Artificial-Intelligence-Enhanced Analysis of In Vivo Confocal Microscopy in Corneal Diseases: A Review
Abstract
1. Introduction
2. Convolutional Neural Network Architecture
3. Artificial Intelligence Issues
3.1. Low Quality of Images
3.2. Overfitting
3.3. Unrepresentative Training Set
3.4. Limited Dataset Size
3.5. “Black-Box” Problem
4. Methods and Materials
5. Evaluation of Individual Disease Articles
5.1. Keratitis
5.1.1. Fungal Keratitis
5.1.2. Bacterial Keratitis
5.1.3. Acanthamoeba Keratitis
5.1.4. Viral Keratitis
5.2. Dry Eye Disease
5.3. Diabetic Corneal Neuropathy
6. Conclusions
7. Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- FDA. Artificial Intelligence and Machine Learning (AI/ML)-Enabled Medical Devices. Available online: https://www.fda.gov/medical-devices/software-medical-device-samd/artificial-intelligence-and-machine-learning-aiml-enabled-medical-devices (accessed on 10 October 2023).
- Popescu Patoni, S.I.; Muşat, A.A.M.; Patoni, C.; Popescu, M.N.; Munteanu, M.; Costache, I.B.; Pîrvulescu, R.A.; Mușat, O. Artificial intelligence in ophthalmology. Rom. J. Ophthalmol. 2023, 67, 207–213. [Google Scholar] [CrossRef]
- Jin, K.; Ye, J. Artificial intelligence and deep learning in ophthalmology: Current status and future perspectives. Adv. Ophthalmol. Pract. Res. 2022, 2, 100078. [Google Scholar] [CrossRef]
- Nagendran, M.; Chen, Y.; A Lovejoy, C.; Gordon, A.C.; Komorowski, M.; Harvey, H.; Topol, E.J.; Ioannidis, J.P.A.; Collins, G.S.; Maruthappu, M. Artificial intelligence versus clinicians: Systematic review of design, reporting standards, and claims of deep learning studies. BMJ 2020, 368, m689. [Google Scholar] [CrossRef]
- Valueva, M.; Nagornov, N.; Lyakhov, P.; Valuev, G.; Chervyakov, N. Application of the residue number system to reduce hardware costs of the convolutional neural network implementation. Math. Comput. Simul. 2020, 177, 232–243. [Google Scholar] [CrossRef]
- Raghav, P. Understanding of Convolutional Neural Network (CNN)—Deep Learning. Available online: https://medium.com/@RaghavPrabhu/understanding-of-convolutional-neural-network-cnn-deep-learning-99760835f148 (accessed on 4 March 2018).
- Goswami, S.I.A.K.; Mishra, S.P.; Asopa, P. Conceptual Understanding of Convolutional Neural Network—A Deep Learning Approach. Procedia Comput. Sci. 2018, 132, 679–688. [Google Scholar]
- Eric, W. Convolution. From MathWorld—A Wolfram Web Resource. Available online: https://mathworld.wolfram.com/Convolution.html (accessed on 1 January 2022).
- Eckle, K.; Schmidt-Hieber, J. A comparison of deep networks with ReLU activation function and linear spline-type methods. Neural Netw. 2019, 110, 232–242. [Google Scholar] [CrossRef]
- Brownlee, J. A Gentle Introduction to Pooling Layers for Convolutional Neural Networks. Available online: https://machinelearningmastery.com/pooling-layers-for-convolutional-neural-networks/ (accessed on 5 July 2019).
- Nirthika, R.; Manivannan, S.; Ramanan, A.; Wang, R. Pooling in convolutional neural networks for medical image analysis: A survey and an empirical study. Neural Comput. Appl. 2022, 34, 5321–5347. [Google Scholar] [CrossRef]
- Zhuo, Z.; Zhou, Z. Low Dimensional Discriminative Representation of Fully Connected Layer Features Using Extended LargeVis Method for High-Resolution Remote Sensing Image Retrieval. Sensors 2020, 20, 4718. [Google Scholar] [CrossRef]
- Yamashita, R.; Nishio, M.; Do, R.K.G.; Togashi, K. Convolutional neural networks: An overview and application in radiology. Insights Imaging 2018, 9, 611–629. [Google Scholar] [CrossRef]
- Tsuneki, M. Deep learning models in medical image analysis. J. Oral Biosci. 2022, 64, 312–320. [Google Scholar] [CrossRef]
- Dorfman, E. How Much Data Is Required for Machine Learning? Available online: https://postindustria.com/how-much-data-is-required-for-machine-learning/ (accessed on 25 March 2022).
- Smolic, H. How Much Data Is Needed For Machine Learning? Available online: https://graphite-note.com/how-much-data-is-needed-for-machine-learning (accessed on 15 December 2022).
- Avanzo, M.; Wei, L.; Stancanello, J.; Vallières, M.; Rao, A.; Morin, O.; Mattonen, S.A.; El Naqa, I. Machine and deep learning methods for radiomics. Med. Phys. 2020, 47, e185–e202. [Google Scholar] [CrossRef]
- Hao, R.; Namdar, K.; Liu, L.; Haider, M.A.; Khalvati, F. A Comprehensive Study of Data Augmentation Strategies for Prostate Cancer Detection in Diffusion-Weighted MRI Using Convolutional Neural Networks. J. Digit. Imaging 2021, 34, 862–876. [Google Scholar] [CrossRef]
- Kabir, M.M.; Ohi, A.Q.; Rahman, M.S.; Mridha, M.F. An Evolution of CNN Object Classifiers on Low-Resolution Images. arXiv 2021, arXiv:2101.00686. [Google Scholar] [CrossRef]
- Michał, K.; Bogusław, C. Impact of Low Resolution on Image Recognition with Deep Neural Networks: An Experimental Study. Int. J. Appl. Math. Comput. Sci. 2018, 28, 735–744. [Google Scholar] [CrossRef]
- Cai, D.; Chen, K.; Qian, Y.; Kämäräinen, J.-K. Convolutional low-resolution fine-grained classification. Pattern Recognit. Lett. 2019, 119, 166–171. [Google Scholar] [CrossRef]
- Qu, J.; Qin, X.; Peng, R.; Xiao, G.; Gu, S.; Wang, H.; Hong, J. Assessing abnormal corneal endothelial cells from in vivo confocal microscopy images using a fully automated deep learning system. Eye Vis. 2023, 10, 20. [Google Scholar] [CrossRef]
- Hosseini, M.; Powell, M.; Collins, J.; Callahan-Flintoft, C.; Jones, W.; Bowman, H.; Wyble, B. I tried a bunch of things: The dangers of unexpected overfitting in classification of brain data. Neurosci. Biobehav. Rev. 2020, 119, 456–467. [Google Scholar] [CrossRef]
- Demšar, J.; Zupan, B. Hands-on training about overfitting. PLOS Comput. Biol. 2021, 17, e1008671. [Google Scholar] [CrossRef]
- Eertink, J.J.; Heymans, M.W.; Zwezerijnen, G.J.C.; Zijlstra, J.M.; de Vet, H.C.W.; Boellaard, R. External validation: A simulation study to compare cross-validation versus holdout or external testing to assess the performance of clinical prediction models using PET data from DLBCL patients. EJNMMI Res. 2022, 12, 58. [Google Scholar] [CrossRef]
- Eche, T.; Schwartz, L.H.; Mokrane, F.Z.; Dercle, L. Toward Generalizability in the Deployment of Artificial Intelligence in Ra-diology: Role of Computation Stress Testing to Overcome Underspecification. Radiol Artif. Intell. 2021, 3, e210097. [Google Scholar] [CrossRef]
- Ting, D.S.J.; Ho, C.S.; Deshmukh, R.; Said, D.G.; Dua, H.S. Infectious keratitis: An update on epidemiology, causative microorganisms, risk factors, and antimicrobial resistance. Eye, 1084; 35. [Google Scholar] [CrossRef]
- Essalat, M.; Abolhosseini, M.; Le, T.H.; Moshtaghion, S.M.; Kanavi, M.R. Interpretable deep learning for diagnosis of fungal and acanthamoeba keratitis using in vivo confocal microscopy images. Sci. Rep. 2023, 13, 8953. [Google Scholar] [CrossRef]
- Liu, Z.; Cao, Y.; Li, Y.; Xiao, X.; Qiu, Q.; Yang, M.; Zhao, Y.; Cui, L. Automatic diagnosis of fungal keratitis using data augmentation and image fusion with deep convolutional neural network. Comput. Methods Programs Biomed. 2020, 187, 105019. [Google Scholar] [CrossRef]
- Tougui, I.; Jilbab, A.; El Mhamdi, J. Impact of the Choice of Cross-Validation Techniques on the Results of Machine Learning-Based Diagnostic Applications. Healthc. Inform. Res. 2021, 27, 189–199. [Google Scholar] [CrossRef]
- Bradshaw, T.J.; Huemann, Z.; Hu, J.; Rahmim, A. A Guide to Cross-Validation for Artificial Intelligence in Medical Imaging. Radiol. Artif. Intell. 2023, 5, e220232. [Google Scholar] [CrossRef]
- Chlap, P.; Min, H.; Vandenberg, N.; Dowling, J.; Holloway, L.; Haworth, A. A review of medical image data augmentation techniques for deep learning applications. J. Med. Imaging Radiat. Oncol. 2021, 65, 545–563. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M.; Furht, B. Text Data Augmentation for Deep Learning. J. Big Data 2021, 8, 101. [Google Scholar] [CrossRef]
- Shorten, C.; Khoshgoftaar, T.M. A survey on Image Data Augmentation for Deep Learning. J. Big Data 2019, 6, 1–48. [Google Scholar] [CrossRef]
- Kebaili, A.; Lapuyade-Lahorgue, J.; Ruan, S. Deep Learning Approaches for Data Augmentation in Medical Imaging: A Review. J. Imaging 2023, 9, 81. [Google Scholar] [CrossRef]
- Nakagawa, K.; Moukheiber, L.; Leo, A. Celi, Malhar Patel, Faisal Mahmood, Dibson Gondim, Michael Hogarth and Richard Levenson, AI in Pathology: What could possibly go wrong? Semin. Diagn. Pathol. 2023, 40, 100–101. [Google Scholar] [CrossRef]
- de Hond, A.A.H.; Leeuwenberg, A.M.; Hooft, L.; Kant, I.M.J.; Nijman, S.W.J.; van Os, H.J.A.; Aardoom, J.J.; Debray, T.P.A.; Schuit, E.; van Smeden, M.; et al. Guidelines and quality criteria for artificial intelli-gence-based prediction model in healthcare: A scoping review. NPJ Digit. Med. 2022, 5, 2. [Google Scholar] [CrossRef]
- Géron, A. Hands-On Machine Learning with Scikit-Learn, Keras, and TensorFlow: Concepts, Tools, and Techniques to Build Intel-ligent Systems, 3rd ed.; O’Reilly Media: Sebastopol, CA, USA, 2023; ISBN 978-83-832-2424-4. [Google Scholar]
- Pillai, M.; Adapa, K.; Shumway, J.W.; Dooley, J.; Das, S.K.; Chera, B.S.; Mazur, L. Feature Engineering for Interpretable Machine Learning for Quality Assurance in Radiation Oncology. IOS 2021, 29, 460–464. [Google Scholar] [CrossRef]
- Ye, Z.; Yang, G.; Jin, X.; Liu, Y.; Huang, K. Rebalanced Zero-Shot Learning. IEEE Trans. Image Process. 2023, 32, 4185–4198. [Google Scholar] [CrossRef]
- London, A.J. Artificial Intelligence and Black-Box Medical Decisions: Accuracy versus Explainability. Hast. Cent. Rep. 2019, 49, 15–21. [Google Scholar] [CrossRef]
- Ayhan, M.S.; Kümmerle, L.B.; Kühlewein, L.; Inhoffen, W.; Aliyeva, G.; Ziemssen, F.; Berens, P. Clinical validation of saliency maps for understanding deep neural networks in ophthalmology. Med. Image Anal. 2022, 77, 102364. [Google Scholar] [CrossRef]
- Maity, A. Improvised Salient Object Detection and Manipulation. arXiv 2015. [Google Scholar] [CrossRef]
- Stapleton, F. The epidemiology of infectious keratitis. Ocul. Surf. 2023, 8, 351–363. [Google Scholar]
- Donovan, C.; Arenas, E.; Ayyala, R.S.; E Margo, C.; Espana, E.M. Fungal keratitis: Mechanisms of infection and management strategies. Surv. Ophthalmol. 2022, 67, 758–769. [Google Scholar] [CrossRef]
- Brown, L.; Leck, A.K.; Gichangi, M.; Burton, M.J.; Denning, D.W. The global incidence and diagnosis of fungal keratitis. Lancet Infect. Dis. 2020, 21, e49–e57. [Google Scholar] [CrossRef]
- Zemba, M.; Dumitrescu, O.-M.; Dimirache, A.-E.; Branisteanu, D.C.; Balta, F.; Burcea, M.; Moraru, A.D.; Gradinaru, S. Diagnostic methods for the etiological assessment of infectious corneal pathology (Review). Exp. Ther. Med. 2022, 23, 137. [Google Scholar] [CrossRef]
- Thomas, P.A.; Kaliamurthy, J. Mycotic keratitis: Epidemiology, diagnosis and management. Clin. Microbiol. Infect. 2013, 19, 210–220. [Google Scholar] [CrossRef] [PubMed]
- Bakken, I.M.; Jackson, C.J.; Utheim, T.P.; Villani, E.; Hamrah, P.; Kheirkhah, A.; Nielsen, E.; Hau, S.; Lagali, N.S. The use of in vivo confocal microscopy in fungal keratitis—Progress and challenges. Ocul. Surf. 2022, 24, 103–118. [Google Scholar] [CrossRef]
- Ting, D.S.J.; Cairns, J.; Gopal, B.P.; Ho, C.S.; Krstic, L.; Elsahn, A.; Lister, M.; Said, D.G.; Dua, H.S. Risk Factors, Clinical Outcomes, and Prognostic Factors of Bacterial Keratitis: The Nottingham Infectious Keratitis Study. Front. Med. 2021, 8, 715118. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, J.J.; Dart, J.K.G.; De, S.K.; Carnt, N.; Cleary, G.; Hau, S. Comparison of culture, confocal microscopy and PCR in routine hospital use for microbial keratitis diagnosis. Eye 2022, 36, 2172–2178. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.E.; Tepelus, T.C.; Vickers, L.A.; Baghdasaryan, E.; Gui, W.; Huang, P.; Irvine, J.A.; Sadda, S.; Hsu, H.Y.; Lee, O.L. Role of in vivo confocal microscopy in the diagnosis of infectious keratitis. Int. Ophthalmol. 2019, 39, 2865–2874. [Google Scholar] [CrossRef] [PubMed]
- Curro-Tafili, K.; Verbraak, F.D.; de Vries, R.; van Nispen, R.M.A.; Ghyczy, E.A.E. Diagnosing and monitoring the characteristics of Acanthamoeba keratitis using slit scanning and laser scanning in vivo confocal microscopy. Ophthalmic Physiol. Opt. 2023, 44, 131–152. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, X.; Wei, Z.; Cao, K.; Zhang, Z.; Liang, Q. The global epidemiology and clinical diagnosis of Acanthamoeba keratitis. J. Infect. Public Health 2023, 16, 841–852. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Bian, J.; Wang, Y.; Wang, S.; Wang, X.; Shi, W. Clinical features and serial changes of Acanthamoeba keratitis: An in vivo confocal microscopy study. Eye 2019, 34, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Koganti, R.; Yadavalli, T.; Naqvi, R.A.; Shukla, D.; Naqvi, A.R. Pathobiology and treatment of viral keratitis. Exp. Eye Res. 2021, 205, 108483. [Google Scholar] [CrossRef]
- Chaloulis, S.K.; Mousteris, G.; Tsaousis, K.T. Incidence and Risk Factors of Bilateral Herpetic Keratitis: 2022 Update. Trop. Med. Infect. Dis. 2022, 7, 92. [Google Scholar] [CrossRef]
- Poon, S.H.L.; Wong, W.H.L.; Lo, A.C.Y.; Yuan, H.; Chen, C.-F.; Jhanji, V.; Chan, Y.K.; Shih, K.C. A systematic review on advances in diagnostics for herpes simplex keratitis. Surv. Oph-Thalmol. 2021, 66, 514–530. [Google Scholar] [CrossRef]
- Mok, E.; Kam, K.W.; Young, A.L. Corneal nerve changes in herpes zoster ophthalmicus: A prospective longitudinal in vivo confocal microscopy study. Eye 2023, 37, 3033–3040. [Google Scholar] [CrossRef]
- Mangan, M.S.; Yildiz-Tas, A.; Yildiz, M.B.; Yildiz, E.; Sahin, A. In Vivo confocal microscopy findings after COVID-19 infection. Ocul. Immunol. Inflamm. 2021, 30, 1866–1868. [Google Scholar] [CrossRef]
- Subaşı, S.; Yüksel, N.; Toprak, M.; Tuğan, B.Y. In Vivo Confocal Microscopy Analysis of the Corneal Layers in Adenoviral Epidemic Keratoconjunctivitis. Turk. J. Ophthalmol. 2018, 48, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Kalpathy-Cramer, J.; Patel, J.B.; Bridge, C.; Chang, K. Basic Artificial Intelligence Techniques. Radiol. Clin. North Am. 2021, 59, 941–954. [Google Scholar] [CrossRef] [PubMed]
- Lincke, A.; Roth, J.; Macedo, A.F.; Bergman, P.; Löwe, W.; Lagali, N.S. AI-Based Decision-Support System for Diagnosing Acanthamoeba Keratitis Using In Vivo Confocal Microscopy Images. Transl. Vis. Sci. Technol. 2023, 12, 29. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Tao, Y.; Qiu, Q.; Wu, X. Application of image recognition-based automatic hyphae detection in fungal keratitis. Australas. Phys. Eng. Sci. Med. 2018, 41, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Soltoggio, A.; Lock, R.; Carter, S. A Structure-Aware Convolutional Neural Network for Automatic Diagnosis of Fungal Keratitis with In Vivo Confocal Microscopy Images. Neural Netw. 2019, 109, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Zhang, K.; Chen, Q.; Chen, Q.; Huang, W.; Cui, L.; Li, M.; Li, J.; Chen, L.; Shen, C.; et al. Deep learning-based automated diagnosis of fungal keratitis with in vivo confocal microscopy images. Ann. Transl. Med. 2020, 8, 706. [Google Scholar] [CrossRef] [PubMed]
- Alam, U.; Anson, M.; Meng, Y.; Preston, F.; Kirthi, V.; Jackson, T.L.; Nderitu, P.; Cuthbertson, D.J.; Malik, R.A.; Zheng, Y.; et al. Artificial Intelligence and Corneal Confocal Microscopy: The Start of a Beautiful Relationship. J. Clin. Med. 2022, 11, 6199. [Google Scholar] [CrossRef]
- Xu, F.; Jiang, L.; He, W.; Huang, G.; Hong, Y.; Tang, F.; Lv, J.; Lin, Y.; Qin, Y.; Lan, R.; et al. The Clinical Value of Explainable Deep Learning for Diagnosing Fungal Keratitis Using in vivo Confocal Microscopy Images. Front. Med. 2021, 8, 797616. [Google Scholar] [CrossRef]
- Tang, N.; Huang, G.; Lei, D.; Jiang, L.; Chen, Q.; He, W.; Tang, F.; Hong, Y.; Lv, J.; Qin, Y.; et al. An artificial intelligence approach to classify pathogenic fungal genera of fungal keratitis using corneal confocal microscopy images. Int. Ophthalmol. 2023, 43, 2203–2214. [Google Scholar] [CrossRef]
- Zhuang, F.; Qi, Z.; Duan, K.; Xi, D.; Zhu, Y.; Zhu, H.; Xiong, H.; He, Q. A Comprehensive Survey on Transfer Learning. Proc. IEEE 2021, 109, 43–76. [Google Scholar] [CrossRef]
- Tang, N.; Huang, G.; Lei, D.; Jiang, L.; Chen, Q.; He, W.; Tang, F.; Hong, Y.; Lv, J.; Qin, Y.; et al. A Hybrid System for Automatic Identification of Corneal Layers on In Vivo Confocal Microscopy Images. Transl. Vis. Sci. Technol. 2023, 12, 8. [Google Scholar] [CrossRef]
- Almasri, M.M.; Alajlan, A.M. Artificial Intelligence-Based Multimodal Medical Image Fusion Using Hybrid S2 Optimal CNN. Electronics 2022, 11, 2124. [Google Scholar] [CrossRef]
- Xu, F.; Qin, Y.; He, W.; Huang, G.; Lv, J.; Xie, X.; Diao, C.; Tang, F.; Jiang, L.; Lan, R.; et al. A deep transfer learning framework for the automated assessment of corneal inflammation on in vivo confocal microscopy images. PLoS ONE 2021, 16, e0252653. [Google Scholar] [CrossRef]
- Yan, Y.; Jiang, W.; Zhou, Y.; Yu, Y.; Huang, L.; Wan, S.; Zheng, H.; Tian, M.; Wu, H.; Huang, L.; et al. Evaluation of a computer-aided diagnostic model for corneal diseases by analyzing in vivo confocal microscopy images. Front. Med. 2023, 10, 1164188. [Google Scholar] [CrossRef]
- Akpek, E.K.; Bunya, V.Y.; Saldanha, I.J. Sjögren’s Syndrome: More Than Just Dry Eye. Cornea 2019, 38, 658–661. [Google Scholar] [CrossRef]
- Caban, M.; Omulecki, W.; Latecka-Krajewska, B. Dry eye in Sjögren’s syndrome—Characteristics and therapy. Eur. J. Ophthalmol. 2022, 32, 3174–3184. [Google Scholar] [CrossRef]
- Yu, K.; Bunya, V.; Maguire, M.; Asbell, P.; Ying, G.S. Dry Eye Assessment and Management Study Research Group. Systemic Conditions Associated with Severity of Dry Eye Signs and Symptoms in the Dry Eye Assessment and Management Study. Ophthalmology. 2021, 128, 1384–1392. [Google Scholar] [CrossRef]
- Sikder, S.; Gire, A.; Selter, J.H. The relationship between Graves’ ophthalmopathy and dry eye syndrome. Clin. Ophthalmol. 2014, 9, 57–62. [Google Scholar] [CrossRef][Green Version]
- Shetty, R.; Dua, H.S.; Tong, L.; Kundu, G.; Khamar, P.; Gorimanipalli, B.; D’souza, S. Role of in vivo confocal microscopy in dry eye disease and eye pain. Indian J. Ophthalmol. 2023, 71, 1099–1104. [Google Scholar] [CrossRef]
- Stan, C.; Diaconu, E.; Hopirca, L.; Petra, N.; Rednic, A.; Stan, C. Ocular cicatricial pemphigoid. Rom. J. Ophthalmol. 2020, 64, 226–230. [Google Scholar] [CrossRef]
- Sobolewska, B.; Schaller, M.; Zierhut, M. Rosacea and Dry Eye Disease. Ocul. Immunol. Inflamm. 2022, 30, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Ungureanu, L.; Chaudhuri, K.R.; Diaconu, S.; Falup-Pecurariu, C. Dry eye in Parkinson’s disease: A narrative review. Front. Neurol. 2023, 14, 1236366. [Google Scholar] [CrossRef] [PubMed]
- Tiemstra, J.D.; Khatkhate, N. Bell’s palsy: Diagnosis and management. Am. Fam. Physician 2007, 76, 997–1002. [Google Scholar] [PubMed]
- Sambhi, R.-D.S.; Mather, R.; Malvankar-Mehta, M.S. Dry eye after refractive surgery: A meta-analysis. Can. J. Ophthalmol. 2020, 55, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Kato, K.; Miyake, K.; Hirano, K.; Kondo, M. Management of Postoperative Inflammation and Dry Eye After Cataract Surgery. Cornea 2019, 38, S25–S33. [Google Scholar] [CrossRef] [PubMed]
- Tariq, M.; Amin, H.; Ahmed, B.; Ali, U.; Mohiuddin, A. Association of dry eye disease with smoking: A systematic review and meta-analysis. Indian J. Ophthalmol. 2022, 70, 1892–1904. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, Y.; Ibrahim, O.M.A. Application of In Vivo Confocal Microscopy in Dry Eye Disease. Investig. Opthalmology Vis. Sci. 2018, 59, DES41–DES47. [Google Scholar] [CrossRef]
- Wei, S.; Shi, F.; Wang, Y.; Chou, Y.; Li, X. A Deep Learning Model for Automated Sub-Basal Corneal Nerve Segmentation and Evaluation Using In Vivo Confocal Microscopy. Transl. Vis. Sci. Technol. 2020, 9, 32. [Google Scholar] [CrossRef]
- Jing, D.; Liu, Y.; Chou, Y.; Jiang, X.; Ren, X.; Yang, L.; Su, J.; Li, X. Change patterns in the corneal sub-basal nerve and corneal aberrations in patients with dry eye disease: An artificial intelligence analysis. Exp. Eye Res. 2021, 215, 108851. [Google Scholar] [CrossRef]
- Jing, D.; Jiang, X.; Chou, Y.; Wei, S.; Hao, R.; Su, J.; Li, X. In vivo Confocal Microscopic Evaluation of Previously Neglected Oval Cells in Corneal Nerve Vortex: An Inflammatory Indicator of Dry Eye Disease. Front. Med. 2022, 9, 906219. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.C.B.; Tran, V.; Wolffsohn, J.S. The impact of dry eye disease on corneal nerve parameters: A systematic review and meta-analysis. Ophthalmic Physiol. Opt. 2023, 43, 1079–1091. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Lin, Z.-X.; Yang, H.-Q.; Zhao, L.; Liu, D.-C.; Pan, Z.-Q. Changes in corneal nerve morphology and function in patients with dry eyes having type 2 diabetes. World J. Clin. Cases 2022, 10, 3014–3026. [Google Scholar] [CrossRef] [PubMed]
- Kundu, G.; Shetty, R.; D’souza, S.; Khamar, P.; Nuijts, R.M.M.A.; Sethu, S.; Roy, A.S. A novel combination of corneal confocal microscopy, clinical features and artificial intelligence for evaluation of ocular surface pain. PLoS ONE 2022, 17, e0277086. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhang, J.; Pereira, E.; Zheng, Y.; Su, P.; Xie, J.; Zhao, Y.; Shi, Y.; Qi, H.; Liu, J.; et al. Automated Tortuosity Analysis of Nerve Fibers in Corneal Confocal Microscopy. IEEE Trans. Med. Imaging 2020, 39, 2725–2737. [Google Scholar] [CrossRef] [PubMed]
- Lecca, M.; Gianini, G.; Serapioni, R.P. Mathematical insights into the Original Retinex Algorithm for Image Enhancement. J. Opt. Soc. Am. A 2022, 39, 2063–2072. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Xie, J.; Yang, T.; Su, P.; Liu, R.; Sun, T.; Zhou, Y.; Wang, H.; Feng, X.; Ma, S.; et al. Quantification of Increased Corneal Subbasal Nerve Tortuosity in Dry Eye Disease and Its Correlation With Clinical Parameters. Transl. Vis. Sci. Technol. 2021, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Fernández, I.; Vázquez, A.; Calonge, M.; Maldonado, M.J.; de la Mata, A.; López-Miguel, A. New Method for the Au-tomated Assessment of Corneal Nerve Tortuosity Using Confocal Microscopy Imaging. Appl. Sci. 2022, 12, 10450. [Google Scholar] [CrossRef]
- Zhang, Y.-Y.; Zhao, H.; Lin, J.-Y.; Wu, S.-N.; Liu, X.-W.; Zhang, H.-D.; Shao, Y.; Yang, W.-F. Artificial Intelligence to Detect Meibomian Gland Dysfunction From in-vivo Laser Confocal Microscopy. Front. Med. 2021, 8, 774344. [Google Scholar] [CrossRef]
- Maruoka, S.; Tabuchi, H.; Nagasato, D.; Masumoto, H.; Chikama, T.; Kawai, A.C.; Oishi, N.C.; Maruyama, T.; Kato, Y.; Hayashi, T.; et al. Deep Neural Network-Based Method for Detecting Obstructive Meibomian Gland Dysfunction With in Vivo Laser Confocal Microscopy. Cornea 2020, 39, 720–725. [Google Scholar] [CrossRef]
- Sim, R.; Yong, K.; Liu, Y.-C.; Tong, L. In Vivo Confocal Microscopy in Different Types of Dry Eye and Meibomian Gland Dysfunction. J. Clin. Med. 2022, 11, 2349. [Google Scholar] [CrossRef] [PubMed]
- Levine, H.; Tovar, A.; Cohen, A.K.; Cabrera, K.; Locatelli, E.; Galor, A.; Feuer, W.; O’Brien, R.; Goldhagen, B.E. Automated identification and quantification of activated dendritic cells in central cornea using artificial intelligence. Ocul. Surf. 2023, 29, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Setu, A.K.; Schmidt, S.; Musial, G.; Stern, M.E.; Steven, P. Segmentation and Evaluation of Corneal Nerves and Dendritic Cells From In Vivo Confocal Microscopy Images Using Deep Learning. Transl. Vis. Sci. Technol. 2022, 11, 24. [Google Scholar] [CrossRef] [PubMed]
- So, W.Z.; Wong, N.S.Q.; Tan, H.C.; Lin, M.T.Y.; Lee, I.X.Y.; Mehta, J.S.; Liu, Y.-C. Diabetic corneal neuropathy as a surrogate marker for diabetic peripheral neuropathy. Neural Regen. Res. 2022, 17, 2172–2178. [Google Scholar] [PubMed]
- Mansoor, H.; Tan, H.C.; Lin, M.T.-Y.; Mehta, J.S.; Liu, Y.-C. Diabetic Corneal Neuropathy. J. Clin. Med. 2020, 9, 3956. [Google Scholar] [CrossRef] [PubMed]
- Dabbah, M.A.; Graham, J.; Petropoulos, I.; Tavakoli, M.; Malik, R.A. Dual-model automatic detection of nerve-fibres in corneal confocal microscopy images. Med. Image Comput. Comput. Assist Interv. 2010, 13 Pt 1, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Dabbah, M.; Graham, J.; Petropoulos, I.; Tavakoli, M.; Malik, R. Automatic analysis of diabetic peripheral neuropathy using multi-scale quantitative morphology of nerve fibres in corneal confocal microscopy imaging. Med. Image Anal. 2011, 15, 738–747. [Google Scholar] [CrossRef]
- Petropoulos, I.N.; Alam, U.; Fadavi, H.; Marshall, A.; Asghar, O.; Dabbah, M.A.; Chen, X.; Graham, J.; Ponirakis, G.; Boulton, A.J.M.; et al. Rapid Automated Diagnosis of Diabetic Peripheral Neuropathy With In Vivo Corneal Confocal Microscopy. Investig. Opthalmology Vis. Sci. 2014, 55, 2071–2078. [Google Scholar] [CrossRef]
- Chen, X.; Graham, J.; Dabbah, M.A.; Petropoulos, I.N.; Tavakoli, M.; Malik, R.A. An Automatic Tool for Quantification of Nerve Fibers in Corneal Confocal Microscopy Images. IEEE Trans. Biomed. Eng. 2017, 64, 786–794. [Google Scholar] [CrossRef]
- Chen, X.; Graham, J.; Petropoulos, I.N.; Ponirakis, G.; Asghar, O.; Alam, U.; Marshall, A.; Ferdousi, M.; Azmi, S.; Efron, N.; et al. Corneal Nerve Fractal Dimension: A Novel Corneal Nerve Metric for the Diagnosis of Diabetic Sensorimotor Polyneuropathy. Investig. Opthalmology Vis. Sci. 2018, 59, 1113–1118. [Google Scholar] [CrossRef]
- Tang, W.; Chen, X.; Yuan, J.; Meng, Q.; Shi, F.; Xiang, D.; Chen, Z.; Zhu, W. Multi-scale and local feature guidance network for corneal nerve fiber segmentation. Phys. Med. Biol. 2023, 68, 095026. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Chen, C.; Yun, J.; Sun, Y.; Tian, J.; Hao, Z.; Yu, H.; Ma, H. Multi-Scale Feature Fusion Convolutional Neural Network for Indoor Small Target Detection. Front. Neurorobotics 2022, 16, 881021. [Google Scholar] [CrossRef] [PubMed]
- Salahouddin, T.; Petropoulos, I.N.; Ferdousi, M.; Ponirakis, G.; Asghar, O.; Alam, U.; Kamran, S.; Mahfoud, Z.R.; Efron, N.; Malik, R.A.; et al. Artificial Intelligence-Based Classification of Diabetic Peripheral Neuropathy From Corneal Confocal Mi-croscopy Images. Diabetes Care 2021, 44, e151–e153. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Preston, F.G.; Ferdousi, M.; Azmi, S.; Petropoulos, I.N.; Kaye, S.; Malik, R.A.; Alam, U.; Zheng, Y. Artificial Intelligence Based Analysis of Corneal Confocal Microscopy Images for Diagnosing Peripheral Neuropathy: A Binary Classification Model. J. Clin. Med. 2023, 12, 1284. [Google Scholar] [CrossRef] [PubMed]
- Preston, F.G.; Meng, Y.; Burgess, J.; Ferdousi, M.; Azmi, S.; Petropoulos, I.N.; Kaye, S.; Malik, R.A.; Zheng, Y.; Alam, U. Artificial intelligence utilising corneal confocal microscopy for the diagnosis of peripheral neuropathy in diabetes mellitus and prediabetes. Diabetologia 2022, 65, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.M.; Borroni, D.; Liu, R.; Zhao, Y.; Zhang, J.; Lim, J.; Ma, B.; Romano, V.; Qi, H.; Ferdousi, M.; et al. An artificial intelligence-based deep learning algorithm for the diagnosis of diabetic neuropathy using corneal confocal microscopy: A development and validation study. Diabetologia 2019, 63, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, E.; Arslan, A.T.; Tas, A.Y.; Acer, A.F.; Demir, S.; Sahin, A.; Barkana, D.E. Generative Adversarial Network Based Automatic Segmentation of Corneal Subbasal Nerves on In Vivo Confocal Microscopy Images. Transl. Vis. Sci. Technol. 2021, 10, 33. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Li, T.; Li, F.; Zhang, C. NerveStitcher: Corneal confocal microscope images stitching with neural networks. Comput. Biol. Med. 2022, 151, 106303. [Google Scholar] [CrossRef]
- Elbita, A.; Qahwaji, R.; Ipson, S.; Sharif, M.S.; Ghanchi, F. Preparation of 2D sequences of corneal images for 3D model building. Comput. Methods Programs Biomed. 2014, 114, 194–205. [Google Scholar] [CrossRef][Green Version]

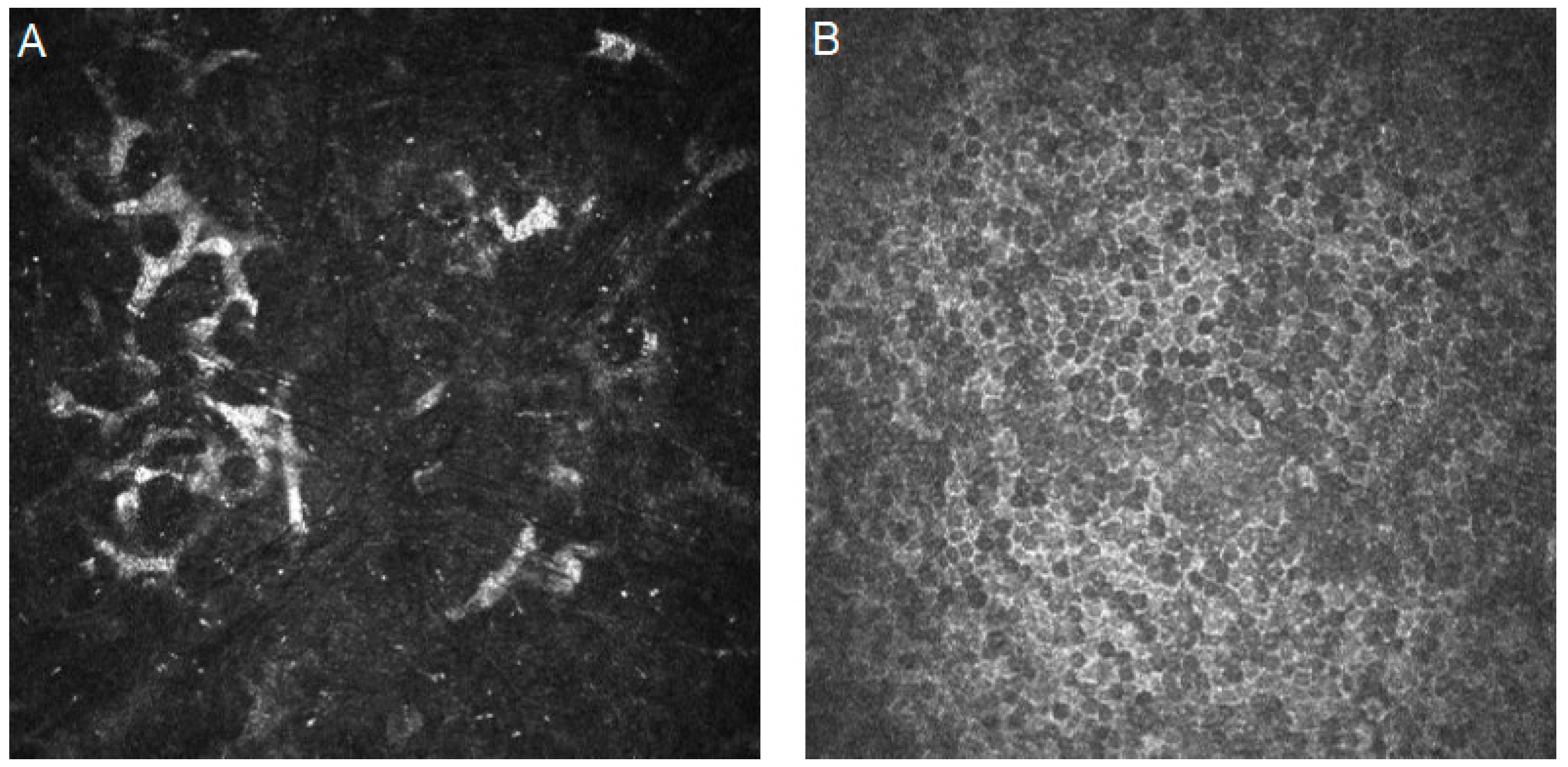
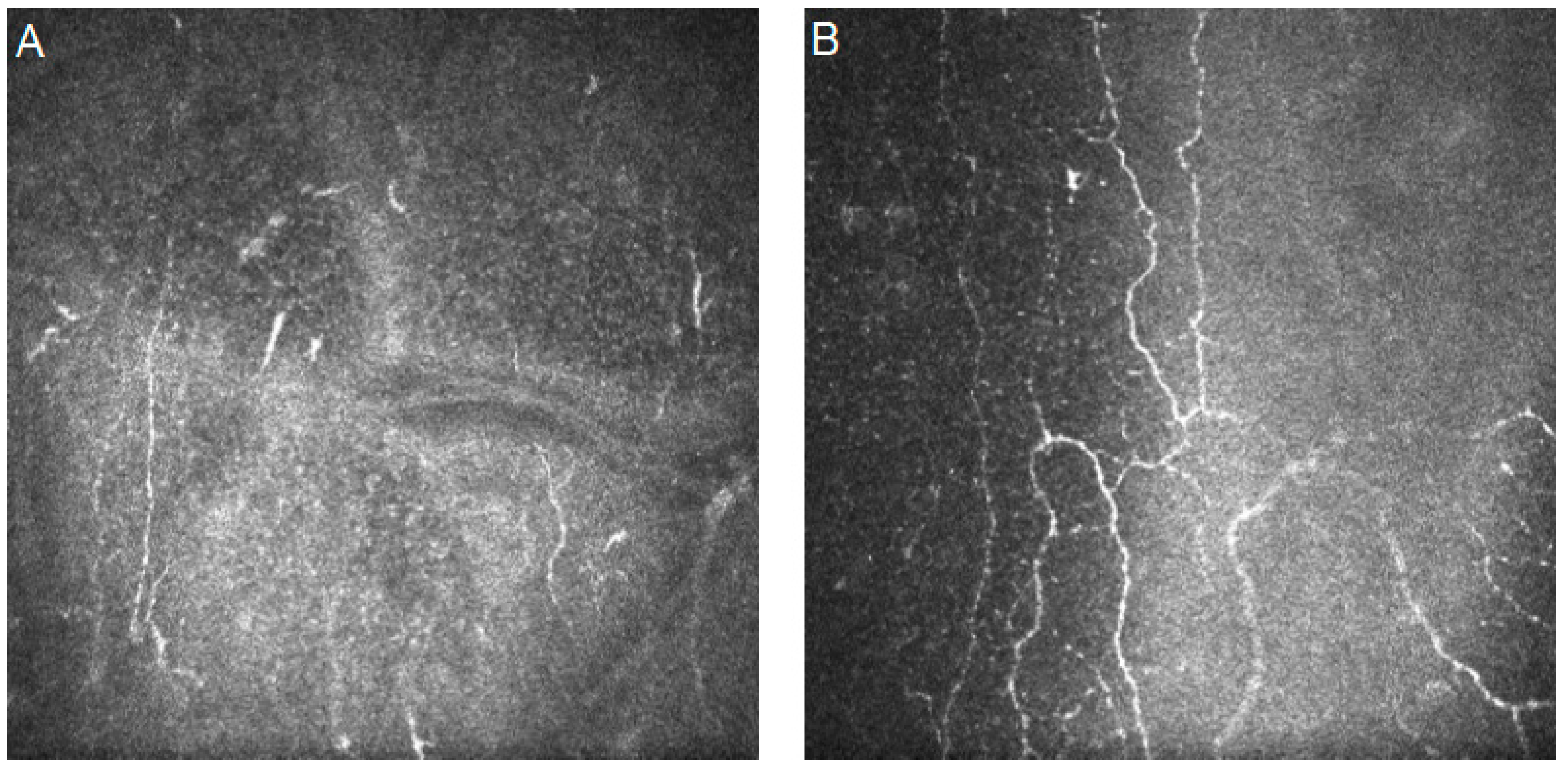
| Authors | Year | Dataset | Artificial Intelligence Method | Results | Additional Techniques and Novelties | |
|---|---|---|---|---|---|---|
| Essalat et al. [28] | 2023 | 4001 images | CNN—Densenet161 | Accuracy 93.55% Precision 92.52% Recall 94.77% F1 score 96.93% | Saliency maps. | |
| Alisa Lincke et al. [63]. | 2023 | 68,970 images | CNN—ResNet101V2 | Healthy/diseased—95% accuracy | Transfer learning. | |
| Xuelian Wu et al. [64] | 2017 | 82 patients | Adaptive robust binary pattern | The accuracy of the model was superior to the corneal smear examination (p < 0.05) approach. | Support vector machine. | |
| Sensitivity 89.29% | ||||||
| Specificity 95.65% | ||||||
| AUC 0.946 | ||||||
| Shanshan Liang et al. [55] | 2023 | 7278 images | SACNN—GoogLeNet and VGGNet | Accuracy 97.73% | Two-stream convolutional network. | |
| Precision 98.68% | ||||||
| Sensitivity 97.02% | ||||||
| Specificity 98.54%, | ||||||
| F1 score 97.84% | ||||||
| Jian Lv et al. [66] | 2020 | 2088 images | CNN—ResNet | Accuracy 96.26% | ||
| Specificity 98.34% | ||||||
| Sensitivity 91.86% | ||||||
| AUC 0.9875 | ||||||
| Jian Lv et al. [67] | 2021 | 1089 images | CNN—ResNet | Accuracy 96.5% | Grad-CAM and guided Grad-CAM to generate explanation maps and pixel explanations. | |
| Sensitivity 93.6% | ||||||
| Specificity 98.2% | ||||||
| AUC 0.983 | ||||||
| Ningning Tang et al. [71] | 2023 | 3364 images | CNN—ResNet | Fusarium | Aspergillus |
Decision tree classifier and CNN-based classifier Grad-CAM and guided Grad-CAM to generate explanation maps and pixel explanation. |
| AUC 0.887 | AUC 0.827 | |||||
| Ningning Tang et al. [69] | 2023 | 7957 images |
CNN—Inception-ResNet V2 and K nearest neighbor | Precision 90.96% | Two classifiers (CNN- and KNN-based) and two hybrid strategies (weighted voting method and LightGBM) were used to fuse the results. | |
| Recall 91.45% | ||||||
| F1 score 91.11% | ||||||
| AUC 0.9841 | ||||||
| Liu Zhi et al. [29] | 2020 | 1213 images | CNN—AlexNet and VGGNet | Accuracy 99.95% |
Sub-area contrast stretching algorithm and histogram matching fusion algorithm. | |
| Sensitivity 99.90% | ||||||
| Specificity 100% | ||||||
| Fan Xu et al. [68] | 2021 | 3453 images | CNN—Inception-ResNet V2 | Activated dendritic cells | Inflammatory cells | Transfer learning technique. |
| Accuracy 93.19% | Accuracy 97.67% | |||||
| Sensitivity 81.71% | Sensitivity 91.74% | |||||
| Specificity 95.17% | Specificity 99.31% | |||||
| G mean 88.72% | G mean 95.45% | |||||
| AUC 0.9646 | AUC 0.9901 | |||||
| Authors | Year | Dataset | Artificial Intelligence Method | Results | Additional Techniques and Novelties | |
|---|---|---|---|---|---|---|
| Yulin Yan et al. [74] | 2023 | 19,612 images | CNN–ResNet50 |
Internal test: Accuracy 91.4%, 95.7%, 96.7%, and 95% for the recognition of each layer. Accuracy 96.1%, 93.2%, 94.5%, and 95.9% for normal/abnormal images recognition (for each layer). | ||
|
External test: Accuracy 96.0%, 96.5%, 96.6%, and 96.4% for the recognition of each layer. Accuracy 98.3%, 97.2%, 94.0%, and 98.2% for normal/abnormal image recognition (for each layer). | ||||||
| Shanshan Wei et al. [88] | 2020 | 5221 images | CNN—ResNet34 | AUC 0.96 | CNS-Net established. | |
| Dalan Jing et al. [90] | 2022 | ~2290 images | CNN—CNS-Net | The corneal nerve morphology (the average density and maximum length) were significantly correlated with the corneal intrinsic aberrations. | The corneal sub-basal nerve morphology and corneal intrinsic aberrations were investigated with CNS-Net. | |
| Gairik Kundu et al. [93] | 2022 | 120 images | CCMetrics for nerve fiber characteristics and Random Forest classifier |
AUC 0.736 Accuracy 86% F1 score 85.9% Precision 85.6% Recall 86.3% | Correlation investigation was conducted between the various clinical symptoms and imaging parameters of ocular surface pain. | |
| Yitian Zhao et al. [94] | 2020 | 322 images | CS-NET The infinite perimeter active contour with hybrid region | Accuracy 81.8% for the first dataset. Accuracy 87.5% for the second dataset. | A Retinex model advanced exponential curvature estimation method with a linear support vector machine. | |
| Baikai Ma et al. [96] | 2021 | 1501 images | kNN-DOWA The infinite perimeter active contour with hybrid region information |
The tortuosity was higher in patients with DED than in healthy volunteers (
p
< 0.001). The tortuosity was positively correlated with the ocular surface disease index (
r
= 0.418,
p
= 0.003) and negatively correlated with tear breakup time (
r
= −0.398 and
p
= 0.007). No correlation was found between the tortuosity and visual analog scale scores, corneal fluorescein staining scores, or the Schirmer I test. | ||
| Fernandez et al. [97] | 2022 | 43 images | Watershed algorithm |
The tortuosity index was significantly higher in post-LASIK patients with ocular pain than in the control patients. No significant differences were detected with manual measurements. The tortuosity quantification was positively correlated with the ocular surface disease index (OSDI) and a numeric rating scale (NRS) assessing pain. | ||
| Ye-Ye Zhang et al. [98] | 2021 | 8311 images | CNN—DenseNet169 | OMGD | AMGD | |
|
AUC 97.3% Sensitivity 88.8% Specificity 95.4% |
AUC 98.6% Sensitivity 89.4% Specificity 98.4% | |||||
| Sachiko Maruoka et al. [99] | 2020 | 380 images | CNNs—DenseNet-201, VGG16, DenseNet-169, and InceptionV3 | The single DL model: AUC 0.966 Sensitivity 94.2% Specificity 82.1% The ensemble DL model (VGG16 + DenseNet-169 + DenseNet-201 + InceptionV3) AUC 0.981 Sensitivity 92.1% Specificity 98.8% | Transfer learning. | |
| Harry Levine et. al. [101] | 2023 | 173 images | CNNs—CSPDarknet53 and YOLOv3 |
The mean number of aDCs in the central cornea were quantified automatically: 0.83 ± 1.33 cells/image. The mean number of aDCs in the central cornea were quantified manually: 1.03 ± 1.65 cells/image. | Transfer learning. | |
| Md Asif Khan Setu et al. [102] | 2022 | 1219 images | CNN—U-Net and Mask R-CNN | The CNFs model | The DCs model | |
|
Sensitivity 86.1% Specificity 90.1% | Precision 89.37% Recall 94.43% F1 score 91.83% | |||||
| Authors | Year | Dataset | Artificial Intelligence Method | Results | Additional Techniques and Novelties | |||
|---|---|---|---|---|---|---|---|---|
| Dabbah et al. [105] | 2010 | 525 images | 2D Gabor wavelet and a Gaussian envelope | The automatic analysis is consistent with the manual analysis at a correlation of (r = 0.92). | ||||
| Dabbah et al. [106] | 2011 | 521 images | 2D Gabor wavelet and a Gaussian envelope | The model had the lowest equal error rate of 15.44%. | ||||
| Ioannis N. Petropoulos et al. [107] | 2014 | 186 patients | 2D Gabor wavelet and a Gaussian envelope |
The manual and automated analysis methods were highly correlated for the following: CNFD (r = 0.9, p < 0.0001) CNFL (r = 0.89, p < 0.0001) CNBD (r = 0.75, p < 0.0001) | ||||
| Xin Chen et al. [108] | 2017 | 888 images | 2D Gabor wavelet and a Gaussian envelope with dual-tree complex wavelet transforms |
Nerve fiber detection: Sensitivity 91.7% Specificity 91.3% | ||||
| Xin Chen et al. [109] | 2018 | 176 patients | 2D Gabor wavelet and a Gaussian envelope with dual-tree complex wavelet transforms |
The AUC for identifying DSPN were comparable: 0.77 for automated CNFD 0.74 for automated CNFL 0.69 for automated CNBD 0.74 for automated ACNFrD. | ||||
| Wei Tang et al. [110] | 2023 | 524 images | CNN—MLFGNet | Dice coefficients were 89.33%, 89.41%, and 88.29%. | A multiscale progressive guidance module, a local feature-guided attention module, and a multiscale deep supervision module. | |||
| Tooba Salahouddin et al. [112] | 2021 | 108 patients | CNN—U-Net |
DPN from the control subjects: AUC 0.86 Sensitivity 84% Specificity 71% | ||||
|
DPN from the DPN+: AUC 0.95 Sensitivity 92% Specificity 80% | ||||||||
|
Control subjects from the DPN+: AUC 1.0 Sensitivity 100% Specificity 95% | ||||||||
| Yanda Meng et al. [113] | 2023 | 279 patients | CNN—ResNet50 |
Sensitivity 91% Specificity 93% AUC 0.95 | Grad-CAM and guided Grad-CAM to generate explanation maps and pixel explanations. | |||
| Yanda Meng et al. [114] | 2022 | 228 patients | CNN—ResNet50 | HV | PN− | PN+ | Grad-CAM and guided Grad-CAM to generate explanation maps and pixel explanations.Occlusion sensitivity. | |
|
Recall 100% Precision 83% F1 score 91% |
Recall 85% Precision 92% F1 score 88% | Recall 83% Precision 100% F1 score 91% | ||||||
| Williams et al. [115] | 2020 | 1698 images | CNN—U-Net |
Intraclass correlations: Total corneal nerve fiber length 0.933 Mean length per segment 0.656 Number of branch points 0.891 | ||||
| Erdost Yıldız et al. [116] | 2021 | 510 images | CNN—U-Net and GAN | U-Net | GAN | |||
| AUC 0.8934 | AUC 0.9439 | |||||||
| Guangxu Li et al. [117] | 2022 | 30 images sets | CNN—VGGNet | The stitching method can evaluate the corneal nerve of patients more accurately and reliably compared to a single image. | ||||
| Abdulhakim Elbita et al. [118] | 2014 | 356 images | Back propagation neural network | Accuracy 99.4% | DCT filter, Gaussian smoothing, contrast standardized, and Otsu’s threshold. | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kryszan, K.; Wylęgała, A.; Kijonka, M.; Potrawa, P.; Walasz, M.; Wylęgała, E.; Orzechowska-Wylęgała, B. Artificial-Intelligence-Enhanced Analysis of In Vivo Confocal Microscopy in Corneal Diseases: A Review. Diagnostics 2024, 14, 694. https://doi.org/10.3390/diagnostics14070694
Kryszan K, Wylęgała A, Kijonka M, Potrawa P, Walasz M, Wylęgała E, Orzechowska-Wylęgała B. Artificial-Intelligence-Enhanced Analysis of In Vivo Confocal Microscopy in Corneal Diseases: A Review. Diagnostics. 2024; 14(7):694. https://doi.org/10.3390/diagnostics14070694
Chicago/Turabian StyleKryszan, Katarzyna, Adam Wylęgała, Magdalena Kijonka, Patrycja Potrawa, Mateusz Walasz, Edward Wylęgała, and Bogusława Orzechowska-Wylęgała. 2024. "Artificial-Intelligence-Enhanced Analysis of In Vivo Confocal Microscopy in Corneal Diseases: A Review" Diagnostics 14, no. 7: 694. https://doi.org/10.3390/diagnostics14070694
APA StyleKryszan, K., Wylęgała, A., Kijonka, M., Potrawa, P., Walasz, M., Wylęgała, E., & Orzechowska-Wylęgała, B. (2024). Artificial-Intelligence-Enhanced Analysis of In Vivo Confocal Microscopy in Corneal Diseases: A Review. Diagnostics, 14(7), 694. https://doi.org/10.3390/diagnostics14070694








