A Multimodal Transformer Model for Recognition of Images from Complex Laparoscopic Surgical Videos
Abstract
1. Introduction
2. Related Works
3. Materials and Methods
3.1. Video-Audio-Text Transformer (VATT)
3.2. Training the VATT
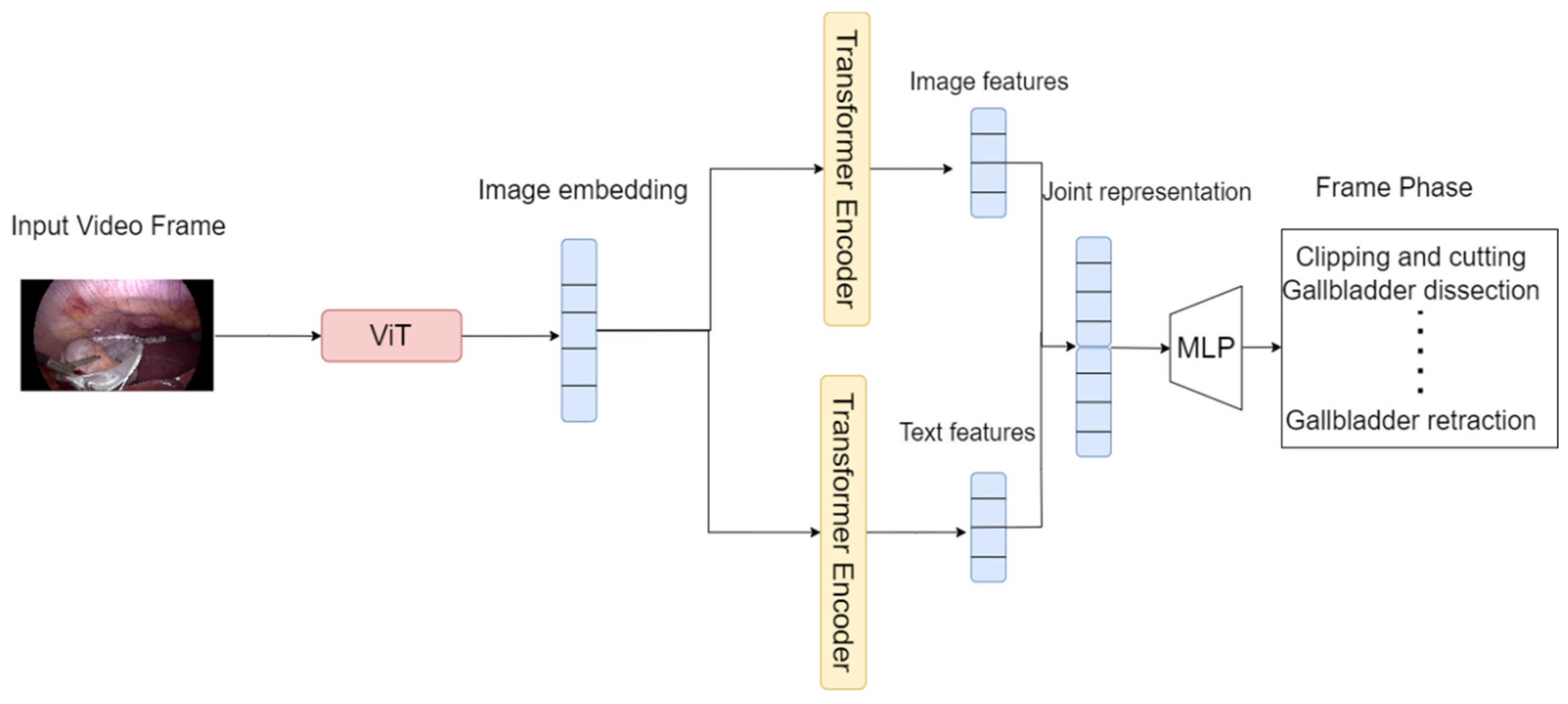
3.3. The Proposed Model
4. Experimental Setup
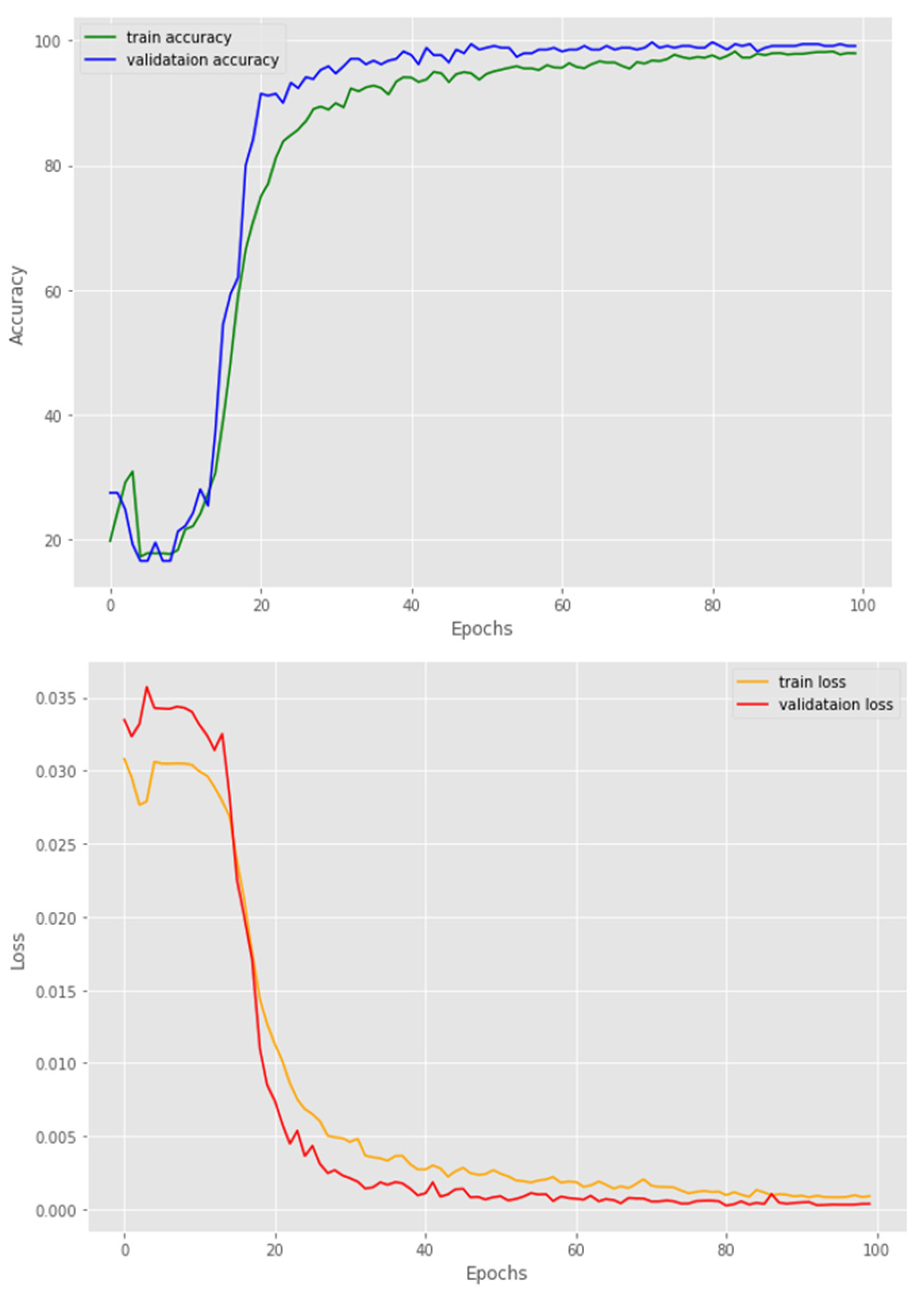
4.1. Implementation Details
4.2. Cholec80 Dataset
5. Results and Discussion
5.1. The Text and Image Embedding Extraction Models
5.2. The Transformer Encoders
5.3. Comparison
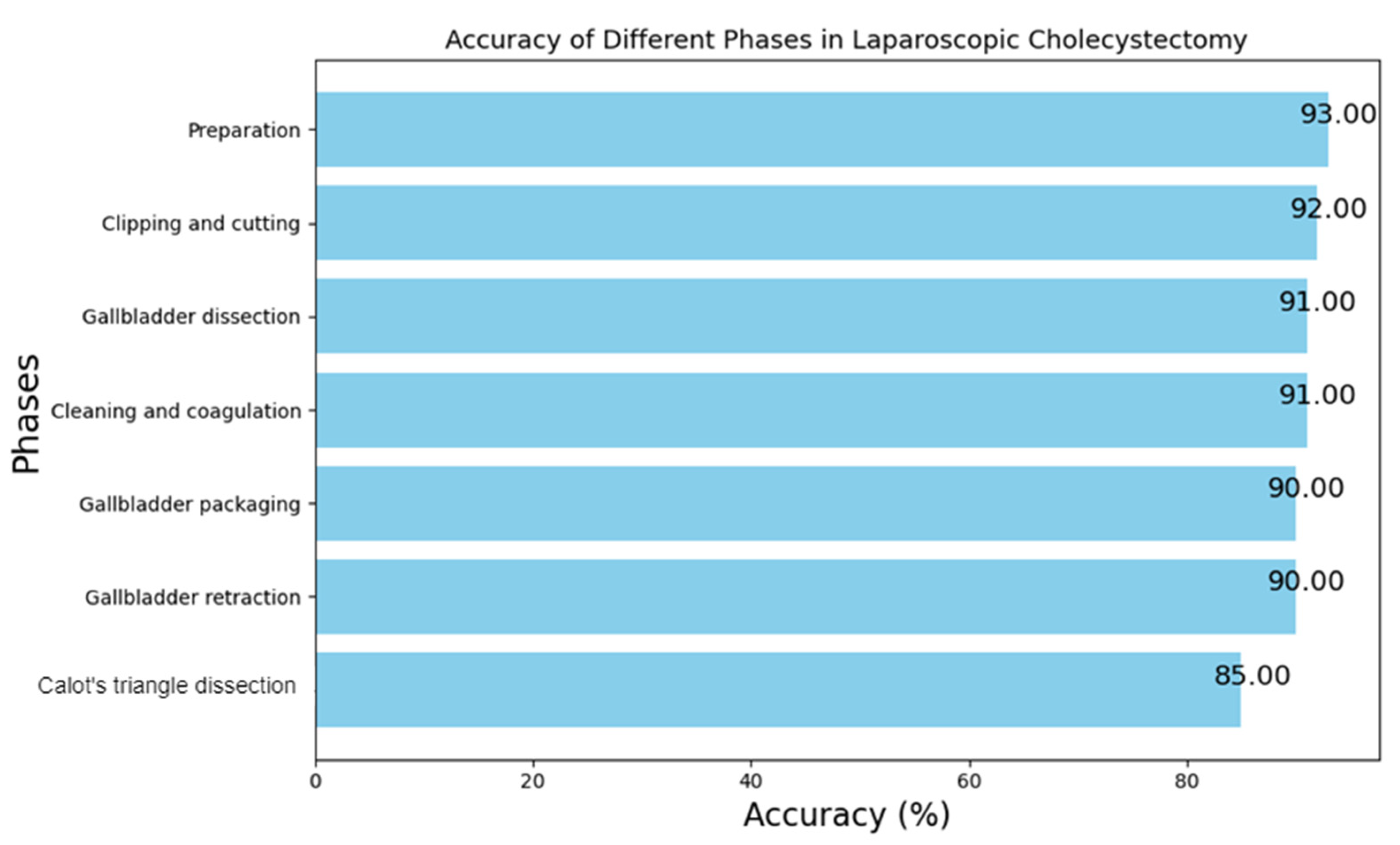
5.4. Inference
5.5. Discussion
6. Conclusions and Future Works
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lavanchy, J.L.; Vardazaryan, A.; Mascagni, P.; Mutter, D.; Padoy, N. Preserving privacy in surgical video analysis using a deep learning classifier to identify out-of-body scenes in endoscopic videos. Sci. Rep. 2023, 13, 9235. [Google Scholar] [CrossRef] [PubMed]
- Bonrath, E.M.; Gordon, L.E.; Grantcharov, T.P. Characterising ‘near miss’ events in complex laparoscopic surgery through video analysis. BMJ Qual. Saf. 2015, 24, 516–521. [Google Scholar] [CrossRef] [PubMed]
- Twinanda, A.P.; Shehata, S.; Mutter, D.; Marescaux, J.; De Mathelin, M.; Padoy, N. Endonet: A deep architecture for recognition tasks on laparoscopic videos. IEEE Trans. Med. Imaging 2016, 36, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Bai, J. Deep learning-based intraoperative video analysis for supporting surgery. Concurr. Comput. Pract. Exp. 2023, 35, e7837. [Google Scholar] [CrossRef]
- Miyawaki, F.; Tsunoi, T.; Namiki, H.; Yaginuma, T.; Yoshimitsu, K.; Hashimoto, D.; Fukui, Y. Development of Automatic Acquisition System of Surgical-Instrument Informantion in Endoscopic and Laparoscopic Surgey. In Proceedings of the 2009 4th IEEE Conference on Industrial Electronics and Applications, Xi’an, China, 25–27 May 2009; IEEE: Piscataway, NJ, USA, 2009; pp. 3058–3063. [Google Scholar]
- Abdulbaki Alshirbaji, T.; Jalal, N.A.; Docherty, P.D.; Neumuth, T.; Möller, K. Robustness of Convolutional Neural Networks for Surgical Tool Classification in Laparoscopic Videos from Multiple Sources and of Multiple Types: A Systematic Evaluation. Electronics 2022, 11, 2849. [Google Scholar] [CrossRef]
- Doignon, C.; Graebling, P.; De Mathelin, M. Real-time segmentation of surgical instruments inside the abdominal cavity using a joint hue saturation color feature. Real-Time Imaging 2005, 11, 429–442. [Google Scholar] [CrossRef]
- Primus, M.J.; Schoeffmann, K.; Böszörmenyi, L. Temporal segmentation of laparoscopic videos into surgical phases. In Proceedings of the 2016 14th International Workshop on Content-Based Multimedia Indexing (CBMI), Bucharest, Romania, 15–17 June 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 1–6. [Google Scholar]
- Helwan, A.; Azar, D.; Ma’aitah, M.K.S. Conventional and deep learning methods in heart rate estimation from RGB face videos. Physiol. Meas. 2024, 45, 02TR01. [Google Scholar] [CrossRef] [PubMed]
- Mishra, K.; Sathish, R.; Sheet, D. Learning latent temporal connectionism of deep residual visual abstractions for identifying surgical tools in laparoscopy procedures. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Honolulu, HI, USA, 21–26 July 2017; pp. 58–65. [Google Scholar]
- Nwoye, C.I.; Mutter, D.; Marescaux, J.; Padoy, N. Weakly supervised convolutional LSTM approach for tool tracking in laparoscopic videos. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 1059–1067. [Google Scholar] [CrossRef]
- Namazi, B.; Sankaranarayanan, G.; Devarajan, V. A contextual detector of surgical tools in laparoscopic videos using deep learning. Surg. Endosc. 2022, 36, 679–688. [Google Scholar] [CrossRef]
- Wang, S.; Xu, Z.; Yan, C.; Huang, J. Graph Convolutional Nets for Tool Presence Detection in Surgical Videos. In Proceedings of the International Conference on Information Processing in Medical Imaging, Hong Kong, China, 2–7 June 2019; Springer: Berlin/Heidelberg, Germany, 2019; pp. 467–478. [Google Scholar]
- Bar, O.; Neimark, D.; Zohar, M.; Hager, G.D.; Girshick, R.; Fried, G.M.; Wolf, T.; Asselmann, D. Impact of data on generalization of AI for surgical intelligence applications. Sci. Rep. 2020, 10, 22208. [Google Scholar] [CrossRef]
- Akbari, H.; Yuan, L.; Qian, R.; Chuang, W.H.; Chang, S.F.; Cui, Y.; Gong, B. Vatt: Transformers for multimodal self-supervised learning from raw video, audio and text. Adv. Neural Inf. Process. Syst. 2021, 34, 24206–24221. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Houlsby, N. An image is worth 16x16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Darwish, M.; Altabel, M.Z.; Abiyev, R.H. Enhancing Cervical Pre-Cancerous Classification Using Advanced Vision Transformer. Diagnostics 2023, 13, 2884. [Google Scholar] [CrossRef]
- Kotei, E.; Thirunavukarasu, R. A Systematic Review of Transformer-Based Pre-Trained Language Models through Self-Supervised Learning. Information 2023, 14, 187. [Google Scholar] [CrossRef]
- Ma, Z.; Collins, M. Noise contrastive estimation and negative sampling for conditional models: Consistency and statistical efficiency. arXiv 2018, arXiv:1809.01812. [Google Scholar]
- Yang, M.; Zhou, P.; Li, S.; Zhang, Y.; Hu, J.; Zhang, A. Multi-Head multimodal deep interest recommendation network. Knowl.-Based Syst. 2023, 276, 110689. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Polosukhin, I. Attention is all you need. Adv. Neural Inf. Process. Syst. 2017, 30. [Google Scholar]
- Ren, Y.; Li, Y. On the Importance of Contrastive Loss in Multimodal Learning. arXiv 2023, arXiv:2304.03717. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. IJCV 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Gotmare, A.; Keskar, N.S.; Xiong, C.; Socher, R. A closer look at deep learning heuristics: Learning rate restarts, warmup and distillation. arXiv 2018, arXiv:1810.13243. [Google Scholar]
- Jin, Y.; Li, H.; Dou, Q.; Chen, H.; Qin, J.; Fu, C.W.; Heng, P.A. Multi-task recurrent convolutional network with correlation loss for surgical video analysis. Med. Image Anal. 2020, 59, 101572. [Google Scholar] [CrossRef] [PubMed]
- Twinanda, A.P.; Mutter, D.; Marescaux, J.; de Mathelin, M.; Padoy, N. Single-and multi-task architectures for surgical workflow challenge at M2CAI 2016. arXiv 2016, arXiv:1610.08844. [Google Scholar]
- Jin, Y.; Dou, Q.; Chen, H.; Yu, L.; Qin, J.; Fu, C.W.; Heng, P.A. SV-RCNet: Workflow recognition from surgical videos using recurrent convolutional network. IEEE Trans. Med. Imaging 2017, 37, 1114–1126. [Google Scholar] [CrossRef] [PubMed]
- Yi, F.; Jiang, T. Hard Frame Detection and Online Mapping for Surgical Phase Recognition. In Proceedings of the Medical Image Computing and Computer Assisted Intervention—MICCAI 2019: 22nd International Conference, Shenzhen, China, 13–17 October 2019; Proceedings, Part V 22. Springer International Publishing: Cham, Switzerland, 2019; pp. 449–457. [Google Scholar]
- Gao, X.; Jin, Y.; Long, Y.; Dou, Q.; Heng, P.A. Trans-svnet: Accurate Phase Recognition from Surgical Videos via Hybrid Embedding Aggregation Transformer. In Proceedings of the Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, 27 September–1 October 2021; Proceedings, Part IV 24. Springer International Publishing: Cham, Switzerland, 2021; pp. 593–603. [Google Scholar]



| Author | Method | Task | Dataset |
|---|---|---|---|
| Twinanda et al. [3] | CNN | Surgical phase recognition | Cholec80 [3] |
| Miyawaki et al. [5] | RFID technology | Surgical tool recognition | - |
| Alshirbaji et al. [6] | CNN (DenseNet-121) | Surgical tool recognition | Cholec80 [3], EndovisChole [6], and Gyna08 [6] |
| Doignon et al. [7] | Discriminant color feature with respect to intensity variations and specularities. | Surgical tool segmentation | - |
| Primus et al. [8] | Object detection, SVM classifiers and ORB features | Temporal segmentation of surgical phases | Cholec80 [3] |
| Mishra et al. [10] | CNN and Long Short-Term Memory network (LSTM) | Identifying surgical tools | Cholec80 [3] |
| Nwoye et al. [11] | CNN + Convolutional LSTM (ConvLSTM) | Surgical tool tracking | Cholec80 [3] |
| Namazi et al. [12] | Recurrent Convolutional Neural Network (RCNN) | Surgical tool recognition | Cholec80 [3] |
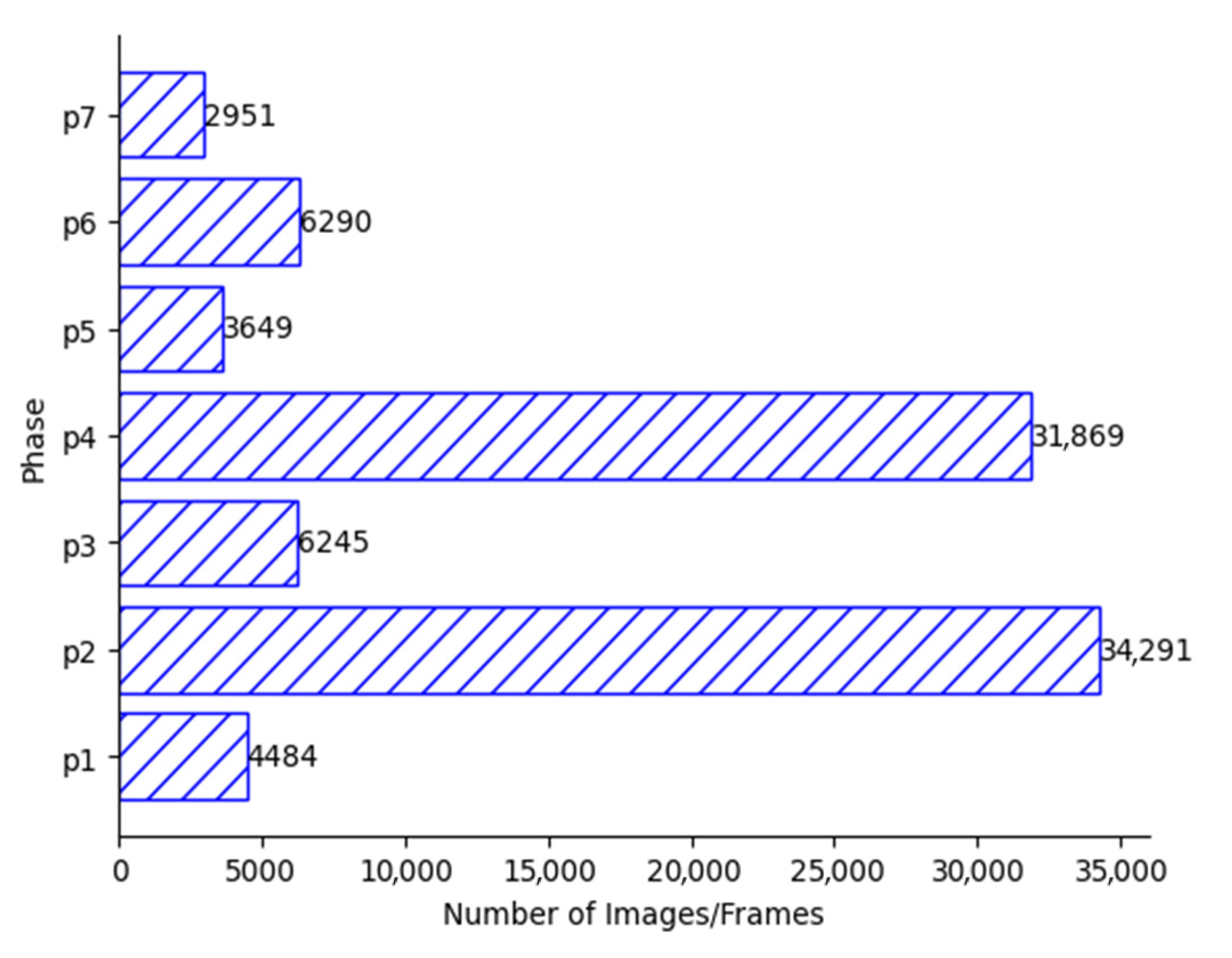
| Cholec80 Dataset | |
|---|---|
| Number of videos | 80 |
| Training | 50 |
| Testing | 30 |
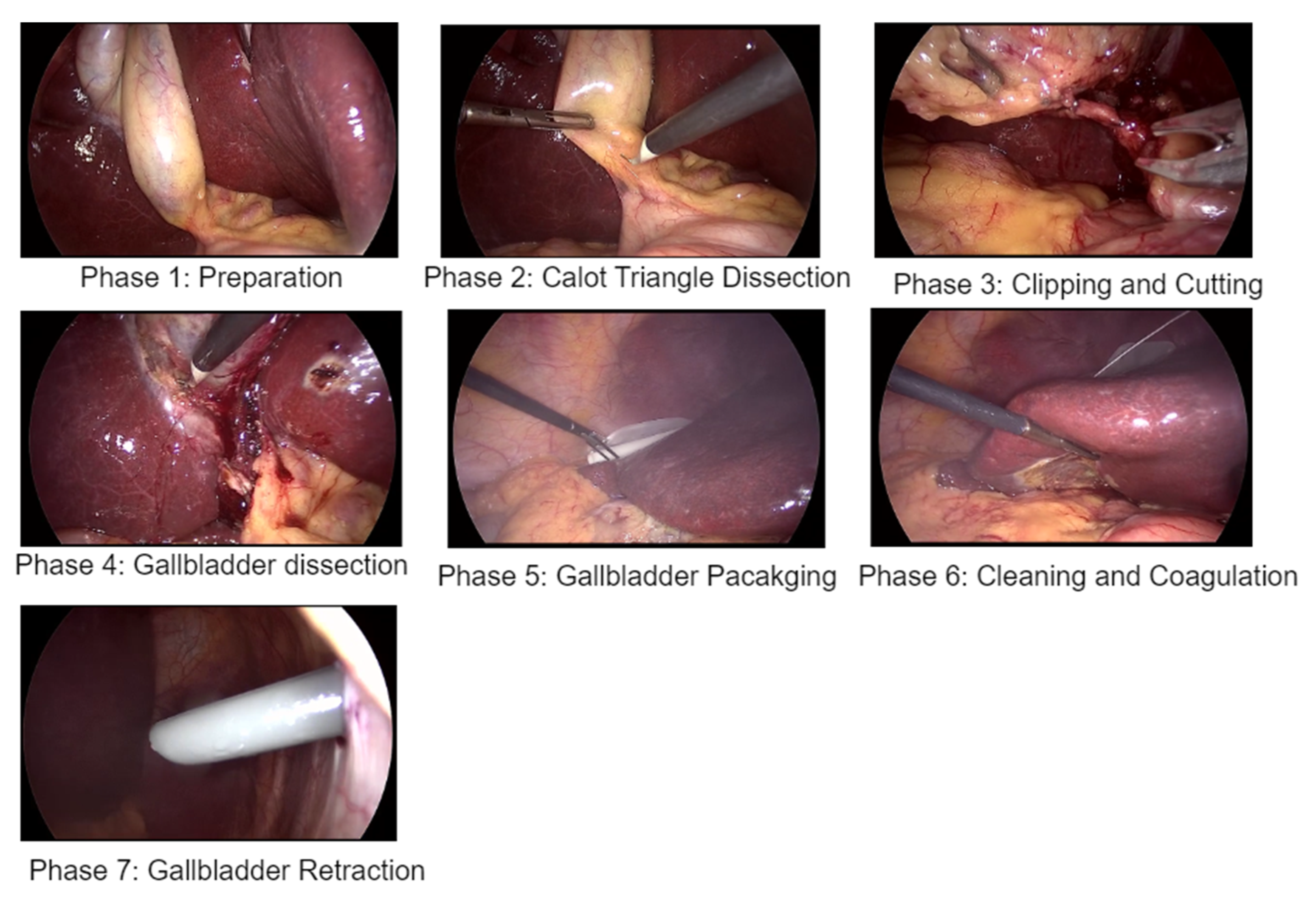
| Phases | Phase Name | Duration (S) |
|---|---|---|
| 1 | Preparation | 125 ± 95 |
| 2 | Calot’s triangle dissection | 954 ± 538 |
| 3 | Clipping and cutting | 168 ± 152 |
| 4 | Gallbladder dissection | 857 ± 551 |
| 5 | Gallbladder packaging | 98 ± 53 |
| 6 | Cleaning and coagulation | 178 ± 166 |
| 7 | Gallbladder retraction | 83 ± 56 |
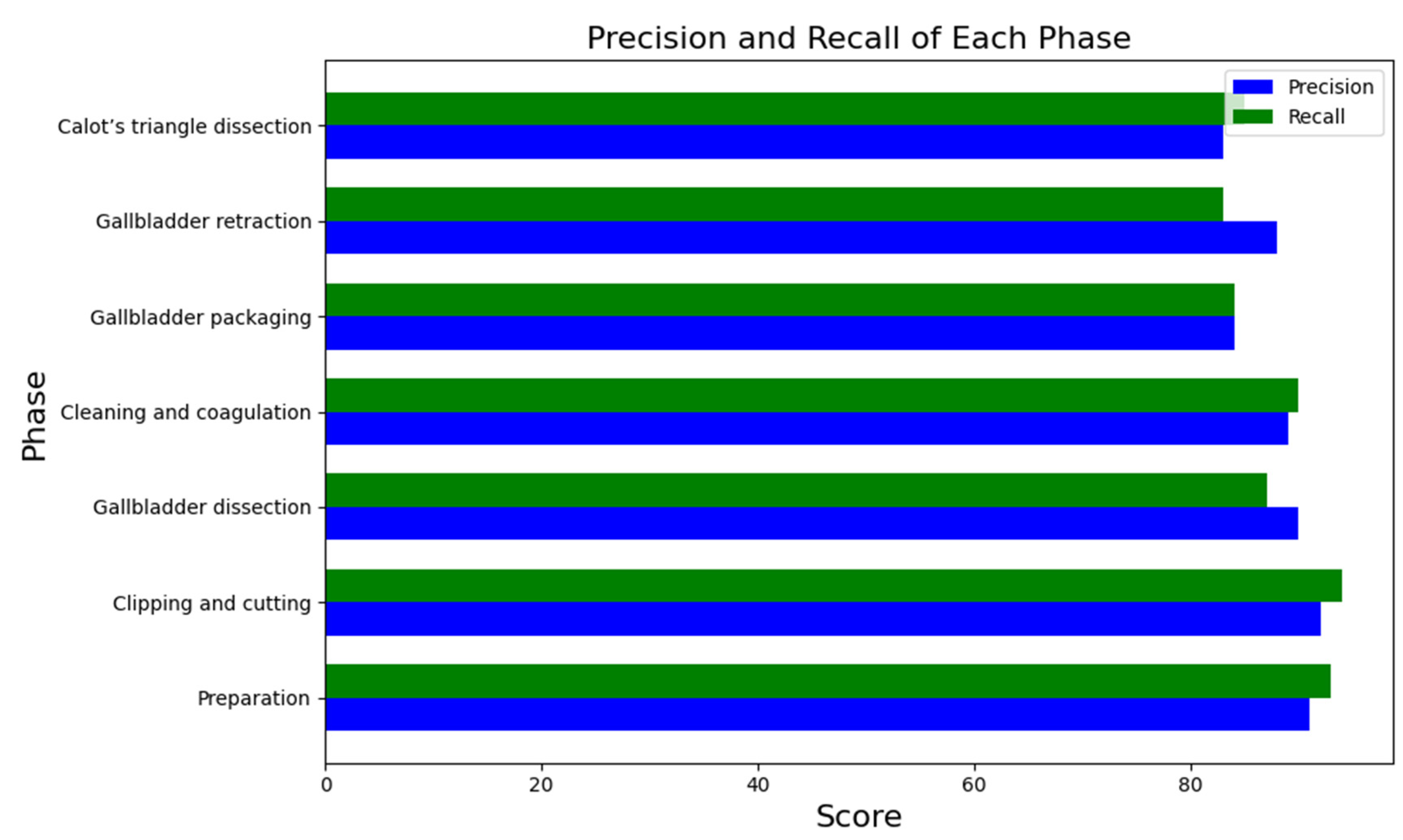
| Mean | Standard Deviation | |
|---|---|---|
| No. of videos | 30 | 30 |
| Accuracy | 0.91 | 0.07 |
| Precision | 0.81 | 0.10 |
| Recall | 0.83 | 0.09 |
| Two-Transformer Encoders Model | One-Transformer Encoder Model | |
|---|---|---|
| No. of videos | 30 | 30 |
| Accuracy | 0.91 | 0.88 |
| Method | Accuracy (%) | |
|---|---|---|
| Cholec90 Dataset | MTRCNet-CL [25] | 89.2 |
| EndoNet [3] | 81.7 | |
| PhaseNet [26] | 78.8 | |
| SV-RCNet [27] | 85.3 | |
| OHFM [28] | 87.3 | |
| Trans-SVNet [29] | 90.3 | |
| Ours | 91.0 |
| Model 1 | Model 2 | |
|---|---|---|
| Accuracy (%) | 0.91 | 0.88 |
| Paried t-test | 6.12 | |
| Alpha | 0.05 | |
| p-value | 0.02 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abiyev, R.H.; Altabel, M.Z.; Darwish, M.; Helwan, A. A Multimodal Transformer Model for Recognition of Images from Complex Laparoscopic Surgical Videos. Diagnostics 2024, 14, 681. https://doi.org/10.3390/diagnostics14070681
Abiyev RH, Altabel MZ, Darwish M, Helwan A. A Multimodal Transformer Model for Recognition of Images from Complex Laparoscopic Surgical Videos. Diagnostics. 2024; 14(7):681. https://doi.org/10.3390/diagnostics14070681
Chicago/Turabian StyleAbiyev, Rahib H., Mohamad Ziad Altabel, Manal Darwish, and Abdulkader Helwan. 2024. "A Multimodal Transformer Model for Recognition of Images from Complex Laparoscopic Surgical Videos" Diagnostics 14, no. 7: 681. https://doi.org/10.3390/diagnostics14070681
APA StyleAbiyev, R. H., Altabel, M. Z., Darwish, M., & Helwan, A. (2024). A Multimodal Transformer Model for Recognition of Images from Complex Laparoscopic Surgical Videos. Diagnostics, 14(7), 681. https://doi.org/10.3390/diagnostics14070681







