 7786. [Google Scholar] [CrossRef]
- Beduk, D.; de Oliveira Filho, J.I.; Beduk, T.; Harmanci, D.; Zihnioglu, F.; Cicek, C.; Sertoz, R.; Arda, B.; Goksel, T.; Turhan, K.; et al. ‘All In One’ SARS-CoV-2 variant recognition platform: Machine learning-enabled point of care diagnostics. Biosens. Bioelectron. X 2022, 10, 100105. [Google Scholar] [CrossRef]
- Tschoellitsch, T.; Dünser, M.; Böck, C.; Schwarzbauer, K.; Meier, J. Machine Learning Prediction of SARS-CoV-2 Polymerase Chain Reaction Results with Routine Blood Tests. Lab. Med. 2021, 52, 146–149. [Google Scholar] [CrossRef]
- Brinati, D.; Campagner, A.; Ferrari, D.; Locatelli, M.; Banfi, G.; Cabitza, F. Detection of COVID-19 Infection from Routine Blood Exams with Machine Learning: A Feasibility Study. J. Med. Syst. 2020, 44, 135. [Google Scholar] [CrossRef]
- Yang, H.S.; Hou, Y.; Vasovic, L.V.; Steel, P.A.D.; Chadburn, A.; Racine-Brzostek, S.E.; Velu, P.; Cushing, M.M.; Loda, R.; Kaushal, R.; et al. Routine Laboratory Blood Tests Predict SARS-CoV-2 Infection Using Machine Learning. Clin. Chem. 2020, 66, 1396–1404. [Google Scholar] [CrossRef]
- Abayomi-Alli, O.O.; Damaševičius, R.; Maskeliūnas, R.; Misra, S. An Ensemble Learning Model for COVID-19 Detection from Blood Test Samples. Sensors 2022, 22, 2224. [Google Scholar] [CrossRef]
- Rocca, M.F.; Zintgraff, J.C.; Dattero, M.E.; Santos, L.S.; Ledesma, M.; Vay, C.; Prieto, M.; Benedetti, E.; Avaro, M.; Russo, M.; et al. A combined approach of MALDI-TOF mass spectrometry and multivariate analysis as a potential tool for the detection of SARS-CoV-2 virus in nasopharyngeal swabs. J. Virol. Methods 2020, 286, 113991. [Google Scholar] [CrossRef]
- Le, A.T.; Wu, M.; Khan, A.; Phillips, N.; Rajpurkar, P.; Garland, M.; Magid, K.; Sibai, M.; Huang, C.; Sahoo, M.K.; et al. Targeted plasma metabolomics combined with machine learning for the diagnosis of severe acute respiratory syndrome virus type 2. Front. Microbiol. 2023, 13, 1059289. [Google Scholar] [CrossRef] [PubMed]
- Rosado, J.; Pelleau, S.; Cockram, C.; Merkling, S.H.; Nekkab, N.; Demeret, C.; Meola, A.; Kerneis, S.; Terrier, B.; Fafi-Kremer, S.; et al. Multiplex assays for the identification of serological signatures of SARS-CoV-2 infection: An antibody-based diagnostic and machine learning study. Lancet Microbe 2021, 2, e60–e69. [Google Scholar] [CrossRef]
- Nachtigall, F.M.; Pereira, A.; Trofymchuk, O.S.; Santos, L.S. Detection of SARS-CoV-2 in nasal swabs using MALDI-MS. Nat. Biotechnol. 2020, 38, 1168–1173, Erratum in Nat. Biotechnol. 2020, 38, 1211–1212. [Google Scholar] [CrossRef]
- Costa, M.M.; Martin, H.; Estellon, B.; Dupé, F.X.; Saby, F.; Benoit, N.; Tissot-Dupont, H.; Million, M.; Pradines, B.; Granjeaud, S.; et al. Exploratory Study on Application of MALDI-TOF-MS to Detect SARS-CoV-2 Infection in Human Saliva. J. Clin. Med. 2022, 11, 295. [Google Scholar] [CrossRef] [PubMed]
- Cobre, A.d.F.; Surek, M.; Stremel, D.P.; Fachi, M.M.; Borba, H.H.L.; Tonin, F.S.; Pontarolo, R. Diagnosis and prognosis of COVID-19 employing analysis of patients′ plasma and serum via LC-MS and machine learning. Comput. Biol. Med. 2022, 146, 105659. [Google Scholar] [CrossRef]
- Ikponmwoba, E.; Ukorigho, O.; Moitra, P.; Pan, D.; Gartia, M.R.; Owoyele, O. A Machine Learning Framework for Detecting COVID-19 Infection Using Surface-Enhanced Raman Scattering. Biosensors 2022, 12, 589. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Apostolopoulos, I.D.; Mpesiana, T.A. Covid-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys. Eng. Sci. Med. 2020, 43, 635–640. [Google Scholar] [CrossRef]
- Horry, M.J.; Chakraborty, S.; Paul, M.; Ulhaq, A.; Pradhan, B.; Saha, M.; Shukla, N. COVID-19 Detection Through Transfer Learning Using Multimodal Imaging Data. IEEE Access 2020, 8, 149808–149824. [Google Scholar] [CrossRef]
- Ozturk, T.; Talo, M.; Yildirim, E.A.; Baloglu, U.B.; Yildirim, O.; Acharya, U.R. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020, 121, 103792. [Google Scholar] [CrossRef]
- Wu, X.; Hui, H.; Niu, M.; Li, L.; Wang, L.; He, B.; Yang, X.; Li, L.; Li, H.; Tian, J.; et al. Deep learning-based multi-view fusion model for screening 2019 novel coronavirus pneumonia: A multicentre study. Eur. J. Radiol. 2020, 128, 109041. [Google Scholar] [CrossRef]
- Zhao, W.; Jiang, W.; Qiu, X. Deep learning for COVID-19 detection based on CT images. Sci. Rep. 2021, 11, 14353. [Google Scholar] [CrossRef]
- Hamza, A.; Khan, M.A.; Wang, S.-H.; Alhaisoni, M.; Alharbi, M.; Hussein, H.S.; Alshazly, H.; Kim, Y.J.; Cha, J. COVID-19 classification using chest X-ray images based on fusion-assisted deep Bayesian optimization and Grad-CAM visualization. Front. Public Health 2022, 10, 1046296. [Google Scholar] [CrossRef]
- Singh, D.; Kumar, V.; Vaishali; Kaur, M. Classification of COVID-19 patients from chest CT images using multi-objective differential evolution–based convolutional neural networks. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 1379–1389. [Google Scholar] [CrossRef]
- Mei, X.; Lee, H.-C.; Diao, K.-Y.; Huang, M.; Lin, B.; Liu, C.; Xie, Z.; Ma, Y.; Robson, P.M.; Chung, M.; et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID-19. Nat. Med. 2020, 26, 1224–1228. [Google Scholar] [CrossRef]
- Yousefzadeh, M.; Esfahanian, P.; Movahed, S.M.S.; Gorgin, S.; Rahmati, D.; Abedini, A.; Nadji, S.A.; Haseli, S.; Bakhshayesh Karam, M.; Kiani, A.; et al. ai-corona: Radiologist-assistant deep learning framework for COVID-19 diagnosis in chest CT scans. PLoS ONE 2021, 16, e0250952, Erratum in PLoS ONE 2021, 16, e0257119. [Google Scholar]
- Tsiknakis, N.; Trivizakis, E.; Vassalou, E.E.; Papadakis, G.Z.; Spandidos, D.A.; Tsatsakis, A.; Sánchez-García, J.; López-González, R.; Papanikolaou, N.; Karantanas, A.H.; et al. Interpretable artificial intelligence framework for COVID-19 screening on chest X-rays. Exp. Ther. Med. 2020, 20, 727–735. [Google Scholar] [CrossRef]
- Demir, F. DeepCoroNet: A deep LSTM approach for automated detection of COVID-19 cases from chest X-ray images. Appl. Soft Comput. 2021, 103, 107160. [Google Scholar] [CrossRef]
- Akbulut, Y. Automated Pneumonia Based Lung Diseases Classification with Robust Technique Based on a Customized Deep Learning Approach. Diagnostics 2023, 13, 260. [Google Scholar] [CrossRef]
- Jia, L.-L.; Zhao, J.-X.; Pan, N.-N.; Shi, L.-Y.; Zhao, L.-P.; Tian, J.-H.; Huang, G. Artificial intelligence model on chest imaging to diagnose COVID-19 and other pneumonias: A systematic review and meta-analysis. Eur. J. Radiol. Open 2022, 9, 100438. [Google Scholar] [CrossRef]
- Tzeng, I.-S.; Hsieh, P.-C.; Su, W.-L.; Hsieh, T.-H.; Chang, S.-C. Artificial Intelligence-Assisted Chest X-ray for the Diagnosis of COVID-19: A Systematic Review and Meta-Analysis. Diagnostics 2023, 13, 584. [Google Scholar] [CrossRef]
- Chang, S.; Ge, C.; Zhang, L.; Xie, L.; Kong, R.; Zhang, H. COVID-19 Imaging-based AI Research—A Literature Review. Curr. Med. Imaging 2022, 18, 496–508. [Google Scholar] [CrossRef]
- Lasker, A.; Obaidullah, S.M.; Chakraborty, C.; Roy, K. Application of Machine Learning and Deep Learning Techniques for COVID-19 Screening Using Radiological Imaging: A Comprehensive Review. SN Comput. Sci. 2022, 4, 65. [Google Scholar] [CrossRef]
- Wang, Q.; Ma, J.; Zhang, L.; Xie, L. Diagnostic performance of corona virus disease 2019 chest computer tomography image recognition based on deep learning: Systematic review and meta-analysis. Medicine 2022, 101, e31346. [Google Scholar] [CrossRef]
- Wang, J.; Yang, X.; Zhou, B.; Sohn, J.J.; Zhou, J.; Jacob, J.T.; Higgins, K.A.; Bradley, J.D.; Liu, T. Review of Machine Learning in Lung Ultrasound in COVID-19 Pandemic. J. Imaging 2022, 8, 65. [Google Scholar] [CrossRef]
- Suri, J.S.; Agarwal, S.; Gupta, S.K.; Puvvula, A.; Biswas, M.; Saba, L.; Bit, A.; Tandel, G.S.; Agarwal, M.; Patrick, A.; et al. A narrative review on characterization of acute respiratory distress syndrome in COVID-19-infected lungs using artificial intelligence. Comput. Biol. Med. 2021, 130, 104210. [Google Scholar] [CrossRef]
- Ozsahin, I.; Sekeroglu, B.; Musa, M.S.; Mustapha, M.T.; Ozsahin, D.U. Review on Diagnosis of COVID-19 from Chest CT Images Using Artificial Intelligence. Comput. Math. Methods Med. 2020, 2020, 9756518. [Google Scholar] [CrossRef]
- Smith, K.; Kirby, J. Image analysis and artificial intelligence in infectious disease diagnostics. Clin. Microbiol. Infect. 2020, 26, 1318–1323. [Google Scholar] [CrossRef]
- Loh, D.R.; Yong, W.X.; Yapeter, J.; Subburaj, K.; Chandramohanadas, R. A deep learning approach to the screening of malaria infection: Automated and rapid cell counting, object detection and instance segmentation using Mask R-CNN. Comput. Med. Imaging Graph. 2021, 88, 101845. [Google Scholar] [CrossRef]
- Holmström, O.; Stenman, S.; Suutala, A.; Moilanen, H.; Kücükel, H.; Ngasala, B.; Mårtensson, A.; Mhamilawa, L.; Aydin-Schmidt, B.; Lundin, M.; et al. A novel deep learning-based point-of-care diagnostic method for detecting Plasmodium falciparum with fluorescence digital microscopy. PLoS ONE 2020, 15, e0242355. [Google Scholar] [CrossRef]
- Oliveira, A.d.S.; Costa, M.G.F.; Barbosa, M.d.G.V.; Filho, C.F.F.C. A new approach for malaria diagnosis in thick blood smear images. Biomed. Signal Process. Control 2022, 78, 103931. [Google Scholar] [CrossRef]
- Sengar, N.; Burget, R.; Dutta, M.K. A vision transformer based approach for analysis of Plasmodium vivax life cycle for malaria prediction using thin blood smear microscopic images. Comput. Methods Programs Biomed. 2022, 224, 106996. [Google Scholar] [CrossRef]
- Park, H.S.; Rinehart, M.T.; Walzer, K.A.; Chi, J.T.; Wax, A. Automated Detection of P. falciparum Using Machine Learning Algorithms with Quantitative Phase Images of Unstained Cells. PLoS ONE 2016, 11, e0163045. [Google Scholar] [CrossRef]
- Kassim, Y.M.; Yang, F.; Yu, H.; Maude, R.J.; Jaeger, S. Diagnosing Malaria Patients with Plasmodium falciparum and vivax Using Deep Learning for Thick Smear Images. Diagnostics 2021, 11, 1994. [Google Scholar] [CrossRef]
- Dey, S.; Nath, P.; Biswas, S.; Nath, S.; Ganguly, A. Malaria detection through digital microscopic imaging using Deep Greedy Network with transfer learning. J. Med. Imaging 2021, 8, 054502. [Google Scholar] [CrossRef]
- Ufuktepe, D.K.; Yang, F.; Kassim, Y.M.; Yu, H.; Maude, R.J.; Palaniappan, K.; Jaeger, S. Deep Learning-Based Cell Detection and Extraction in Thin Blood Smears for Malaria Diagnosis. In Proceedings of the 2021 IEEE Applied Imagery Pattern Recognition Workshop (AIPR), Washington, DC, USA, 12–14 October 2021; pp. 1–6. [Google Scholar]
- Hemachandran, K.; Alasiry, A.; Marzougui, M.; Ganie, S.M.; Pise, A.A.; Alouane, M.T.-H.; Chola, C. Performance Analysis of Deep Learning Algorithms in Diagnosis of Malaria Disease. Diagnostics 2023, 13, 534. [Google Scholar] [CrossRef]
- Holmström, O.; Linder, N.; Ngasala, B.; Mårtensson, A.; Linder, E.; Lundin, M.; Moilanen, H.; Suutala, A.; Diwan, V.; Lundin, J. Point-of-care mobile digital microscopy and deep learning for the detection of soil-transmitted helminths and Schistosoma haematobium. Glob. Health Action 2017, 10, 1337325. [Google Scholar] [CrossRef]
- Kuok, C.; Horng, M.; Liao, Y.; Chow, N.; Sun, Y. An effective and accurate identification system of Mycobacterium tuberculosis using convolution neural networks. Microsc. Res. Tech. 2019, 82, 709–719. [Google Scholar] [CrossRef]
- Yang, M.; Nurzynska, K.; Walts, A.E.; Gertych, A. A CNN-based active learning framework to identify mycobacteria in digitized Ziehl-Neelsen stained human tissues. Comput. Med. Imaging Graph. 2020, 84, 101752. [Google Scholar] [CrossRef]
- Ibrahim, A.U.; Guler, E.; Guvenir, M.; Suer, K.; Serte, S.; Ozsoz, M. Automated detection of Mycobacterium tuberculosis using transfer learning. J. Infect. Dev. Ctries. 2021, 15, 678–686. [Google Scholar] [CrossRef]
- Xiong, Y.; Ba, X.; Hou, A.; Zhang, K.; Chen, L.; Li, T. Automatic detection of mycobacterium tuberculosis using artificial intelligence. J. Thorac. Dis. 2018, 10, 1936–1940. [Google Scholar] [CrossRef]
- Horvath, L.; Hänselmann, S.; Mannsperger, H.; Degenhardt, S.; Last, K.; Zimmermann, S.; Burckhardt, I. Machine-assisted interpretation of auramine stains substantially increases through-put and sensitivity of microscopic tuberculosis diagnosis. Tuberculosis 2020, 125, 101993. [Google Scholar] [CrossRef]
- Smith, K.P.; Kang, A.D.; Kirby, J.E. Automated Interpretation of Blood Culture Gram Stains by Use of a Deep Convolutional Neural Network. J. Clin. Microbiol. 2018, 56, e01521-17. [Google Scholar] [CrossRef]
- Hoorali, F.; Khosravi, H.; Moradi, B. Automatic Bacillus anthracis bacteria detection and segmentation in microscopic images using UNet++. J. Microbiol. Methods 2020, 177, 106056. [Google Scholar] [CrossRef]
- Kang, R.; Park, B.; Chen, K. Identifying non-O157 Shiga toxin-producing Escherichia coli (STEC) using deep learning methods with hyperspectral microscope images. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2020, 224, 117386. [Google Scholar] [CrossRef]
- Oyamada, Y.; Ozuru, R.; Masuzawa, T.; Miyahara, S.; Nikaido, Y.; Obata, F.; Saito, M.; Villanueva, S.Y.A.M.; Fujii, J. A machine learning model of microscopic agglutination test for diagnosis of leptospirosis. PLoS ONE 2021, 16, e0259907. [Google Scholar] [CrossRef]
- Zieliński, B.; Plichta, A.; Misztal, K.; Spurek, P.; Brzychczy-Włoch, M.; Ochońska, D. Deep learning approach to bacterial colony classification. PLoS ONE 2017, 12, e0184554. [Google Scholar] [CrossRef]
- Ahmad, F.; Khan, M.U.G.; Tahir, A.; Masud, F. Deep ensemble approach for pathogen classification in large-scale images using patch-based training and hyper-parameter optimization. BMC Bioinform. 2023, 24, 273. [Google Scholar] [CrossRef]
- Van, T.T.; Mata, K.; Bard, J.D. Automated Detection of Streptococcus pyogenes Pharyngitis by Use of Colorex Strep A CHROMagar and WASPLab Artificial Intelligence Chromogenic Detection Module Software. J. Clin. Microbiol. 2019, 57, e00811-19. [Google Scholar] [CrossRef]
- Gammel, N.; Ross, T.L.; Lewis, S.; Olson, M.; Henciak, S.; Harris, R.; Hanlon, A.; Carroll, K.C. Comparison of an Automated Plate Assessment System (APAS Independence) and Artificial Intelligence (AI) to Manual Plate Reading of Methicillin-Resistant and Methicillin-Susceptible Staphylococcus aureus CHROMagar Surveillance Cultures. J. Clin. Microbiol. 2021, 59, e0097121. [Google Scholar] [CrossRef]
- Rattray, J.B.; Lowhorn, R.J.; Walden, R.; Márquez-Zacarías, P.; Molotkova, E.; Perron, G.; Solis-Lemus, C.; Alarcon, D.P.; Brown, S.P. Machine learning identification of Pseudomonas aeruginosa strains from colony image data. PLoS Comput. Biol. 2023, 19, e1011699. [Google Scholar] [CrossRef]
- Zhang, B.; Zhou, Z.; Cao, W.; Qi, X.; Xu, C.; Wen, W. A New Few-Shot Learning Method of Bacterial Colony Counting Based on the Edge Computing Device. Biology 2022, 11, 156. [Google Scholar] [CrossRef]
- Koo, T.; Kim, M.H.; Jue, M.-S. Automated detection of superficial fungal infections from microscopic images through a regional convolutional neural network. PLoS ONE 2021, 16, e0256290. [Google Scholar] [CrossRef]
- Ma, H.; Yang, J.; Chen, X.; Jiang, X.; Su, Y.; Qiao, S.; Zhong, G. Deep convolutional neural network: A novel approach for the detection of Aspergillus fungi via stereomicroscopy. J. Microbiol. 2021, 59, 563–572. [Google Scholar] [CrossRef]
- Liu, L.; Yuan, Y.; Zhang, J.; Lei, H.; Wang, Q.; Liu, J.; Du, X.; Ni, G.; Liu, Y. Automatic identification of fungi under complex microscopic fecal images. J. Biomed. Opt. 2015, 20, 76004. [Google Scholar] [CrossRef]
- Maeda, Y.; Sugiyama, Y.; Lim, T.-K.; Harada, M.; Yoshino, T.; Matsunaga, T.; Tanaka, T. Rapid discrimination of fungal species by the colony fingerprinting. Biosens. Bioelectron. 2019, 146, 111747. [Google Scholar] [CrossRef]
- Khan, S.; Ullah, R.; Khan, A.; Ashraf, R.; Ali, H.; Bilal, M.; Saleem, M. Analysis of hepatitis B virus infection in blood sera using Raman spectroscopy and machine learning. Photodiagn. Photodyn. Ther. 2018, 23, 89–93. [Google Scholar] [CrossRef]
- Rohaim, M.A.; Clayton, E.; Sahin, I.; Vilela, J.; Khalifa, M.E.; Al-Natour, M.Q.; Bayoumi, M.; Poirier, A.C.; Branavan, M.; Tharmakulasingam, M.; et al. Artificial Intelligence-Assisted Loop Mediated Isothermal Amplification (AI-LAMP) for Rapid Detection of SARS-CoV-2. Viruses 2020, 12, 972. [Google Scholar] [CrossRef]
- Ito, E.; Sato, T.; Sano, D.; Utagawa, E.; Kato, T. Virus Particle Detection by Convolutional Neural Network in Transmission Electron Microscopy Images. Food Environ. Virol. 2018, 10, 201–208. [Google Scholar] [CrossRef]
- Tong, D.; Chen, C.; Zhang, J.; Lv, G.; Zheng, X.; Zhang, Z.; Lv, X. Application of Raman spectroscopy in the detection of hepatitis B virus infection. Photodiagn. Photodyn. Ther. 2019, 28, 248–252. [Google Scholar] [CrossRef]
- Tabarov, A.; Vitkin, V.; Andreeva, O.; Shemanaeva, A.; Popov, E.; Dobroslavin, A.; Kurikova, V.; Kuznetsova, O.; Grigorenko, K.; Tzibizov, I.; et al. Detection of A and B Influenza Viruses by Surface-Enhanced Raman Scattering Spectroscopy and Machine Learning. Biosensors 2022, 12, 1065. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Lee, T.-Y.; Tseng, Y.-J.; Liu, T.-P.; Huang, K.-Y.; Chang, Y.-T.; Chen, C.-H.; Lu, J.-J. A new scheme for strain typing of methicillin-resistant Staphylococcus aureus on the basis of matrix-assisted laser desorption ionization time-of-flight mass spectrometry by using machine learning approach. PLoS ONE 2018, 13, e0194289. [Google Scholar] [CrossRef]
- Cohen, S.; Rokach, L.; Motro, Y.; Moran-Gilad, J.; Veksler-Lublinsky, I. minMLST: Machine learning for optimization of bacterial strain typing. Bioinformatics 2021, 37, 303–311. [Google Scholar] [CrossRef]
- Fiannaca, A.; La Paglia, L.; La Rosa, M.; Bosco, G.L.; Renda, G.; Rizzo, R.; Gaglio, S.; Urso, A. Deep learning models for bacteria taxonomic classification of metagenomic data. BMC Bioinform. 2018, 19, 61–76. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Hung, C.-C.; Chen, C.-H.; Lee, T.-Y.; Huang, K.-Y.; Ning, H.-C.; Lai, N.-C.; Tsai, M.-H.; Lu, L.-C.; Tseng, Y.-J.; et al. Increase Trichomonas vaginalis detection based on urine routine analysis through a machine learning approach. Sci. Rep. 2019, 9, 11074. [Google Scholar] [CrossRef]
- Kukar, M.; Gunčar, G.; Vovko, T.; Podnar, S.; Černelč, P.; Brvar, M.; Zalaznik, M.; Notar, M.; Moškon, S.; Notar, M. COVID-19 diagnosis by routine blood tests using machine learning. Sci. Rep. 2021, 11, 10738. [Google Scholar] [CrossRef]
- Mentis, A.-F.A.; Garcia, I.; Jiménez, J.; Paparoupa, M.; Xirogianni, A.; Papandreou, A.; Tzanakaki, G. Artificial Intelligence in Differential Diagnostics of Meningitis: A Nationwide Study. Diagnostics 2021, 11, 602. [Google Scholar] [CrossRef]
- Arango-Argoty, G.; Garner, E.; Pruden, A.; Heath, L.S.; Vikesland, P.; Zhang, L. DeepARG: A deep learning approach for predicting antibiotic resistance genes from metagenomic data. Microbiome 2018, 6, 23. [Google Scholar] [CrossRef]
- Kuang, X.; Wang, F.; Hernandez, K.M.; Zhang, Z.; Grossman, R.L. Accurate and rapid prediction of tuberculosis drug resistance from genome sequence data using traditional machine learning algorithms and CNN. Sci. Rep. 2022, 12, 2427. [Google Scholar] [CrossRef]
- Green, A.G.; Yoon, C.H.; Chen, M.L.; Ektefaie, Y.; Fina, M.; Freschi, L.; Gröschel, M.I.; Kohane, I.; Beam, A.; Farhat, M. A convolutional neural network highlights mutations relevant to antimicrobial resistance in Mycobacterium tuberculosis. Nat. Commun. 2022, 13, 3817. [Google Scholar] [CrossRef]
- Miglietta, L.; Moniri, A.; Pennisi, I.; Malpartida-Cardenas, K.; Abbas, H.; Hill-Cawthorne, K.; Bolt, F.; Jauneikaite, E.; Davies, F.; Holmes, A.; et al. Coupling Machine Learning and High Throughput Multiplex Digital PCR Enables Accurate Detection of Carbapenem-Resistant Genes in Clinical Isolates. Front. Mol. Biosci. 2021, 8, 775299. [Google Scholar] [CrossRef]
- Nguyen, M.; Long, S.W.; McDermott, P.F.; Olsen, R.J.; Olson, R.; Stevens, R.L.; Tyson, G.H.; Zhao, S.; Davis, J.J. Using Machine Learning To Predict Antimicrobial MICs and Associated Genomic Features for Nontyphoidal Salmonella. J. Clin. Microbiol. 2019, 57, e01260-18. [Google Scholar] [CrossRef]
- Ciloglu, F.U.; Hora, M.; Gundogdu, A.; Kahraman, M.; Tokmakci, M.; Aydin, O. SERS-based sensor with a machine learning based effective feature extraction technique for fast detection of colistin-resistant Klebsiella pneumoniae. Anal. Chim. Acta 2022, 1221, 340094. [Google Scholar] [CrossRef]
- Ciloglu, F.U.; Caliskan, A.; Saridag, A.M.; Kilic, I.H.; Tokmakci, M.; Kahraman, M.; Aydin, O. Drug-resistant Staphylococcus aureus bacteria detection by combining surface-enhanced Raman spectroscopy (SERS) and deep learning techniques. Sci. Rep. 2021, 11, 18444. [Google Scholar] [CrossRef]
- Fu, Q.; Zhang, Y.; Wang, P.; Pi, J.; Qiu, X.; Guo, Z.; Huang, Y.; Zhao, Y.; Li, S.; Xu, J. Rapid identification of the resistance of urinary tract pathogenic bacteria using deep learning–based spectroscopic analysis. Anal. Bioanal. Chem. 2021, 413, 7401–7410. [Google Scholar] [CrossRef]
- Anahtar, M.N.; Yang, J.H.; Kanjilal, S. Applications of Machine Learning to the Problem of Antimicrobial Resistance: An Emerging Model for Translational Research. J. Clin. Microbiol. 2021, 59, e0126020. [Google Scholar] [CrossRef]
- Lv, J.; Deng, S.; Zhang, L. A review of artificial intelligence applications for antimicrobial resistance. Biosaf. Health 2021, 3, 22–31. [Google Scholar] [CrossRef]
- Popa, S.L.; Pop, C.; Dita, M.O.; Brata, V.D.; Bolchis, R.; Czako, Z.; Saadani, M.M.; Ismaiel, A.; Dumitrascu, D.I.; Grad, S.; et al. Deep Learning and Antibiotic Resistance. Antibiotics 2022, 11, 1674. [Google Scholar] [CrossRef]
- Jeon, K.; Kim, J.-M.; Rho, K.; Jung, S.H.; Park, H.S.; Kim, J.-S. Performance of a Machine Learning-Based Methicillin Resistance of Staphylococcus aureus Identification System Using MALDI-TOF MS and Comparison of the Accuracy according to SCCmec Types. Microorganisms 2022, 10, 1903. [Google Scholar] [CrossRef]
- Liu, G.; Stokes, J.M. A brief guide to machine learning for antibiotic discovery. Curr. Opin. Microbiol. 2022, 69, 102190. [Google Scholar] [CrossRef]
- Wang, L.; Le, X.; Li, L.; Ju, Y.; Lin, Z.; Gu, Q.; Xu, J. Discovering New Agents Active against Methicillin-Resistant Staphylococcus aureus with Ligand-Based Approaches. J. Chem. Inf. Model. 2014, 54, 3186–3197. [Google Scholar] [CrossRef]
- Stokes, J.M.; Yang, K.; Swanson, K.; Jin, W.; Cubillos-Ruiz, A.; Donghia, N.M.; MacNair, C.R.; French, S.; Carfrae, L.A.; Bloom-Ackermann, Z.; et al. A Deep Learning Approach to Antibiotic Discovery. Cell 2020, 180, 688–702.e13, Erratum in Cell 2020, 181, 475–483. [Google Scholar] [CrossRef]
- Liu, G.; Catacutan, D.B.; Rathod, K.; Swanson, K.; Jin, W.; Mohammed, J.C.; Chiappino-Pepe, A.; Syed, S.A.; Fragis, M.; Rachwalski, K.; et al. Deep learning-guided discovery of an antibiotic targeting Acinetobacter baumannii. Nat. Chem. Biol. 2023, 19, 1342–1350. [Google Scholar] [CrossRef]
- Puentes, P.R.; Henao, M.C.; Torres, C.E.; Gómez, S.C.; Gómez, L.A.; Burgos, J.C.; Arbeláez, P.; Osma, J.F.; Muñoz-Camargo, C.; Reyes, L.H.; et al. Design, Screening, and Testing of Non-Rational Peptide Libraries with Antimicrobial Activity: In Silico and Experimental Approaches. Antibiotics 2020, 9, 854. [Google Scholar] [CrossRef]
- Das, P.; Sercu, T.; Wadhawan, K.; Padhi, I.; Gehrmann, S.; Cipcigan, F.; Chenthamarakshan, V.; Strobelt, H.; dos Santos, C.; Chen, P.-Y.; et al. Accelerated antimicrobial discovery via deep generative models and molecular dynamics simulations. Nat. Biomed. Eng. 2021, 5, 613–623. [Google Scholar] [CrossRef]
- Wang, C.; Garlick, S.; Zloh, M. Deep Learning for Novel Antimicrobial Peptide Design. Biomolecules 2021, 11, 471. [Google Scholar] [CrossRef]
- Talat, A.; Khan, A.U. Artificial intelligence as a smart approach to develop antimicrobial drug molecules: A paradigm to combat drug-resistant infections. Drug Discov. Today 2023, 28, 103491. [Google Scholar] [CrossRef]
- Lluka, T.; Stokes, J.M. Antibiotic discovery in the artificial intelligence era. Ann. N. Y. Acad. Sci. 2023, 1519, 74–93. [Google Scholar] [CrossRef]
- David, L.; Brata, A.M.; Mogosan, C.; Pop, C.; Czako, Z.; Muresan, L.; Ismaiel, A.; Dumitrascu, D.I.; Leucuta, D.C.; Stanculete, M.F.; et al. Artificial Intelligence and Antibiotic Discovery. Antibiotics 2021, 10, 1376. [Google Scholar] [CrossRef]
- Giuffrè, M.; Moretti, R.; Tiribelli, C. Gut Microbes Meet Machine Learning: The Next Step towards Advancing Our Understanding of the Gut Microbiome in Health and Disease. Int. J. Mol. Sci. 2023, 24, 5229. [Google Scholar] [CrossRef]
- Marcos-Zambrano, L.J.; Karaduzovic-Hadziabdic, K.; Turukalo, T.L.; Przymus, P.; Trajkovik, V.; Aasmets, O.; Berland, M.; Gruca, A.; Hasic, J.; Hron, K.; et al. Applications of Machine Learning in Human Microbiome Studies: A Review on Feature Selection, Biomarker Identification, Disease Prediction and Treatment. Front. Microbiol. 2021, 12, 634511. [Google Scholar] [CrossRef]
- Li, P.; Luo, H.; Ji, B.; Nielsen, J. Machine learning for data integration in human gut microbiome. Microb. Cell Factories 2022, 21, 241. [Google Scholar] [CrossRef]
- Medina, R.H.; Kutuzova, S.; Nielsen, K.N.; Johansen, J.; Hansen, L.H.; Nielsen, M.; Rasmussen, S. Machine learning and deep learning applications in microbiome research. ISME Commun. 2022, 2, 98. [Google Scholar] [CrossRef]
- Zhou, Y.-H.; Gallins, P. A Review and Tutorial of Machine Learning Methods for Microbiome Host Trait Prediction. Front. Genet. 2019, 10, 579. [Google Scholar] [CrossRef]
- Namkung, J. Machine learning methods for microbiome studies. J. Microbiol. 2020, 58, 206–216. [Google Scholar] [CrossRef]
- Monegro, A.F.; Muppidi, V.; Regunath, H. Hospital-Acquired Infections; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Brusselaers, N.; Vogelaers, D.; Blot, S. The rising problem of antimicrobial resistance in the intensive care unit. Ann. Intensive Care 2011, 1, 47. [Google Scholar] [CrossRef]
- Dadi, N.C.T.; Radochová, B.; Vargová, J.; Bujdáková, H. Impact of Healthcare-Associated Infections Connected to Medical Devices—An Update. Microorganisms 2021, 9, 2332. [Google Scholar] [CrossRef]
- Ragusa, R.; Giorgianni, G.; Lupo, L.; Sciacca, A.; Rametta, S.; La Verde, M.; Mulè, S.; Marranzano, M. Healthcare-associated Clostridium difficile infection: Role of correct hand hygiene in cross-infection control. J. Prev. Med. Hyg. 2018, 59, E145–E152. [Google Scholar]
- Boyce, J.M.; Pittet, D. Guideline for Hand Hygiene in Health-Care Settings: Recommendations of the Healthcare Infection Control Practices Advisory Committee and the HICPAC/SHEA/APIC/IDSA Hand Hygiene Task Force. Infect. Control Hosp. Epidemiol. 2002, 23, S3–S40. [Google Scholar] [CrossRef]
- Pittet, D.; Allegranzi, B.; Storr, J.; Nejad, S.B.; Dziekan, G.; Leotsakos, A.; Donaldson, L. Infection control as a major World Health Organization priority for developing countries. J. Hosp. Infect. 2008, 68, 285–292. [Google Scholar] [CrossRef]
- Zimlichman, E.; Henderson, D.; Tamir, O.; Franz, C.; Song, P.; Yamin, C.K.; Keohane, C.; Denham, C.R.; Bates, D.W. Health care-associated infections: A meta-analysis of costs and financial impact on the US health care system. JAMA Intern. Med. 2013, 173, 2039–2046. [Google Scholar] [CrossRef]
- Habehh, H.; Gohel, S. Machine Learning in Healthcare. Curr. Genom. 2021, 22, 291–300. [Google Scholar] [CrossRef]
- Scardoni, A.; Balzarini, F.; Signorelli, C.; Cabitza, F.; Odone, A. Artificial intelligence-based tools to control healthcare associated infections: A systematic review of the literature. J. Infect. Public Health 2020, 13, 1061–1077. [Google Scholar] [CrossRef]
- Hong, N.; Liu, C.; Gao, J.; Han, L.; Chang, F.; Gong, M.; Su, L. State of the Art of Machine Learning–Enabled Clinical Decision Support in Intensive Care Units: Literature Review. JMIR Med. Inform. 2022, 10, e28781. [Google Scholar] [CrossRef]
- De Corte, T.; Van Hoecke, S.; De Waele, J. Artificial Intelligence in Infection Management in the ICU. Crit Care. 2022, 26, 79. [Google Scholar] [CrossRef] [PubMed]
- Wolters, K. Expert Insights, Predicting Hospital Infections: How AI Makes it Possible. Available online: https://www.wolterskluwer.com/en/expert-insights/predicting-hospital-infections-how-ai-makes-it-possible (accessed on 16 December 2023).
- Koenig, S.M.; Truwit, J.D. Ventilator-Associated Pneumonia: Diagnosis, Treatment, and Prevention. Clin. Microbiol. Rev. 2006, 19, 637–657. [Google Scholar] [CrossRef]
- Papazian, L.; Klompas, M.; Luyt, C.-E. Ventilator-associated pneumonia in adults: A narrative review. Intensive Care Med. 2020, 46, 888–906. [Google Scholar] [CrossRef]
- Tejerina, E.; Frutos-Vivar, F.; Restrepo, M.I.; Anzueto, A.; Abroug, F.; Palizas, F.; González, M.; D’Empaire, G.; Apezteguía, C.; Esteban, A. Incidence, risk factors, and outcome of ventilator-associated pneumonia. J. Crit. Care 2006, 21, 56–65. [Google Scholar] [CrossRef]
- Liang, Y.; Zhu, C.; Tian, C.; Lin, Q.; Li, Z.; Li, Z.; Ni, D.; Ma, X. Early prediction of ventilator-associated pneumonia in critical care patients: A machine learning model. BMC Pulm. Med. 2022, 22, 250. [Google Scholar] [CrossRef]
- Giang, C.; Calvert, J.; Rahmani, K.; Barnes, G.; Siefkas, A.S.; Green-Saxena, A.; Hoffman, J.; Mao, Q.; Das, R. Predicting ventilator-associated pneumonia with machine learning. Medicine 2021, 100, e26246. [Google Scholar] [CrossRef]
- Samadani, A.; Wang, T.; van Zon, K.; Celi, L.A. VAP risk index: Early prediction and hospital phenotyping of ventilator-associated pneumonia using machine learning. Artif. Intell. Med. 2023, 146, 102715. [Google Scholar] [CrossRef]
- Jeon, E.-T.; Lee, H.J.; Park, T.Y.; Jin, K.N.; Ryu, B.; Lee, H.W.; Kim, D.H. Machine learning-based prediction of in-ICU mortality in pneumonia patients. Sci. Rep. 2023, 13, 11527. [Google Scholar] [CrossRef]
- Wang, B.; Li, Y.; Tian, Y.; Ju, C.; Xu, X.; Pei, S. Novel pneumonia score based on a machine learning model for predicting mortality in pneumonia patients on admission to the intensive care unit. Respir. Med. 2023, 217, 107363. [Google Scholar] [CrossRef]
- Wang, R.; Cai, L.; Liu, Y.; Zhang, J.; Ou, X.; Xu, J. Machine learning algorithms for prediction of ventilator associated pneumonia in traumatic brain injury patients from the MIMIC-III database. Heart Lung 2023, 62, 225–232. [Google Scholar] [CrossRef]
- Hallam, C.; Jackson, T.; Rajgopal, A.; Russell, B. Establishing catheter-related bloodstream infection surveillance to drive improvement. J. Infect. Prev. 2018, 19, 160–166. [Google Scholar] [CrossRef]
- Rahmani, K.; Garikipati, A.; Barnes, G.; Hoffman, J.; Calvert, J.; Mao, Q.; Das, R. Early prediction of central line associated bloodstream infection using machine learning. Am. J. Infect. Control 2022, 50, 440–445. [Google Scholar] [CrossRef]
- Beeler, C.; Dbeibo, L.; Kelley, K.; Thatcher, L.; Webb, D.; Bah, A.; Monahan, P.; Fowler, N.R.; Nicol, S.; Judy-Malcolm, A.; et al. Assessing patient risk of central line-associated bacteremia via machine learning. Am. J. Infect. Control 2018, 46, 986–991. [Google Scholar] [CrossRef]
- Parreco, J.P.; Hidalgo, A.E.; Badilla, A.D.; Ilyas, O.; Rattan, R. Predicting central line-associated bloodstream infections and mortality using supervised machine learning. J. Crit. Care 2018, 45, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Bonello, K.; Emani, S.; Sorensen, A.; Shaw, L.; Godsay, M.; Delgado, M.; Sperotto, F.; Santillana, M.; Kheir, J. Prediction of impending central-line-associated bloodstream infections in hospitalized cardiac patients: Development and testing of a machine-learning model. J. Hosp. Infect. 2022, 127, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Jenks, P.; Laurent, M.; McQuarry, S.; Watkins, R. Clinical and economic burden of surgical site infection (SSI) and predicted financial consequences of elimination of SSI from an English hospital. J. Hosp. Infect. 2014, 86, 24–33. [Google Scholar] [CrossRef]
- Hu, Z.; Simon, G.J.; Arsoniadis, E.G.; Wang, Y.; Kwaan, M.R.; Melton, G.B. Automated Detection of Postoperative Surgical Site Infections Using Supervised Methods with Electronic Health Record Data. Stud. Health Technol. Inform. 2015, 216, 706–710. [Google Scholar] [PubMed]
- Kuo, P.-J.; Wu, S.-C.; Chien, P.-C.; Chang, S.-S.; Rau, C.-S.; Tai, H.-L.; Peng, S.-H.; Lin, Y.-C.; Chen, Y.-C.; Hsieh, H.-Y.; et al. Artificial neural network approach to predict surgical site infection after free-flap reconstruction in patients receiving surgery for head and neck cancer. Oncotarget 2018, 9, 13768–13782. [Google Scholar] [CrossRef]
- Sohn, S.; Larson, D.W.; Habermann, E.B.; Naessens, J.M.; Alabbad, J.Y.; Liu, H. Detection of clinically important colorectal surgical site infection using Bayesian network. J. Surg. Res. 2017, 209, 168–173. [Google Scholar] [CrossRef]
- Soguero-Ruiz, C.; Fei, W.M.E.; Jenssen, R.; Augestad, K.M.; Álvarez, J.-L.R.; Jiménez, I.M.; Lindsetmo, R.-O.; Skrøvseth, S.O. Data-driven Temporal Prediction of Surgical Site Infection. AMIA Annu. Symp. Proc. 2015, 2015, 1164–1173. [Google Scholar]
- Mamlook, R.E.A.; Wells, L.J.; Sawyer, R. Machine-learning models for predicting surgical site infections using patient pre-operative risk and surgical procedure factors. Am. J. Infect. Control 2023, 51, 544–550. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Kim, Z.; Chung, D.; Cho, B.; Chung, M.; Kim, J.; Jeong, J. Development of machine learning models for the surveillance of colon surgical site infections. J. Hosp. Infect. 2023, 23, 00124-X. [Google Scholar] [CrossRef]
- Petrosyan, Y.; Thavorn, K.; Smith, G.; Maclure, M.; Preston, R.; van Walravan, C.; Forster, A.J. Predicting postoperative surgical site infection with administrative data: A random forests algorithm. BMC Med. Res. Methodol. 2021, 21, 179. [Google Scholar] [CrossRef]
- Wu, G.; Cheligeer, C.; Southern, D.A.; Martin, E.A.; Xu, Y.; Leal, J.; Ellison, J.; Bush, K.; Williamson, T.; Quan, H.; et al. Development of machine learning models for the detection of surgical site infections following total hip and knee arthroplasty: A multicenter cohort study. Antimicrob. Resist. Infect. Control 2023, 12, 88. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Liu, C.; Zhang, Z.; Liang, T.; Zhu, J.; Zhou, C.; Wu, S.; Yao, Y.; Huang, C.; Zhang, B.; et al. Using Machine Learning to Predict Surgical Site Infection After Lumbar Spine Surgery. Infect. Drug Resist. 2023, 16, 5197–5207. [Google Scholar] [CrossRef] [PubMed]
- Agrebi, S.; Larbi, A. Use of artificial intelligence in infectious diseases. In Artificial intelligence in precision health; Academic Press: Cambridge, MA, USA, 2020; pp. 415–438. [Google Scholar]
- Horng, S.; Sontag, D.A.; Halpern, Y.; Jernite, Y.; Shapiro, N.I.; Nathanson, L.A. Creating an automated trigger for sepsis clinical decision support at emergency department triage using machine learning. PLoS ONE 2017, 12, e0174708. [Google Scholar] [CrossRef]
- Nemati, S.; Holder, A.M.; Razmi, F.; Stanley, M.D.; Clifford, G.D.; Buchman, T.G. An Interpretable Machine Learning Model for Accurate Prediction of Sepsis in the ICU. Crit. Care Med. 2018, 46, 547–553. [Google Scholar] [CrossRef]
- Shimabukuro, D.W.; Barton, C.W.; Feldman, M.D.; Mataraso, S.J.; Das, R. Effect of a machine learning-based severe sepsis prediction algorithm on patient survival and hospital length of stay: A randomised clinical trial. BMJ Open Respir. Res. 2017, 4, e000234. [Google Scholar] [CrossRef]
- Bedoya, A.D.; Futoma, J.; Clement, M.E.; Corey, K.; Brajer, N.; Lin, A.; Simons, M.G.; Gao, M.; Nichols, M.; Balu, S.; et al. Machine learning for early detection of sepsis: An internal and temporal validation study. JAMIA Open 2020, 3, 252–260. [Google Scholar] [CrossRef]
- Mao, Q.; Jay, M.; Hoffman, J.L.; Calvert, J.; Barton, C.; Shimabukuro, D.; Shieh, L.; Chettipally, U.; Fletcher, G.; Kerem, Y.; et al. Multicentre validation of a sepsis prediction algorithm using only vital sign data in the emergency department, general ward and ICU. BMJ Open 2018, 8, e017833. [Google Scholar] [CrossRef]
- Barton, C.; Chettipally, U.; Zhou, Y.; Jiang, Z.; Lynn-Palevsky, A.; Le, S.; Calvert, J.; Das, R. Evaluation of a machine learning algorithm for up to 48-hour advance prediction of sepsis using six vital signs. Comput. Biol. Med. 2019, 109, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, J.; Sun, Y.; Ding, X.; Zhang, X.; Liu, S.; Han, B.; Wang, H.; Duan, X.; Sun, T. A Machine Learning Model for Accurate Prediction of Sepsis in ICU Patients. Front. Public Health 2021, 9, 754348. [Google Scholar] [CrossRef] [PubMed]
- Lauritsen, S.M.; Kalør, M.E.; Kongsgaard, E.L.; Lauritsen, K.M.; Jørgensen, M.J.; Lange, J.; Thiesson, B. Early detection of sepsis utilizing deep learning on electronic health record event sequences. Artif. Intell. Med. 2020, 104, 101820. [Google Scholar] [CrossRef]
- Yuan, K.-C.; Tsai, L.-W.; Lee, K.-H.; Cheng, Y.-W.; Hsu, S.-C.; Lo, Y.-S.; Chen, R.-J. The development an artificial intelligence algorithm for early sepsis diagnosis in the intensive care unit. Int. J. Med. Inform. 2020, 141, 104176. [Google Scholar] [CrossRef]
- Fagerström, J.; Bång, M.; Wilhelms, D.; Chew, M.S. LiSep LSTM: A Machine Learning Algorithm for Early Detection of Septic Shock. Sci. Rep. 2019, 9, 15132. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.; Makar, M.; Fusco, C.; McCaffrey, R.; Rao, K.; Ryan, E.E.; Washer, L.; West, L.R.; Young, V.B.; Guttag, J.; et al. A Generalizable, Data-Driven Approach to Predict Daily Risk of Clostridium difficile Infection at Two Large Academic Health Centers. Infect. Control Hosp. Epidemiol. 2018, 39, 425–433. [Google Scholar] [CrossRef]
- Panchavati, S.; Zelin, N.S.; Garikipati, A.; Pellegrini, E.; Iqbal, Z.; Barnes, G.; Hoffman, J.; Calvert, J.; Mao, Q.; Das, R. A comparative analysis of machine learning approaches to predict C. difficile infection in hospitalized patients. Am. J. Infect. Control 2022, 50, 250–257. [Google Scholar] [CrossRef]
- Escobar, G.J.; Baker, J.M.; Kipnis, P.; Greene, J.D.; Mast, T.C.; Gupta, S.B.; Cossrow, N.; Mehta, V.; Liu, V.; Dubberke, E.R. Prediction of Recurrent Clostridium difficile Infection Using Comprehensive Electronic Medical Records in an Integrated Healthcare Delivery System. Infect. Control Hosp. Epidemiol. 2017, 38, 1196–1203. [Google Scholar] [CrossRef]
- Li, B.Y.; Oh, J.; Young, V.B.; Rao, K.; Wiens, J. Using Machine Learning and the Electronic Health Record to Predict Complicated Clostridium difficile Infection. Open Forum Infect. Dis. 2019, 6, ofz186. [Google Scholar] [CrossRef]
- Mora-Jiménez, I.; Tarancón-Rey, J.; Álvarez-Rodríguez, J.; Soguero-Ruiz, C. Artificial Intelligence to Get Insights of Multi-Drug Resistance Risk Factors during the First 48 Hours from ICU Admission. Antibiotics 2021, 10, 239. [Google Scholar] [CrossRef]
- Liang, Q.; Zhao, Q.; Xu, X.; Zhou, Y.; Huang, M. Early prediction of carbapenem-resistant Gram-negative bacterial carriage in intensive care units using machine learning. J. Glob. Antimicrob. Resist. 2022, 29, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Mouajou, V.; Adams, K.; DeLisle, G.; Quach, C. Hand hygiene compliance in the prevention of hospital-acquired infections: A systematic review. J. Hosp. Infect. 2022, 119, 33–48. [Google Scholar] [CrossRef]
- Marques, R.; Gregório, J.; Pinheiro, F.; Póvoa, P.; da Silva, M.M.; Lapão, L.V. How can information systems provide support to nurses’ hand hygiene performance? Using gamification and indoor location to improve hand hygiene awareness and reduce hospital infections. BMC Med. Inform. Decis. Mak. 2017, 17, 15. [Google Scholar] [CrossRef]
- Scheithauer, S.; Bickenbach, J.; Heisel, H.; Fehling, P.; Marx, G.; Lemmen, S. Do WiFi-based hand hygiene dispenser systems increase hand hygiene compliance? Am. J. Infect. Control 2018, 46, 1192–1194. [Google Scholar] [CrossRef]
- Boyce, J.M.; Laughman, J.A.; Ader, M.H.; Wagner, P.T.; Parker, A.E.; Arbogast, J.W. Impact of an automated hand hygiene monitoring system and additional promotional activities on hand hygiene performance rates and healthcare-associated infections. Infect. Control Hosp. Epidemiol. 2019, 40, 741–747. [Google Scholar] [CrossRef]
- Geilleit, R.; Hen, Z.; Chong, C.; Loh, A.; Pang, N.; Peterson, G.; Ng, K.; Huis, A.; de Korne, D. Feasibility of a real-time hand hygiene notification machine learning system in outpatient clinics. J. Hosp. Infect. 2018, 100, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Kwok, Y.L.A.; Callard, M.; McLaws, M.-L. An automated hand hygiene training system improves hand hygiene technique but not compliance. Am. J. Infect. Control 2015, 43, 821–825. [Google Scholar] [CrossRef] [PubMed]
- Lacey, G.; Zhou, J.; Li, X.; Craven, C.; Gush, C. The impact of automatic video auditing with real-time feedback on the quality and quantity of handwash events in a hospital setting. Am. J. Infect. Control 2020, 48, 162–166. [Google Scholar] [CrossRef]
- Greco, A.; Percannella, G.; Ritrovato, P.; Saggese, A.; Vento, M. A deep learning based system for handwashing procedure evaluation. Neural Comput. Appl. 2022, 35, 15981–15996. [Google Scholar] [CrossRef]
- Nagar, A.; Kumar, M.A.; Vaegae, N.K. Hand hygiene monitoring and compliance system using convolution neural networks. Multimed. Tools Appl. 2022, 81, 44431–44444. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Huang, J.; Wang, G.; Lu, H.; He, M.; Wang, W. A Pilot Study of Deep Learning Models for Camera based Hand Hygiene Monitoring in ICU. In Proceedings of the 2023 45th Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Sydney, Australia, 24–27 July 2023; pp. 1–5. [Google Scholar]
- Singh, A.; Haque, A.; Alahi, A.; Yeung, S.; Guo, M.; Glassman, J.R.; Beninati, W.; Platchek, T.; Fei-Fei, L.; Milstein, A. Automatic detection of hand hygiene using computer vision technology. J. Am. Med. Inform. Assoc. 2020, 27, 1316–1320. [Google Scholar] [CrossRef] [PubMed]
- Kutafina, E.; Laukamp, D.; Jonas, S.M. Wearable Sensors in Medical Education: Supporting Hand Hygiene Training with a Forearm EMG. Stud. Health Technol. Inform. 2015, 211, 286–291. [Google Scholar] [PubMed]
- Band, S.S.; Yarahmadi, A.; Hsu, C.-C.; Biyari, M.; Sookhak, M.; Ameri, R.; Dehzangi, I.; Chronopoulos, A.T.; Liang, H.-W. Application of explainable artificial intelligence in medical health: A systematic review of interpretability methods. Inform. Med. Unlocked 2023, 40, 101286. [Google Scholar] [CrossRef]
- Zhang, Y.; Weng, Y.; Lund, J. Applications of Explainable Artificial Intelligence in Diagnosis and Surgery. Diagnostics 2022, 12, 237. [Google Scholar] [CrossRef]
- Loh, H.W.; Ooi, C.P.; Seoni, S.; Barua, P.D.; Molinari, F.; Acharya, U.R. Application of explainable artificial intelligence for healthcare: A systematic review of the last decade (2011–2022). Comput. Methods Programs Biomed. 2022, 226, 107161. [Google Scholar] [CrossRef]
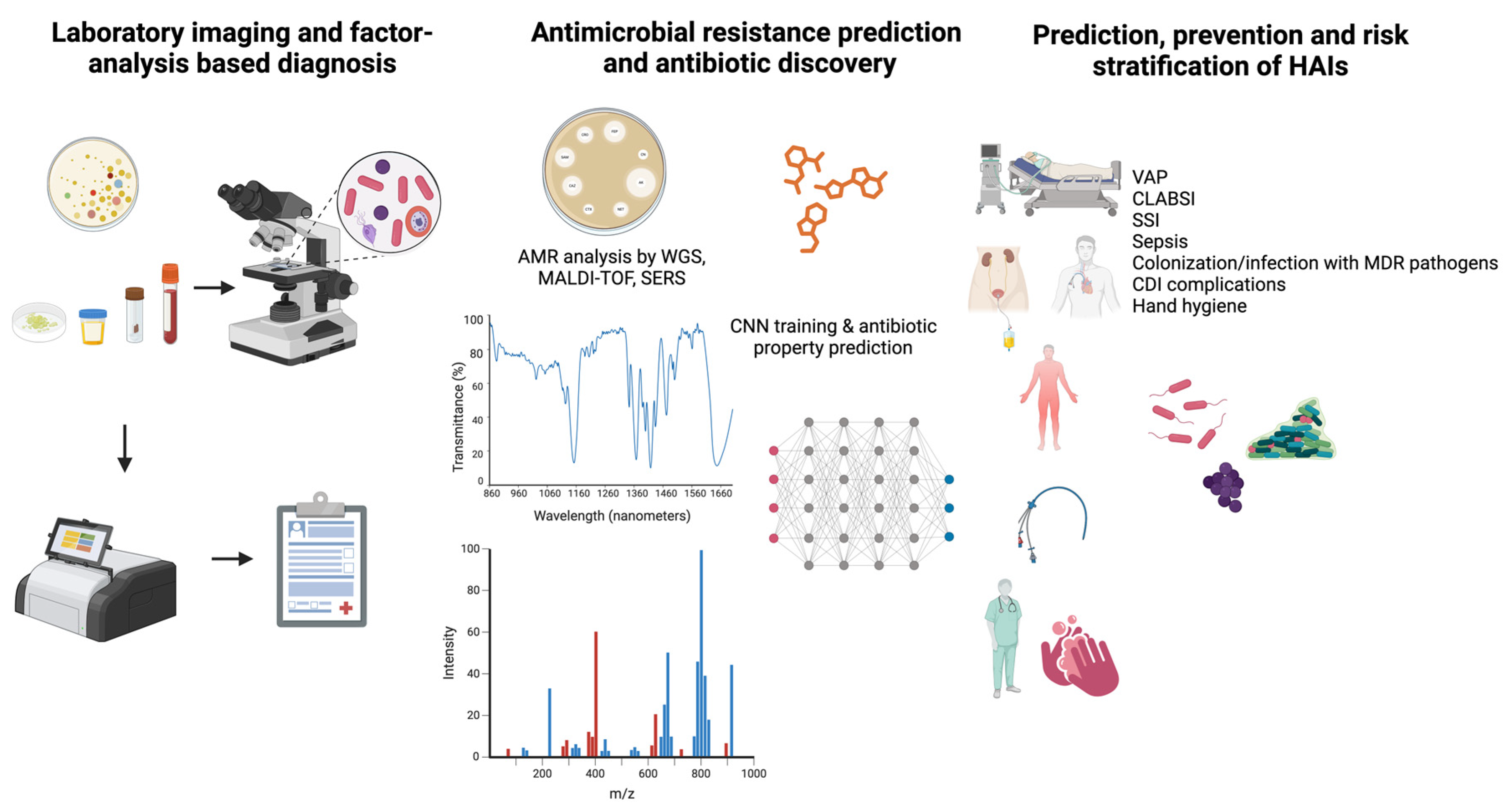

| Application of AI in Infectious Diseases | Approaches | Challenges |
|---|---|---|
| Laboratory and imaging diagnosis | Digital culture plate reading Pathogen detection and identification via microscopy images Analysis of RT-PCR data Analysis of MALDI-TOF MS, SERS spectra Clinical radiography imaging analysis Feature/factor analysis of clinical laboratory data | Data standardization among laboratories and centers Availability of big data Data quality and management Risk of bias Legal issues |
| Antimicrobial resistance analysis | Detection of MDR pathogens Antimicrobial susceptibility analysis Analysis of genomic, sequencing and spectral data | |
| Antimicrobial discovery | Molecule screening Chemical library mining Design of antimicrobial peptides | |
| Microbiome analysis | Mining of metagenomic, metatranscriptomic and metabolomic data | |
| Clinical decision support | Infection prediction and risk stratification Tracking infection epidemiology Tracking human behavior and adherence |
| Authors (Year) | n | Diagnosis Method | Input | Model/Analysis | Objective |
|---|---|---|---|---|---|
| Alouani et al. [7] (2021) | 50,146 | Real-time PCR (RT-PCR) | Fluorescent readings | Deep convolutional neural network-based software (qPCRdeepNet) https://github.com/davidalouani/qPCRdeepNet, accessed date (18 February 2024) | Detection of false positive results and improvement of test specificity, a quality assurance tool |
| Lee et al. [8] (2022) | 5810 | Real-time PCR (RT-PCR) | Fluorescence values | Long-term short memory (LSTM) | Improvement of the speed of COVID-19 RT-PCR diagnosis |
| Özbilge et al. [9] (2022) | 560 | Real-time PCR (RT-PCR) | Amplification curves | MobileNetV2 DCNN | Rapid and reliable diagnosis |
| Villarreal-González et al. [10] (2020) | 14,230 | RT-PCR | RT-PCR curves | K-neighbor classifier, support vector machine for classification (SVC), decision tree classifier, random forest classifier (RFC) | Detecting atypical profiles in PCR curves caused by contamination or artifacts |
| Alvargonzález et al. [11] (2023) | 20,418 | rRT-PCR | Ct values | Support vector machine (SVM) and neural network (NN) | Detection of a Ct pattern that is characteristic of virus variants |
| Beduk et al. [12] (2022) | 63 | Laser-scribed graphene (LSG) sensors coupled with gold nanoparticles (AuNPs) | Electrochemical sensor data | Dense neural network (DNN) | Utilization of point-of-care device as biosensing platform for new variants |
| Tschoellitsch et al. [13] (2021) | 1357 | SARS-CoV-2 RT-PCR test and blood tests | RT-PCR and blood tests results | Random forest algorithm | Prediction of SARS-CoV-2 PCR results with routine blood tests |
| Brinati et al. [14] (2020) | 279 | Routine blood tests and COVID-19 RT-PCR tests | Blood test parameters and COVID-19 RT-PCR test results | Decision tree (DT); extremely randomized trees (ETs), k-nearest neighbor (KNN) Logistic regression (LR), naïve Bayes (NB), random forest (RF), support vector machine (SVM) | Discrimination between SARS-CoV-2 positive and negative patients |
| Yang et al. [15] (2020) | 3,356 | Routine blood tests, COVID-19 RT-PCR tests | Blood parameters, COVID-19 RT-PCR test results | Gradient boosting decision tree (GBDT), random tree (RT), logistic regression (LR), decision tree (DT) | Diagnosis of COVID-19 using the results of routine laboratory tests |
| Abayomi-Alli et al. [16] (2022) | 279 | Routine blood tests | Hematochemical values | KNN, linear SVM, RBF SVM, random forest, decision tree, neural network (multilayer perceptron), AdaBoost, extremely randomized trees (ExtraTrees), naïve Bayes, LDA, QDA, logistic regression, passive classifier, ridge classifier, and stochastic gradient descent classifier (SGDC) | Effective detection of COVID-19 using routine laboratory blood test results |
| Rocca et al. [17] (2020) | 311 | MALDI-TOF MS AND RT-PCR | Main spectra profiles | ClinPro Tools, GA/k-nearest neighbor algorithm | Identification of biomarker patterns for COVID-19 |
| Le et al. [18] (2023) | 200 | LC/MS-MS | Mass spectra | SHapley Additive exPlanations (SHAP), gradient boosted decision trees, scikit-learn v0.23.2 for random forest, stratified k-fold cross-validation, grid search | Development of an alternative diagnostic strategy for SARS-CoV-2 diagnosis |
| Rosado et al. [19] (2021) | 550 | Multiplex serological assay, RT-PCR | IgG and IgM antibody responses, RT-qPCR results | Random forest algorithm | Development of accurate serological diagnostics |
| Nachtigall et al. [20] (2020) | 3621 | MALDI-MS, RT-PCR | Mass spectra | Decision tree, DT; k-nearest neighbors, KNN; naive Bayes, NB; random forest, RF; support vector machine with a linear kernel, SVM-L; support vector machine with a radial kernel, SVM-R) | Alternative detection of SARS-CoV-2 in nasal swabs |
| Costa et al. [21] (2022) | 360 | MALDI-TOF MS | Mass spectra | Support vector machine with linear kernel (SVM-LK), support vector machine with radial basis function kernel (SVM-RK), random forest (RF) and k-nearest neighbors (K-NN), and linear discriminant analysis (LDA) | Alternative method for detection of SARS-CoV-2 in nasal swabs |
| de Fátima Cobre et al. [22] (2022) | 192 | LC-MS | Mass spectra | PLS-DA, ANNDA, XGBoostDA, SIMCA, SVM, LREG and KNN | Prediction of COVID-19 diagnosis, severity, and fatality |
| Ikponmwoba et al. [23] (2022) | 20 | SERS | Spectra | Gaussian process classifier (GPC), k-fold cross-validation | Predictive diagnosis of COVID-19 in biological samples |
| Authors (Year) | n | Diagnosis Method | Input | Model/Analysis | Objective |
|---|---|---|---|---|---|
| Loh et al. [46] (2021) | 297 | Blood smear microscopy | Microscopic smear images | Mask R-CNN | Alternative method for automated rapid malaria screening |
| Holmström et al. [47] (2020) | 125 | Thin blood and Giemsa-stained thick smear microscopy | Microscopic smear images | Cloud-based machine-learning platform (Aiforia Cloud and Create), GoogLeNet network | Digitalization of blood smears, application of deep learning (DL) algorithms to detect Plasmodium falciparum |
| Oliveira et al. [48] (2022) | 676 | Thick blood smear films | Microscopy images | Multilayer perceptron (MLP) and decision tree (DT) | Automated malaria diagnosis |
| Sengar et al. [49] (2022) | 2329 | Thin blood smears | Microscopic images | Generative adversarial network (GAN), Vision Transformers (ViTs) | Automated, non-invasive multi-class Plasmodium vivax life cycle classification and malaria diagnosis |
| Park et al. [50] (2016) | 413 | Quantitative phase spectroscopy | Quantitative phase images of unstained cells | Linear discriminant classification (LDC), logistic regression (LR), and k-nearest neighbor classification (KNC), | Automated analysis for detection and staging of red blood cells infected with Plasmodium falciparum at trophozoite or schizont stage |
| Kassim et al. [51] (2021) | 5972 | Thick smear films | Annotated thick smear microscopy images | Mask regional–convolutional neural network (Mask R-CNN), ResNet50 classifier | Application of PlasmodiumVF-Net for automated malaria diagnosis on both image and patient level |
| Dey et al. [52] (2021) | 27,558 | Thick blood films | Blood smear cell microscopy images | ResNet 152 model integrated with the deep greedy network | Automating the detection of malaria parasites in thin blood smear images |
| Ufuktepe et al. [53] (2021) | 955 | Thin blood smears | Thin blood smear microscopy | Channel-wise feature pyramid network for medicine (CFPNet-M) | Red blood cell detection, counting infected cells or identifying parasite species |
| Hemachandran et al. [54] (2023) | 27,558 | Blood smears | Blood smear microscopy images | CNN, MobileNetV2, and ResNet50 | Automatic image identification system for parasite-infected RBC detection |
| Holmström et al. [55] (2017) | 7385 | Iodine-stained stool sample smears | Digital images from a mobile microscope and whole slide-scanner | Sequential algorithms | Automated detection of soil-transmitted helminths and Schistosoma haematobium |
| Kuok et al. [56] (2019) | 19,234 | Sputum smears stained by acid-fast staining | Smear microscopy images | Refined Faster region-based CNN, support vector machine (SVM) | Two-stage Mycobacterium tuberculosis identification system |
| Yang et al. [57] (2020) | 167 | Ziehl–Neelsen stained human tissue samples | Digitized images | CNNIN, CNNAL | Automated identification of mycobacteria in human tissues |
| Ibrahim et al. [58] (2021) | 1050 | Acid-fast staining of sputum | Microscopy images | AlexNet model | Automated detection of Mycobacterium tuberculosis using transfer learning |
| Xiong et al. [59] (2018) | 3,088,492 | Acid-fast stained tissue samples | Microscopy images | CIFAR-10 CNN | AI-assisted detection method for acid-fast stained TB bacillus |
| Horvath et al. [60] (2020) | 15,204 | Auramine-stained sputum smears | Slide microscopy images | DNN classifier, Keras, TensorFlow | Machine-assisted interpretation of auramine stains for microscopic tuberculosis diagnosis |
| Smith et al. [61] (2018) | 25,488 | Gram staining of blood cultures | Microscopy images | Inception v3 CNN, Python, TensorFlow | Automated interpretation of blood culture gram stains |
| Hoorali et al. [62] (2020) | 954 | Tissue slides of patients suffering from cutaneous anthrax | Microscopy images | UNet and UNet++, Keras, TensorFlow | Automatic and rapid diagnosis of anthrax via detection and segmentation of Bacillus anthracis |
| Kang et al. [63] (2020) | 84,000 | Hyperspectral microscope imaging (HMI) method | Hyperspectral microscope images | Linear discriminant analysis (LDA), support vector machine (SVM) and soft-max regression (SR) | Identification of non-O157 Shiga toxin-producing Escherichia coli (STEC) using deep learning |
| Oyamada et al. [64] (2021) | 910 | Microscopic Agglutination Test (MAT) | MAT microscopic images | Support vector machine (SVM) | Determine agglutination within microscopic images for the diagnosis of leptospirosis |
| Zieliński et al. [65] (2017) | 660 | Stained clinical samples | DIBas dataset of digital bacterial images | CNN, support vector machine, random forest | Deep learning-based classification of bacterial genera and species |
| Ahmad et al. [66] (2023) | 480 | Stained clinical samples | High-resolution microscopic images from DIBas dataset | InceptionV3, MobileNetV2 | Deep ensemble approach-based pathogen classification in large-scale images |
| Van et al. [67] (2019] | 480 | Clinical throat specimens on CHROMagar confirmed by MALDI-TOF MS | Microscopic images | WASPLab PhenoMATRIX chromogenic detection module | AI-detection of Streptococcus pyogenes using CHROMagar |
| Gammel et al. [68] (2021) | 5913 | Patient samples collected from the nares plated onto BD BBL CHROMagar MRSA II and BD BBL CHROMagar Staph aureus | Digital images | Automated Plate Assessment System (APAS Independence) | Evaluation of an automated plate assessment system |
| Rattray et al. [69] (2023) | 335 | Culture specimens of clinical and environmental P. aeruginosa isolates | Digital colony images | ResNet-50, VGG-19, MobileNetV2 and Xception | Identification of from colony image data |
| Zhang et al. [70] (2022) | 960 | Escherichia coli cultures on agar medium | Digital colony images | Random cover targets algorithm (RCTA), YOLOv3 | Deep learning-based bacterial colony detection |
| Koo et al. [71] (2021) | 3707 | Slides with skin and nail specimens | Microscopy images | YOLO v4 | Automated detection of superficial fungal infections |
| Ma et al. [72] (2021) | 17,142 | Dissecting microscopy (DM)/stereomicroscopy platform | Original colony images | Xception | Validating a novel approach for the detection of Aspergillus fungi via stereomicroscopy |
| Liu et al. [73] (2015) | 1000 | Fecal specimens | Microscopic fecal images | ANN-1, ANN-2 | Automatic identification of fungi in fecal specimens |
| Meeda et al. [74] (2019) | 30 | Fungal cultures, confocal microscopy | Colony fingerprint digital images | Support vector machine (SVM) and random forest (RF) | Rapid discrimination of fungal species by the colony fingerprinting |
| Khan et al. [75] (2018) | 119 | Raman spectroscopy | Spectral images | Support vector machine (SVM) | Analysis of hepatitis B virus infection in blood sera using ML |
| Rohaim et al. [76] (2020) | 199 | Reverse-transcribed loop-mediated isothermal amplification (LAMP) assay | Quantitative measurements using qRT-PCR | CNN model with binary cross-entropy and Adam | Rapid detection of SARS-CoV-2 using AI in loop-mediated isothermal amplification assays |
| Ito et al. [77] (2018) | 35 | Transmission electron microscopy (TEM) | Microscopy images | Cross-point method (CPM), RDP, spectral rings (SR), fully convolutional neural networks (FCN and FCN+) | Automated feline calicivirus particle detection in TEM images |
| Tong et al. [78] (2019) | 600 | Raman spectroscopy of serum samples | Raman spectra | Principal component analysis (PCA), support vector machine (SVM) | AI-aided detection of hepatitis B virus infection using Raman spectroscopy |
| Tabarov et al. [79] (2022) | 90 | Surface-enhanced Raman scattering spectroscopy (SERS) | SERS spectra | Support vector machine (SVM) | Detection of A and B influenza viruses by SERS coupled with ML |
| Authors (Year) | Dataset | Input | Model/Analysis | Objective | Results |
|---|---|---|---|---|---|
| Liang et al. [129] (2022) | Multiparameter Intelligent Monitoring in Intensive Care (MIMIC)-III dataset | 42 VAP risk factors at admission and routinely measured the vital characteristics and laboratory results from 38,515 ventilation sessions | Random forest compared to clinical pulmonary infection score (CPIS)-based model | Early prediction of ventilator-associated pneumonia in critical care patients | AUC of 84% in the validation, 74% sensitivity and 71% specificity 24 h after intubation |
| Giang et al. [130] (2021) | Multiparameter Intelligent Monitoring in Intensive Care (MIMIC)-III dataset | Data from 6126 adult ICU encounters | Five different ML models trained: logistic regression, multilayer perceptron, random forest, support vector machines, and gradient boosted trees | Prediction of ventilator-associated pneumonia with ML | The highest performing model achieved an AUROC value of 0.854 |
| Samadani et al. [131] (2023) | Philips eRI dataset | 9204 presumed VAP events | XGBoost gradient boosting algorithm, random forest, logistic regression, ADABoost, KNN | Early prediction and hospital phenotyping of ventilator-associated pneumonia | The model predicts the development of VAP 24 h in advance with an AUC of 76% and AUPRC of 75% |
| Jeon et al. [132] (2023) | SNU-SMG Boramae Medical Center database | 816 patient data including the period from hospital admission to ICU admission, age, APACHE II scores, PaO2/FiO2 ratio, history of chronic respiratory disease, history of cerebrovascular accident (CVA) or dementia, mechanical ventilation, use of vasopressors | Logistic regression with L2 regularization, gradient-boosted decision tree (LightGBM), multilayer perceptron (MLP) | ML-based prediction of in-ICU mortality in pneumonia patients | ML models significantly outperformed the Simplified Acute Physiology Score II (AU-ROC: 0.650 vs. 0.820 for logistic regression vs 0.827 for LightGBM 0.838 for MLP |
| Wang et al. [133] (2023) | MIMIC-IV and eICU databases | MIMIC-IV (n = 4697) and eICU (n = 13,760) databases, six variables included: metastatic solid tumor, Charlson Comorbidity Index, readmission, congestive heart failure, age, and Acute Physiology Score II | Logistic regression, decision tree, random forest, multilayer perceptron, XGBoost | Prediction of mortality in pneumonia patients on intensive care unit admission | AUC value ranged in predicting 1-year and hospital mortality were 0.784–0.797 and 0.691–0.780, respectively |
| Wang et al. [134] (2023) | Medical Information Mart for Intensive Care-III (MIMIC-III) database | 786 VAP incidences with traumatic brain injury (TBI) patients | Random forest, XGBoost and AdaBoost | Development of algorithms for prediction of ventilator associated pneumonia in traumatic brain injury patients | The random forest performed the best on predicting VAP in the training cohort with AUC of 1.000. AdaBoost performed the best on predicting VAP in the validation cohort with a AUC of 0.706. |
| Rahmani et al. [136] (2022) | National longitudinal electronic health records | Demographics, number of days a patient had been hospitalized before placement of a central line, laboratory and vital values (n = 27,619) | XGBoost, logistic regression, decision tree | Early prediction of central line associated bloodstream infection using ML | XGBoost was the highest performing model with an AUROC of 0.762 for CLABSI risk prediction at 48 h after the recorded time for central line placement |
| Beeler et al. [137] (2018) | Indiana University Health Academic Health Center (IUH AHC) database | Intrinsic and extrinsic risk factors (n = 70,218) | Logistic regression and random forest | ML-based assessment of patient risk for central line-associated bacteremia | Random forest had AUROC of 0.82, while AUROC curve for the logistic regression model was 0.79 |
| Parreco et al. [138] (2018) | Multiparameter Intelligent Monitoring in Intensive Care III database | Variables included six different severities of illness scores calculated on the first day of ICU admission with their components and comorbidities. The outcomes of interest were in-hospital mortality, central line placement, and CLABSI (n = 57,786) | Logistic regression, gradient boosted trees, and deep learning. | Prediction of central line-associated bloodstream infections and mortality using supervised ML | Classifiers using deep learning performed with the highest AUC for mortality, 0.885 and central line placement, 0.816. The classifier using logistic regression for predicting CLABSI performed with an AUC of 0.722 |
| Bonello et al. [139] (2022) | Boston Children’s Hospital database | Patient-level risk factors, encounter-level risk factors, demographics, vital signs measurements from the preceding 24 h, recent course-related risk factors, laboratory values and CVC-associated risk factors (n = 7468) | Generalized linear modeling, random forest, lasso regression | Prediction of impending CLABSI infections in hospitalized cardiac patients | ML predicted 25% of patients with impending CLABSI with an FPR of 0.11% and AUC of 0.82 |
| Hu et al. [141] (2015) | Surgical patient database at the University of Minnesota Medical Center | Clinical data included six data types: demographics, diagnosis codes, orders, lab results, vital signs, and medications. Demographics included each patient’s gender, race, and age at the time of surgery | Single-task learning, Hierarchical classification, offset method, propensity-weighted observations (PWO), multi-task learning with penalties (MTLP), partial least squares regression (PLS) | Automated detection of postoperative complications using EHR data | The models demonstrated high detection performance, which ensures the feasibility of accelerating manual chart review (MCR) |
| Kuo et al. [142] (2018) | Kaohsiung Chang Gung Memorial Hospital database | Dataset including 1836 patients with 1854 free-flap reconstructions and 438 postoperative SSIs | Feed-forward artificial neural network (ANN) and logistic regression (LR) models | Artificial neural network approach to predict surgical site infection after free-flap reconstruction in patients receiving surgery for head and neck cancer | ANN had a significantly higher AUC (0.892) of postoperative prediction and AUC (0.808) of pre-operative prediction than LR |
| Sohn et al. [143] (2017) | American College of Surgeons National Surgical Quality Improvement Program (ACS-NSQIP) cohort | Cohort data | Bayesian network coupled with natural language processing (NLP) | Detection of clinically important colorectal surgical site infection | Bayesian network detected ACS-NSQIP-captured SSIs with a receiver operating characteristic AUC of 0.827 |
| Soguero-Ruiz et al. [144] (2015) | EHR of the Department of Gastrointestinal Surgery at the University Hospital of North Norway | A cohort based on relevant International Classification of Diseases (ICD10) or NOMESCO Classification of Surgical Procedures (NCSP) codes related to severe post-operative complications (101 cases and 904 controls) | Gaussian process (GP) regression, support vector machine (SVM) | Data-driven temporal prediction of surgical site infection | Real-time prediction and identification of patients at risk for developing SSI was shown |
| Mamlook et al. [145] (2023) | American College of Surgeons’ National Surgical Quality Improvement Program (ACS NSQIP) database | Data from 2,882,526 surgical procedures | Logistic regression (LR), naïve Bayes (NB), random forest (RF), decision tree (DT), support vector machine (SVM), artificial neural network (ANN), and deep neural network (DNN) | Prediction of surgical site infections using patient pre-operative risk and surgical procedure factors | DNN model offers the best predictive performance with 10-fold compared to the other 6 approaches considered (area under the curve 0.8518, accuracy 0.8518, precision 0.8517, sensitivity 0.8527, F1-score 0.8518) |
| Cho et al. [146] (2023) | Samsung Medical Center clinical data warehouse (CDW) | Clinical data | Python, Tensorflow, Keras, Scikit-learn libraries, random forest (RF), gradient boosting (GB), and neural network (NN) with or without recursive feature elimination (RFE) | Development of ML models for the surveillance of colon surgical site infections | NN with RFE using 29 variables had the best performance with an AUC of 0.963. PPV of 21.1%, sensitivity of 95% |
| Petrosyan et al. [147] (2021) | The Ottawa hospital database | Patients aged 18 years and older who underwent surgery, included in the American College of Surgeons National Surgical Quality Improvement Program (NSQIP) data collection | Random forest algorithm, high-performance logistic regression | Prediction of postoperative surgical site infection with administrative data | Final model, including hospitalization diagnostic, physician diagnostic and procedure codes, demonstrated excellent discrimination (C statistics, 0.91, 95% CI, 0.90–0.92 |
| Wu et al. [148] (2023) | Calgary, Canada acute care hospital database | Cohort included adult patients (age ≥ 18 years) who underwent primary total elective hip (THA) or knee (TKA) arthroplasty | XGBoost models | ML-aided detection of surgical site infections following total hip and knee arthroplasty | XGBoost models using a combination of administrative data and text data to identify complex SSIs achieved the best performance, with F1 score of 0.788, ROC AUC of 0.906 |
| Chen et al. [149] (2023) | The First Affiliated Hospital of Guangxi Medical University, Department of Spine and Osteopathy Ward database | Patients who underwent lumbar internal fixation surgery at (n = 4019) | Lasso regression analysis, support vector machine, random forest | Application of ML to predict surgical site infection after lumbar spine surgery | C-index of the model was 0.986, ROC AUC curve 0.988 |
| Wang et al. [157] (2021] | Observational cohort from the Intensive Care Unit of the First Affiliated Hospital of Zhengzhou University | Electronic medical record data, a set of 55 features (variables) from 4449 infected patients | Random forest | Application of ML for accurate prediction of sepsis in ICU patients | ROC AUC was 0.91 with 87%, sensitivity, 89% specificity for sepsis prediction |
| Lauritsen et al. [158] (2020) | Retrospective data from multiple Danish hospitals | EHR, including biochemistry, medicine, microbiology, medical imaging, and the patient administration system (PAS) | Combination of a convolutional neural network and a long short-term memory network | Early detection of sepsis utilizing deep learning on EHR event sequences | Model performance ranged from AUROC 0.856 (3 h before sepsis onset) to AUROC 0.756 (24 h before sepsis onset) |
| Yuan et al. [159] (2020) | Prospective open-label cohort study conducted at Taipei Medical University Hospital | Data including the vital signs, laboratory results, examination reports, text data, and image of every ICU patient | Logistic regression, support vector machine, XGBoost, and neural network | Development an AI algorithm for early sepsis diagnosis in the intensive care unit | Established AI algorithm achieved accuracy of 82%, sensitivity of 65%, specificity of 88%, precision = 67%, F1 = 0.66 ± 0.02. AUROC was 0.89 |
| Fagerström et al. [160] (2019) | Medical Information Mart for Intensive Care database | Vital signs, laboratory data, and journal entries (n = 59,000 ICU patients) | LiSep LSTM; a long short-term memory neural network, Keras with a Google TensorFlow | Application of ML algorithm for early detection of septic shock | LiSep LSTM outperforms a less complex model, using the same features and targets, with an AUROC 0.8306 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baddal, B.; Taner, F.; Uzun Ozsahin, D. Harnessing of Artificial Intelligence for the Diagnosis and Prevention of Hospital-Acquired Infections: A Systematic Review. Diagnostics 2024, 14, 484. https://doi.org/10.3390/diagnostics14050484
Baddal B, Taner F, Uzun Ozsahin D. Harnessing of Artificial Intelligence for the Diagnosis and Prevention of Hospital-Acquired Infections: A Systematic Review. Diagnostics. 2024; 14(5):484. https://doi.org/10.3390/diagnostics14050484
Chicago/Turabian StyleBaddal, Buket, Ferdiye Taner, and Dilber Uzun Ozsahin. 2024. "Harnessing of Artificial Intelligence for the Diagnosis and Prevention of Hospital-Acquired Infections: A Systematic Review" Diagnostics 14, no. 5: 484. https://doi.org/10.3390/diagnostics14050484
APA StyleBaddal, B., Taner, F., & Uzun Ozsahin, D. (2024). Harnessing of Artificial Intelligence for the Diagnosis and Prevention of Hospital-Acquired Infections: A Systematic Review. Diagnostics, 14(5), 484. https://doi.org/10.3390/diagnostics14050484








