Breaking Boundaries in Pneumonia Diagnostics: Transitioning from Tradition to Molecular Frontiers with Multiplex PCR
Abstract
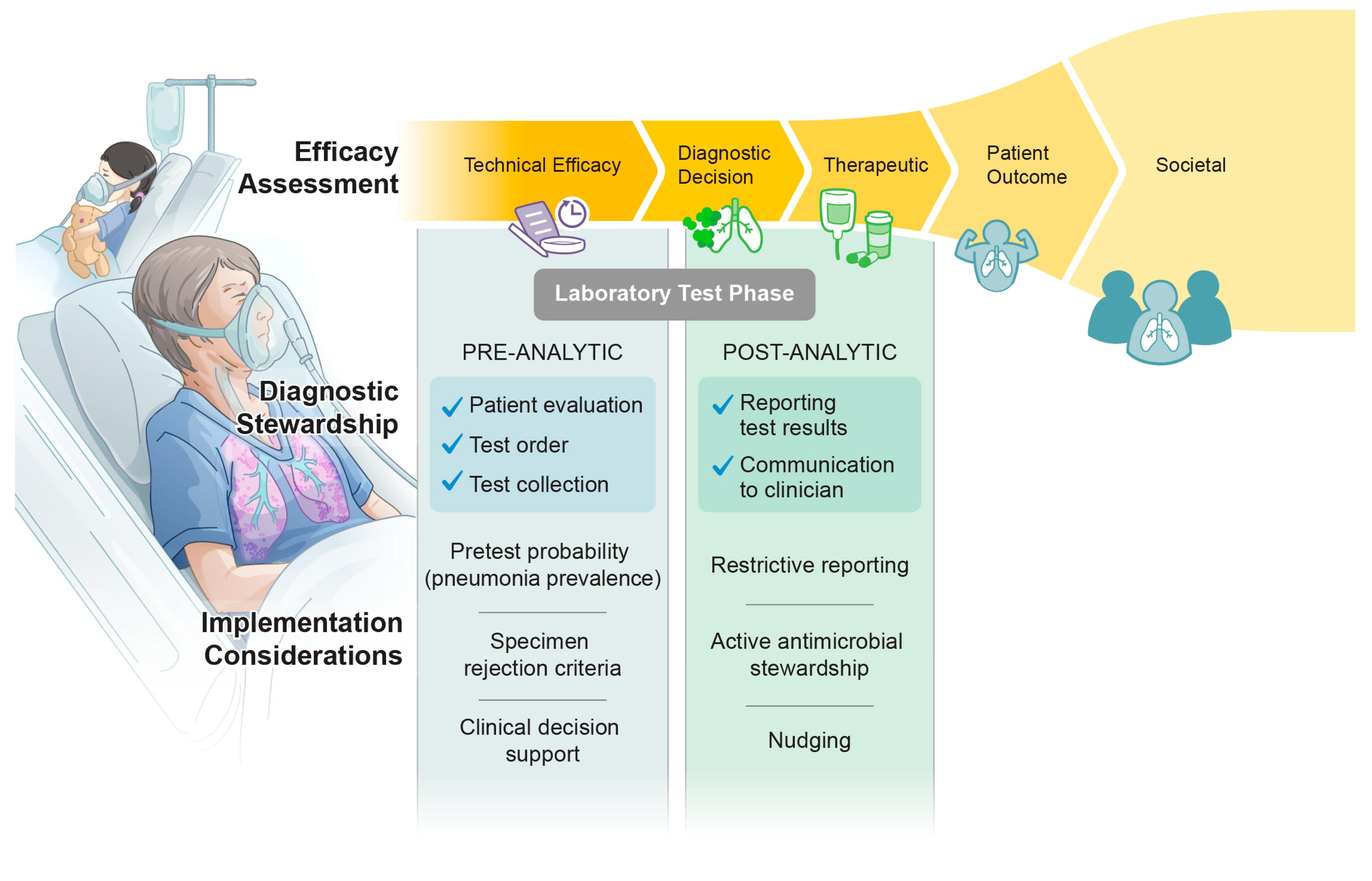
:1. Introduction
2. Performance of mPCR vs. Culture
2.1. mPCR Discordance with Respiratory Culture
2.2. Impact of Antimicrobial Exposure on Diagnostic Yield
2.3. Quantification of Culture and mPCR Results
2.4. The Challenge of a Sub-Optimal Gold Standard
3. Clinical Utility of mPCR
3.1. Clinical Relevance of Additional mPCR Detections
3.2. Potential Impact on Prescribing Behavior and AMS
3.3. Molecular Diagnostics for Pneumonia in Special Populations
3.4. Cost Considerations
4. Guideline Changes, Lessons, and Future Research
5. Current State of Diagnostic Stewardship and Future Directions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hill, B.; Narayanan, N.; Palavecino, E.; Perez, K.K.; Premraj, S.; Streifel, A.; Wrenn, R.H.; Zeitler, K. The Role of an Antimicrobial Stewardship Team in the Use of Rapid Diagnostic Testing in Acute Care: An Official Position Statement of the Society of Infectious Diseases Pharmacists. Infect. Control Hosp. Epidemiol. 2018, 39, 473–475. [Google Scholar] [CrossRef] [PubMed]
- CDC. Core Elements of Hospital Antibiotic Stewardship Programs. US Department of Health and Human Services, CDC. Available online: http://www.cdc.gov/getsmart/healthcare/implementation/core-elements.html (accessed on 2 February 2024).
- Moy, A.C.; Kimmoun, A.; Merkling, T.; Berçot, B.; Caméléna, F.; Poncin, T.; Deniau, B.; Mebazaa, A.; Dudoignon, E.; Dépret, F. Performance Evaluation of a Pcr Panel (Filmarray® Pneumonia Plus) for Detection of Respiratory Bacterial Pathogens in Respiratory Specimens: A Systematic Review and Meta-Analysis. Anaesth. Crit. Care Pain. Med. 2023, 42, 101300. [Google Scholar] [CrossRef] [PubMed]
- Hanson, K.E.; Azar, M.M.; Banerjee, R.; Chou, A.; Colgrove, R.C.; Ginocchio, C.C.; Hayden, M.K.; Holodiny, M.; Jain, S.; Koo, S.; et al. Molecular Testing for Acute Respiratory Tract Infections: Clinical and Diagnostic Recommendations from the Idsa’s Diagnostics Committee. Clin. Infect. Dis. 2020, 71, 2744–2751. [Google Scholar] [CrossRef] [PubMed]
- Fryback, D.G.; Thornbury, J.R. The Efficacy of Diagnostic Imaging. Med. Decis. Mak. 1991, 11, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Buchan, B.W.; Armand-Lefevre, L.; Anderson, N. Molecular Diagnosis of Pneumonia (Including Multiplex Panels). Clin. Chem. 2021, 68, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Enne, V.I.; Aydin, A.; Baldan, R.; Owen, D.R.; Richardson, H.; Ricciardi, F.; Russell, C.; Nomamiukor-Ikeji, B.O.; Swart, A.M.; High, J.; et al. Multicentre Evaluation of Two Multiplex Pcr Platforms for the Rapid Microbiological Investigation of Nosocomial Pneumonia in Uk Icus: The Inhale Wp1 Study. Thorax 2022, 77, 1220–1228. [Google Scholar] [CrossRef] [PubMed]
- Rand, K.H.; Beal, S.G.; Cherabuddi, K.; Couturier, B.; Lingenfelter, B.; Rindlisbacher, C.; Jones, J.; Houck, H.J.; Lessard, K.J.; Tremblay, E.E. Performance of a Semiquantitative Multiplex Bacterial and Viral Pcr Panel Compared with Standard Microbiological Laboratory Results: 396 Patients Studied with the Biofire Pneumonia Panel. Open Forum Infect. Dis. 2021, 8, ofaa560. [Google Scholar] [CrossRef] [PubMed]
- Rabin, E.E.; Walter, J.M.; Wunderink, R.G.; Qi, C.; Pickens, C.I. Clinical Significance of Culture-Negative, Pcr-Positive Bronchoalveolar Lavage Results in Severe Pneumonia. ERJ Open Res. 2023, 9, 00343-2023. [Google Scholar] [CrossRef] [PubMed]
- Buchan, B.W.; Windham, S.; Balada-Llasat, J.M.; Leber, A.; Harrington, A.; Relich, R.; Murphy, C.; Dien Bard, J.; Naccache, S.; Ronen, S.; et al. Practical Comparison of the Biofire Filmarray Pneumonia Panel to Routine Diagnostic Methods and Potential Impact on Antimicrobial Stewardship in Adult Hospitalized Patients with Lower Respiratory Tract Infections. J. Clin. Microbiol. 2020, 58, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Webber, D.M.; Wallace, M.A.; Burnham, C.A.; Anderson, N.W. Evaluation of the BioFire FilmArray Pneumonia Panel for Detection of Viral and Bacterial Pathogens in Lower Respiratory Tract Specimens in the Setting of a Tertiary Care Academic Medical Center. J. Clin. Microbiol. 2020, 58, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Courboules, C.; Dournon, N.; Lawrence, C.; Noussair, L.; Descours, G.; Sivadon-Tardy, V.; Jarraud, S.; Herrmann, J.-L.; Gaillard, J.-L.; Espinasse, F.; et al. Non-Legionella Pneumophila Serogroup 1 Pneumonia: Diagnosis of a Nosocomial Legionellosis with the Biofire Pneumonia Plus Panel. IDCases 2022, 28, e01487. [Google Scholar] [CrossRef] [PubMed]
- Rand, K.H.; Beal, S.G.; Rivera, K.; Allen, B.; Payton, T.; Lipori, G.P. Hourly Effect of Pretreatment with IV Antibiotics on Blood Culture Positivity Rate in Emergency Department Patients. Open Forum Infect. Dis. 2019, 6, ofz179. [Google Scholar] [CrossRef]
- Harris, A.M.; Bramley, A.M.; Jain, S.; Arnold, S.R.; Ampofo, K.; Self, W.H.; Williams, D.J.; Anderson, E.J.; Grijalva, C.G.; McCullers, J.A.; et al. Influence of Antibiotics on the Detection of Bacteria by Culture-Based and Culture-Independent Diagnostic Tests in Patients Hospitalized with Community-Acquired Pneumonia. Open Forum Infect. Dis. 2017, 4, ofx014. [Google Scholar] [CrossRef] [PubMed]
- Fratoni, A.J.; Roberts, A.L.; Nicolau, D.P.; Kuti, J.L. Effects of Clinically Achievable Pulmonary Antibiotic Concentrations on the Recovery of Bacteria: In Vitro Comparison of the Biofire Filmarray Pneumonia Panel Versus Conventional Culture Methods in Bronchoalveolar Lavage Fluid. J. Clin. Microbiol. 2024, 62, e0113323. [Google Scholar] [CrossRef] [PubMed]
- Casadevall, A.; Pirofski, L.A. The Damage-Response Framework of Microbial Pathogenesis. Nat. Rev. Microbiol. 2003, 1, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Falsey, A.R.; Branche, A.R.; Croft, D.P.; Formica, M.A.; Peasley, M.R.; Walsh, E.E. Real-Life Assessment of Biofire Filmarray Pneumonia Panel in Adults Hospitalized with Respiratory Illness. J. Infect. Dis. 2024, 229, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Kyriazopoulou, E.; Karageorgos, A.; Liaskou-Antoniou, L.; Koufargyris, P.; Safarika, A.; Damoraki, G.; Lekakis, V.; Saridaki, M.; Adamis, G.; Giamarellos-Bourboulis, E.J. Biofire® Filmarray® Pneumonia Panel for Severe Lower Respiratory Tract Infections: Subgroup Analysis of a Randomized Clinical Tria. Infect. Dis. Ther. 2021, 10, 1437–1449. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, D.N.; Leggett, J.E.; Wang, L.; Ferdosian, S.; Gelfer, G.D.; Johnston, M.L.; Footer, B.W.; Hendrickson, K.W.; Park, H.S.; White, E.E.; et al. Enhanced Detection of Community-Acquired Pneumonia Pathogens with the Biofire® Pneumonia Filmarray® Panel. Diagn. Microbiol. Infect. Dis. 2021, 99, 115246. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.; Niederman, M.S.; Chastre, J.; Ewig, S.; Fernandez-Vandellos, P.; Hanberger, H.; Kollef, M.; Li Bassi, G.; Luna, C.M.; Martin-Loeches, I.; et al. International Ers/Esicm/Escmid/Alat Guidelines for the Management of Hospital-Acquired Pneumonia and Ventilator-Associated Pneumonia: Guidelines for the Management of Hospital-Acquired Pneumonia (Hap)/Ventilator-Associated Pneumonia (Vap) of the European Respiratory Society (Ers), European Society of Intensive Care Medicine (Esicm), European Society of Clinical Microbiology and Infectious Diseases (Escmid) and Asociación Latinoamericana Del Tórax (Alat). Eur. Respir. J. 2017, 50, 1700582. [Google Scholar] [CrossRef] [PubMed]
- Kalil, A.C.; Metersky, M.L.; Klompas, M.; Muscedere, J.; Sweeney, D.A.; Palmer, L.B.; Napolitano, L.M.; O’Grady, N.P.; Bartlett, J.G.; Carratalà, J.; et al. Management of Adults with Hospital-Acquired and Ventilator-Associated Pneumonia: 2016 Clinical Practice Guidelines by the Infectious Diseases Society of America and the American Thoracic Society. Clin. Infect. Dis. 2016, 63, e61–e111. [Google Scholar] [CrossRef] [PubMed]
- Murphy, C.N.; Fowler, R.; Balada-Llasat, J.M.; Carroll, A.; Stone, H.; Akerele, O.; Buchan, B.; Windham, S.; Hopp, A.; Ronen, S.; et al. Multicenter Evaluation of the BioFire FilmArray Pneumonia/Pneumonia Plus Panel for Detection and Quantification of Agents of Lower Respiratory Tract Infection. J. Clin. Microbiol. 2020, 58. [Google Scholar] [CrossRef] [PubMed]
- Ginocchio, C.C.; Garcia-Mondragon, C.; Mauerhofer, B.; Rindlisbacher, C. Multinational Evaluation of the Biofire® Filmarray® Pneumonia Plus Panel as Compared to Standard of Care Testing. Eur. J. Clin. Microbiol. Infect. Dis. 2021, 40, 1609–1622. [Google Scholar] [CrossRef] [PubMed]
- Gastli, N.; Loubinoux, J.; Daragon, M.; Lavigne, J.P.; Saint-Sardos, P.; Pailhoriès, H.; Lemarié, C.; Benmansour, H.; d’Humières, C.; Broutin, L.; et al. Multicentric Evaluation of Biofire Filmarray Pneumonia Panel for Rapid Bacteriological Documentation of Pneumonia. Clin. Microbiol. Infect. 2021, 27, 1308–1314. [Google Scholar] [CrossRef]
- Ferrer, J.; Clari, M.; Giménez, E.; Carbonell, N.; Torres, I.; Blasco, M.L.; Albert, E.; Navarro, D. The Biofire® Filmarray® Pneumonia Plus Panel for Management of Lower Respiratory Tract Infection in Mechanically-Ventilated Patients in the COVID-19 Era: A Diagnostic and Cost-Benefit Evaluation. Diagn. Microbiol. Infect. Dis. 2023, 105, 115847. [Google Scholar] [CrossRef] [PubMed]
- Posteraro, B.; Cortazzo, V.; Liotti, F.M.; Menchinelli, G.; Ippoliti, C.; De Angelis, G.; La Sorda, M.; Capalbo, G.; Vargas, J.; Antonelli, M.; et al. Diagnosis and Treatment of Bacterial Pneumonia in Critically Ill Patients with COVID-19 Using a Multiplex Pcr Assay: A Large Italian Hospital’s Five-Month Experience. Microbiol. Spectr. 2021, 9, e0069521. [Google Scholar] [CrossRef]
- Prinzi, A.; Parker, S.K.; Thurm, C.; Birkholz, M.; Sick-Samuels, A. Association of Endotracheal Aspirate Culture Variability and Antibiotic Use in Mechanically Ventilated Pediatric Patients. JAMA Netw. Open 2021, 4, e2140378. [Google Scholar] [CrossRef] [PubMed]
- Prinzi, A.M.; Parker, S.K.; Curtis, D.J.; Ziniel, S.I. The Pediatric Endotracheal Aspirate Culture Survey (Petacs): Examining Practice Variation across Pediatric Microbiology Laboratories in the United States. J. Clin. Microbiol. 2021, 59, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Cheung, A.; Dufour, S.; Jones, G.; Kostoulas, P.; Stevenson, M.A.; Singanallur, N.B.; Firestone, S.M. Bayesian Latent Class Analysis When the Reference Test Is Imperfect. Rev. Sci. Tech. 2021, 40, 271–286. [Google Scholar] [CrossRef] [PubMed]
- Rand, K.H.; Beal, S.G.; Cherabuddi, K.; Houck, H.; Lessard, K.; Tremblay, E.E.; Couturier, B.; Lingenfelter, B.; Rindlisbacher, C.; Jones, J. Relationship of Multiplex Molecular Pneumonia Panel Results with Hospital Outcomes and Clinical Variables. Open Forum Infect. Dis. 2021, 8, ofab368. [Google Scholar] [CrossRef] [PubMed]
- Zacharioudakis, I.M.; Zervou, F.N.; Dubrovskaya, Y.; Inglima, K.; See, B.; Aguero-Rosenfeld, M. Evaluation of a Multiplex PCR Panel for the Microbiological Diagnosis of Pneumonia in Hospitalized Patients: Experience from an Academic Medical Center. Int. J. Infect. Dis. 2021, 104, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.M.; Van Schooneveld, T.C.; Stohs, E.J.; Marcelin, J.R.; Alexander, B.T.; Watkins, A.B.; Creager, H.M.; Bergman, S.J. Implementation of a Rapid Multiplex Polymerase Chain Reaction Pneumonia Panel and Subsequent Antibiotic De-Escalation. Open Forum Infect. Dis. 2023, 10, ofad382. [Google Scholar] [CrossRef] [PubMed]
- Poole, S.; Tanner, A.R.; Naidu, V.V.; Borca, F.; Phan, H.; Saeed, K.; Grocott, M.P.W.; Dushianthan, A.; Moyses, H.; Clark, T.W. Molecular point-of-care testing for lower respiratory tract pathogens improves safe antibiotic de-escalation in patients with pneumonia in the ICU: Results of a randomised controlled trial. J Infect. 2022, 85, 625–633. [Google Scholar] [CrossRef] [PubMed]
- Azar, M.M.; Turbett, S.; Gaston, D.; Gitman, M.; Razonable, R.; Koo, S.; Hanson, K.; Kotton, C.; Silveira, F.; Banach, D.B.; et al. A Consensus Conference to Define the Utility of Advanced Infectious Disease Diagnostics in Solid Organ Transplant Recipients. Am. J. Transplant. 2022, 22, 3150–3169. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.C.; Lu, H.W.; Cheng, K.B.; Li, H.P.; Xu, J.F. Evaluation of Pcr in Bronchoalveolar Lavage Fluid for Diagnosis of Pneumocystis Jirovecii Pneumonia: A Bivariate Meta-Analysis and Systematic Review. PLoS ONE 2013, 8, e73099. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.; Vogel, S.; Procop, G.W. Pneumocystis PCR: It Is Time to Make Pcr the Test of Choice. Open Forum Infect. Dis. 2017, 4, ofx193. [Google Scholar] [CrossRef]
- Goterris, L.; Mancebo Fernández, M.A.; Aguilar-Company, J.; Falcó, V.; Ruiz-Camps, I.; Martín-Gómez, M.T. Molecular Diagnosis of Pneumocystis Jirovecii Pneumonia by Use of Oral Wash Samples in Immunocompromised Patients: Usefulness and Importance of the DNA Target. J. Clin. Microbiol. 2019, 57, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Alanio, A.; Hauser, P.M.; Lagrou, K.; Melchers, W.J.; Helweg-Larsen, J.; Matos, O.; Cesaro, S.; Maschmeyer, G.; Einsele, H.; Donnelly, J.P.; et al. ECIL Guidelines for the Diagnosis of Pneumocystis Jirovecii Pneumonia in Patients with Haematological Malignancies and Stem Cell Transplant Recipients. J. Antimicrob. Chemother. 2016, 71, 2386–2396. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, J.P.; Chen, S.C.; Kauffman, C.A.; Steinbach, W.J.; Baddley, J.W.; Verweij, P.E.; Clancy, C.J.; Wingard, J.R.; Lockhart, S.R.; Groll, A.H.; et al. Revision and Update of the Consensus Definitions of Invasive Fungal Disease from the European Organization for Research and Treatment of Cancer and the Mycoses Study Group Education and Research Consortium. Clin. Infect. Dis. 2020, 71, 1367–1376. [Google Scholar] [CrossRef] [PubMed]
- Gits-Muselli, M.; White, P.L.; Mengoli, C.; Chen, S.; Crowley, B.; Dingemans, G.; Fréalle, E.; Gorton, R.L.; Guiver, M.; Hagen, F.; et al. The Fungal Pcr Initiative’s Evaluation of in-House and Commercial Pneumocystis Jirovecii Qpcr Assays: Toward a Standard for a Diagnostics Assay. Med. Mycol. 2020, 58, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Guillotin, F.; Poulain, C.; Gaborit, B.; Bouras, M.; Cinotti, R.; Lakhal, K.; Vourc’h, M.; Rozec, B.; Asehnoune, K.; Vibet, M.A.; et al. Potential Impact of Rapid Multiplex PCR on Antimicrobial Therapy Guidance for Ventilated Hospital-Acquired Pneumonia in Critically Ill Patients, A Prospective Observational Clinical and Economic Study. Front. Cell Infect. Microbiol. 2022, 12, 804611. [Google Scholar] [CrossRef] [PubMed]
- Martin-Loeches, I.; Torres, A.; Nagavci, B.; Aliberti, S.; Antonelli, M.; Bassetti, M.; Bos, L.D.; Chalmers, J.D.; Derde, L.; de Waele, J.; et al. Ers/Esicm/Escmid/Alat Guidelines for the Management of Severe Community-Acquired Pneumonia. Intensive Care Med. 2023, 49, 615–632. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.C.; Lee, Y.T.; Wang, C.H.; Chiu, I.M.; Tsai, W.; Lin, Y.R.; Li, C.H.; Hsu, C.W.; Lai, P.F.; Chen, J.H.; et al. Guidelines for COVID-19 Laboratory Testing for Emergency Departments from the New Diagnostic Technology Team of the Taiwan Society of Emergency Medicine. J. Acute Med. 2022, 12, 45–52. [Google Scholar] [CrossRef]
- Wu, H.Y.; Chang, P.H.; Huang, Y.S.; Tsai, C.S.; Chen, K.Y.; Lin, I.F.; Hsih, W.H.; Tsai, W.L.; Chen, J.A.; Yang, T.L.; et al. Recommendations and Guidelines for the Diagnosis and Management of Coronavirus Disease-19 (COVID-19) Associated Bacterial and Fungal Infections in Taiwanv. J. Microbiol. Immunol. Infect. 2023, 56, 207–235. [Google Scholar] [CrossRef] [PubMed]
- CHEST. Rapid Diagnostics for Infectious Diseases in the ICU. Available online: https://www.chestnet.org/guidelines-and-topic-collections/topic-collections/sepsis/rapid-diagnostics-for-infectious-diseases-in-the-icu (accessed on 25 February 2024).
- Hitchcock, M.M.; Gomez, C.A.; Pozdol, J.; Banaei, N. Effective Approaches to Diagnostic Stewardship of Syndromic Molecular Panels. J. Appl. Lab. Med. 2024, 9, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S. The Rise and Fall and Rise Again of Toxin Testing for the Diagnosis of Clostridioides difficile Infection. Clin. Infect. Dis. 2019, 69, 1675–1677. [Google Scholar] [CrossRef] [PubMed]
- Curry, S.R.; Hecker, M.T.; O’Hagan, J.; Kutty, P.K.; Alhmidi, H.; Ng-Wong, Y.K.; Cadnum, J.L.; Jencson, A.L.; Gonzalez-Orta, M.; Saldana, C.; et al. Natural History of Clostridioides difficile Colonization and Infection Following New Acquisition of Carriage in Healthcare Settings: A Prospective Cohort Study. Clin. Infect. Dis. 2023, 77, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Polage, C.R.; Gyorke, C.E.; Kennedy, M.A.; Leslie, J.L.; Chin, D.L.; Wang, S.; Nguyen, H.H.; Huang, B.; Tang, Y.W.; Lee, L.W.; et al. Overdiagnosis of Clostridium difficile Infection in the Molecular Test Era. JAMA Intern. Med. 2015, 175, 1792–1801. [Google Scholar] [CrossRef] [PubMed]
- Planche, T.D.; Davies, K.A.; Coen, P.G.; Finney, J.M.; Monahan, I.M.; Morris, K.A.; O’Connor, L.; Oakley, S.J.; Pope, C.F.; Wren, M.W.; et al. Differences in Outcome According to Clostridium Difficile Testing Method: A Prospective Multicentre Diagnostic Validation Study of C. difficile Infection. Lancet Infect. Dis. 2013, 13, 936–945. [Google Scholar] [CrossRef] [PubMed]
- Crobach, M.J.; Planche, T.; Eckert, C.; Barbut, F.; Terveer, E.M.; Dekkers, O.M.; Wilcox, M.H.; Kuijper, E.J. European Society of Clinical Microbiology and Infectious Diseases: Update of the diagnostic guidance document for Clostridium difficile infection. Clin. Microbiol. Infect. 2016, 22 (Suppl. S4), S63–S81. [Google Scholar] [CrossRef] [PubMed]
- McDonald, L.C.; Gerding, D.N.; Johnson, S.; Bakken, J.S.; Carroll, K.C.; Coffin, S.E.; Dubberke, E.R.; Garey, K.W.; Gould, C.V.; Kelly, C.; et al. Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin. Infect. Dis. 2018, 66, e1–e48. [Google Scholar] [CrossRef]
- Raman, K.; Nailor, M.D.; Nicolau, D.P.; Aslanzadeh, J.; Nadeau, M.; Kuti, J.L. Early Antibiotic Discontinuation in Patients with Clinically Suspected Ventilator-Associated Pneumonia and Negative Quantitative Bronchoscopy Cultures. Crit. Care Med. 2013, 41, 1656–1663. [Google Scholar] [CrossRef] [PubMed]
- Fabre, V.; Davis, A.; Diekema, D.J.; Granwehr, B.; Hayden, M.K.; Lowe, C.F.; Pfeiffer, C.D.; Sick-Samuels, A.C.; Sullivan, K.V.; Van Schooneveld, T.C.; et al. Principles of Diagnostic Stewardship: A Practical Guide from the Society for Healthcare Epidemiology of America Diagnostic Stewardship Task Force. Infect. Control Hosp. Epidemiol. 2023, 44, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Hueth, K.D.; Prinzi, A.M.; Timbrook, T.T. Diagnostic Stewardship as a Team Sport: Interdisciplinary Perspectives on Improved Implementation of Interventions and Effect Measurement. Antibiotics 2022, 11, 250. [Google Scholar] [CrossRef] [PubMed]
- Ku, T.S.N.; Al Mohajer, M.; Newton, J.A.; Wilson, M.H.; Monsees, E.; Hayden, M.K.; Messacar, K.; Kisgen, J.J.; Diekema, D.J.; Morgan, D.J.; et al. Improving Antimicrobial Use through Better Diagnosis: The Relationship between Diagnostic Stewardship and Antimicrobial Stewardship. Infect. Control Hosp. Epidemiol. 2023, 44, 1901–1908. [Google Scholar] [CrossRef] [PubMed]
- Advani, S.D.; Claeys, K. Behavioral Strategies in Diagnostic Stewardship. Infect. Dis. Clin. N. Am. 2023, 37, 729–747. [Google Scholar] [CrossRef] [PubMed]
- Albin, O.R.; Soper, N.S. Healthcare providers consistently overestimate the diagnostic probability of ventilator-associated pneumonia. Infect. Control Hosp. Epidemiol. 2023, 44, 1927–1931. [Google Scholar] [CrossRef]
- Morgan, D.J.; Pineles, L.; Owczarzak, J.; Magder, L.; Scherer, L.; Brown, J.P.; Pfeiffer, C.; Terndrup, C.; Leykum, L.; Feldstein, D.; et al. Accuracy of Practitioner Estimates of Probability of Diagnosis before and after Testing. JAMA Intern. Med. 2021, 181, 747–755. [Google Scholar] [CrossRef] [PubMed]
- Chiotos, K.; Marshall, D.; Kellom, K.; Whittaker, J.; Wolfe, H.; Woods-Hill, C.; Stinson, H.; Keim, G.; Blumenthal, J.; Piccione, J.; et al. Mixed-Methods Process Evaluation of a Respiratory-Culture Diagnostic Stewardship Intervention. Infect. Control Hosp. Epidemiol. 2023, 44, 191–199. [Google Scholar] [CrossRef]
- Sick-Samuels, A.C.; Koontz, D.W.; Xie, A.; Kelly, D.; Woods-Hill, C.Z.; Aneja, A.; Xiao, S.; Colantuoni, E.A.; Marsteller, J.; Milstone, A.M.; et al. A Survey of PICU Clinician Practices and Perceptions regarding Respiratory Cultures in the Evaluation of Ventilator-Associated Infections in the BrighT STAR Collaborative. Pediatr. Crit. Care Med. 2024, 25, e20–e30. [Google Scholar] [CrossRef] [PubMed]
- Prinzi, A.M.; Wattier, R.L.; Curtis, D.J.; Ziniel, S.I.; Fitzgerald, A.; Pearce, K.; Parker, S.K. Impact of Organism Reporting from Endotracheal Aspirate Cultures on Antimicrobial Prescribing Practices in Mechanically Ventilated Pediatric Patients. J. Clin. Microbiol. 2022, 60, e0093022. [Google Scholar] [CrossRef] [PubMed]
- Vaughn, V.M.; Gupta, A.; Petty, L.A.; Malani, A.N.; Osterholzer, D.; Patel, P.K.; Younas, M.; Bernstein, S.J.; Burdick, S.; Ratz, D.; et al. A Statewide Quality Initiative to Reduce Unnecessary Antibiotic Treatment of Asymptomatic Bacteriuria. JAMA Intern. Med. 2023, 183, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Sick-Samuels, A.C.; Linz, M.; Bergmann, J.; Fackler, J.C.; Berenholtz, S.M.; Ralston, S.L.; Hoops, K.; Dwyer, J.; Colantuoni, E.; Milstone, A.M. Diagnostic Stewardship of Endotracheal Aspirate Cultures in a PICU. Pediatrics 2021, 147, e20201634. [Google Scholar] [CrossRef] [PubMed]
- McCormick, W.L.; Jackson, G.; Andrea, S.B.; Whitehead, V.; Chargualaf, T.L.; Touzard-Romo, F. Impact of Mandatory Nucleic Acid Amplification Test (Naat) Testing Approval on Hospital-Onset Clostridioides difficile Infection (Ho-Cdi) Rates: A Diagnostic Stewardship Intervention. Infect. Control Hosp. Epidemiol. 2024, 45, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Bussell, C.; Vincent, J.; Brust, K. Implementation of a Multidisciplinary Process to Improve Diagnostic Stewardship of Hospital-Onset Clostridioides difficile Infections. Am. J. Infect. Control 2023, 51, 1329–1333. [Google Scholar] [CrossRef] [PubMed]
- Halabi, K.C.; Ross, B.; Acker, K.P.; Cannon, J.M.; Messina, M.; Mangino, D.; Balzer, K.; Hill-Ricciuti, A.; Green, D.A.; Westblade, L.F.; et al. Successful diagnostic stewardship for Clostridioides difficile testing in Pediatrics. Infect. Control Hosp. Epidemiol. 2023, 44, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Berg, M.L.; Ayres, A.M.; Weber, D.R.; McCullough, M.; Crall, V.D.; Lewis, C.L.; Valek, A.L.; Vincent, L.A.; Penzelik, J.; Sasinoski, C.A.; et al. Diagnostic Stewardship for Clostridioides difficile Testing in an Acute Care Hospital: A Quality Improvement Intervention. Antimicrob. Steward. Heal. Epidemiol. 2023, 3, e67. [Google Scholar] [CrossRef] [PubMed]
- Albin, O.R.; Troost, J.P.; Saravolatz, L., 2nd; Thomas, M.P.; Hyzy, R.C.; Konkle, M.A.; Weirauch, A.J.; Dickson, R.P.; Rao, K.; Kaye, K.S. A Quasi-Experimental Study of a Bundled Diagnostic Stewardship Intervention for Ventilator-Associated Pneumonia. Clin. Microbiol. Infect. 2023, 30, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Plattner, A.S.; Lockowitz, C.R.; Dumm, R.; Banerjee, R.; Newland, J.G.; Same, R.G. Practice vs. Potential: The Impact of the Biofire Filmarray Pneumonia Panel on Antibiotic Use in Children. J. Pediatr. Infect. Dis. Soc. 2024, 13, 196–202. [Google Scholar] [CrossRef] [PubMed]
- Feldman, E. Low Diagnostic Utility of Frequent Serial Tracheal Aspirate Cultures (TACs) in the Pediatric Intensive Care Unit. Pediatr. Crit. Care Med. 2023, 24, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Prinzi, A.M.; Baker, C.D. So Tell Me… Does This Tracheal Aspirate Culture Indicate “Infection” or “Colonization”? Pediatr. Pulmonol. 2023, 58, 2439–2441. [Google Scholar] [CrossRef] [PubMed]
- Prinzi, A.M.; Chiotos, K. Repeat Tracheal Aspirate Cultures: A Port in the Storm or a Sinking Ship? Pediatr. Crit. Care Med. 2023, 24, 708–710. [Google Scholar] [CrossRef] [PubMed]
- Langford, B.J.; Leung, E.; Haj, R.; McIntyre, M.; Taggart, L.R.; Brown, K.A.; Downing, M.; Matukas, L.M. Nudging in Microbiology Laboratory Evaluation (Nimble): A Scoping Review. Infect. Control Hosp. Epidemiol. 2019, 40, 1400–1406. [Google Scholar] [CrossRef] [PubMed]
- Musgrove, M.A.; Kenney, R.M.; Kendall, R.E.; Peters, M.; Tibbetts, R.; Samuel, L.; Davis, S.L. Microbiology Comment Nudge Improves Pneumonia Prescribing. Open Forum Infect. Dis. 2018, 5, ofy162. [Google Scholar] [CrossRef]
- Moradi, T.; Bennett, N.; Shemanski, S.; Kennedy, K.; Schlachter, A.; Boyd, S. Use of Procalcitonin and a Respiratory Polymerase Chain Reaction Panel to Reduce Antibiotic Use via an Electronic Medical Record Alert. Clin. Infect. Dis. 2020, 71, 1684–1689. [Google Scholar] [CrossRef] [PubMed]
- Timbrook, T.T.; Morton, J.B.; McConeghy, K.W.; Caffrey, A.R.; Mylonakis, E.; LaPlante, K.L. The Effect of Molecular Rapid Diagnostic Testing on Clinical Outcomes in Bloodstream Infections: A Systematic Review and Meta-Analysis. Clin. Infect. Dis. 2017, 64, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Messacar, K.; Hurst, A.L.; Child, J.; Campbell, K.; Palmer, C.; Hamilton, S.; Dowell, E.; Robinson, C.C.; Parker, S.K.; Dominguez, S.R. Clinical Impact and Provider Acceptability of Real-Time Antimicrobial Stewardship Decision Support for Rapid Diagnostics in Children with Positive Blood Culture Results. J. Pediatr. Infect. Dis. Soc. 2017, 6, 267–274. [Google Scholar] [CrossRef] [PubMed]
- French, S.D.; Green, S.E.; O’Connor, D.A.; McKenzie, J.E.; Francis, J.J.; Michie, S.; Buchbinder, R.; Schattner, P.; Spike, N.; Grimshaw, J.M. Developing Theory-Informed Behaviour Change Interventions to Implement Evidence into Practice: A Systematic Approach Using the Theoretical Domains Framework. Implement. Sci. 2012, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Messacar, K.; Parker, S.K.; Todd, J.K.; Dominguez, S.R. Implementation of Rapid Molecular Infectious Disease Diagnostics: The Role of Diagnostic and Antimicrobial Stewardship. J. Clin. Microbiol. 2017, 55, 715–723. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walker, A.M.; Timbrook, T.T.; Hommel, B.; Prinzi, A.M. Breaking Boundaries in Pneumonia Diagnostics: Transitioning from Tradition to Molecular Frontiers with Multiplex PCR. Diagnostics 2024, 14, 752. https://doi.org/10.3390/diagnostics14070752
Walker AM, Timbrook TT, Hommel B, Prinzi AM. Breaking Boundaries in Pneumonia Diagnostics: Transitioning from Tradition to Molecular Frontiers with Multiplex PCR. Diagnostics. 2024; 14(7):752. https://doi.org/10.3390/diagnostics14070752
Chicago/Turabian StyleWalker, Alyssa M., Tristan T. Timbrook, Benjamin Hommel, and Andrea M. Prinzi. 2024. "Breaking Boundaries in Pneumonia Diagnostics: Transitioning from Tradition to Molecular Frontiers with Multiplex PCR" Diagnostics 14, no. 7: 752. https://doi.org/10.3390/diagnostics14070752
APA StyleWalker, A. M., Timbrook, T. T., Hommel, B., & Prinzi, A. M. (2024). Breaking Boundaries in Pneumonia Diagnostics: Transitioning from Tradition to Molecular Frontiers with Multiplex PCR. Diagnostics, 14(7), 752. https://doi.org/10.3390/diagnostics14070752




