A Neoteric Feature Extraction Technique to Predict the Survival of Gastric Cancer Patients
Abstract
1. Introduction
2. Materials and Dataset
2.1. Image Dataset
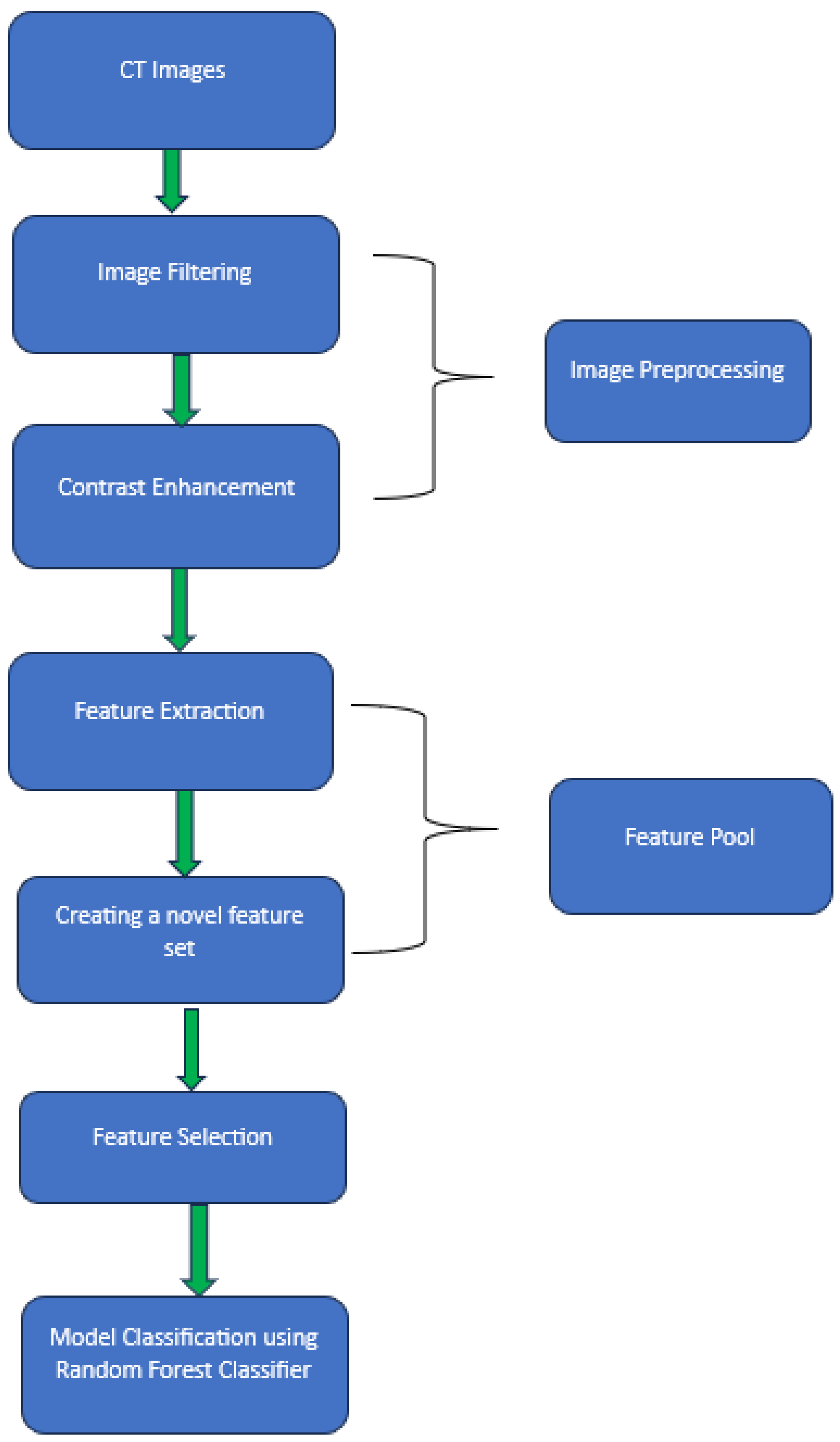
2.2. Image Preprocessing
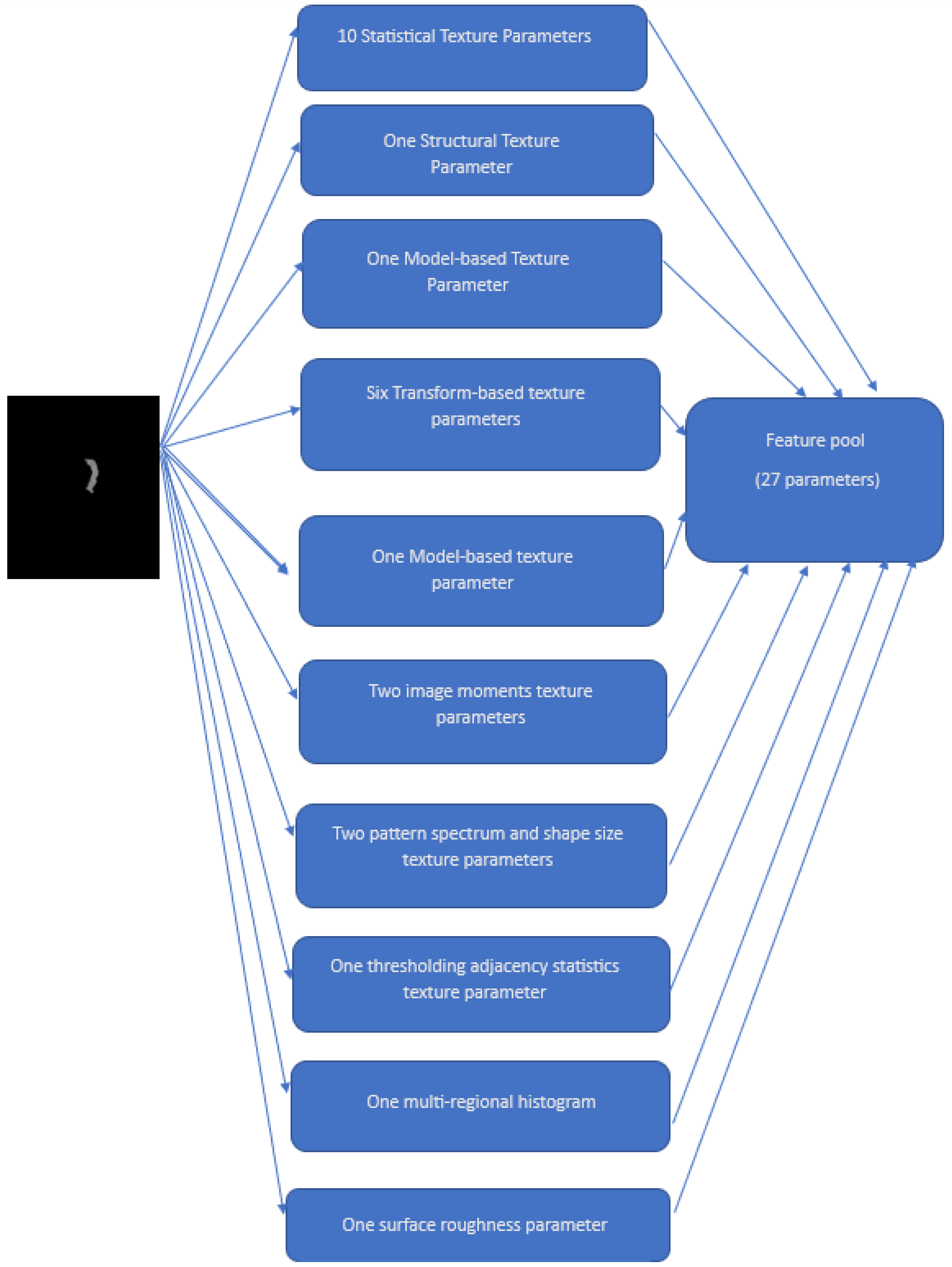
2.3. Feature Extraction
2.3.1. Gray-Level Co-Occurrence Matrix (GLCM)
2.3.2. Gray-Level Run-Length Matrix (GLRLM)
2.3.3. Shape Features
2.3.4. First Order Texture Features
2.3.5. Gray Level Dependence Matrix (GLDM)
- Statistical texture parameters: Histogram, Local Binary Descriptors (LBP), Histogram of Oriented Gradients (HOG), Gray Level Difference Statistics (GLDS), First-Order Statistics (FOS), Correlogram, Statistical Feature Matrices (SFM), and Gray Level Size Zone Matrix (GLSZM)
- One structural texture parameter: Shape-parameter
- One model-based texture parameter: Fractal Dimension Texture Analysis (FDTA)
- Six transform-based texture parameters: Gabor Pattern (GP), Wavelet Packets (WP), Discrete Wavelet Transform (DWT), Stroke Width Transform (SWT), Higher Order Spectra (HOS), and Laws Texture Energy (LTE)
- One model-based texture parameter: Amplitude Modulation-Frequency Modulation (AMFM)
- Two image moments texture parameters: Zero moments and Hu moments
- Two pattern spectrum and shape size texture parameters: Multilevel Binary Morphological Analysis (MultiBNA) and Gray Scale Morphological Analysis (GSMA)
- One thresholding adjacency statistics (TAS) texture parameter
- One multi-regional histogram (multiregional)
- One surface roughness parameter
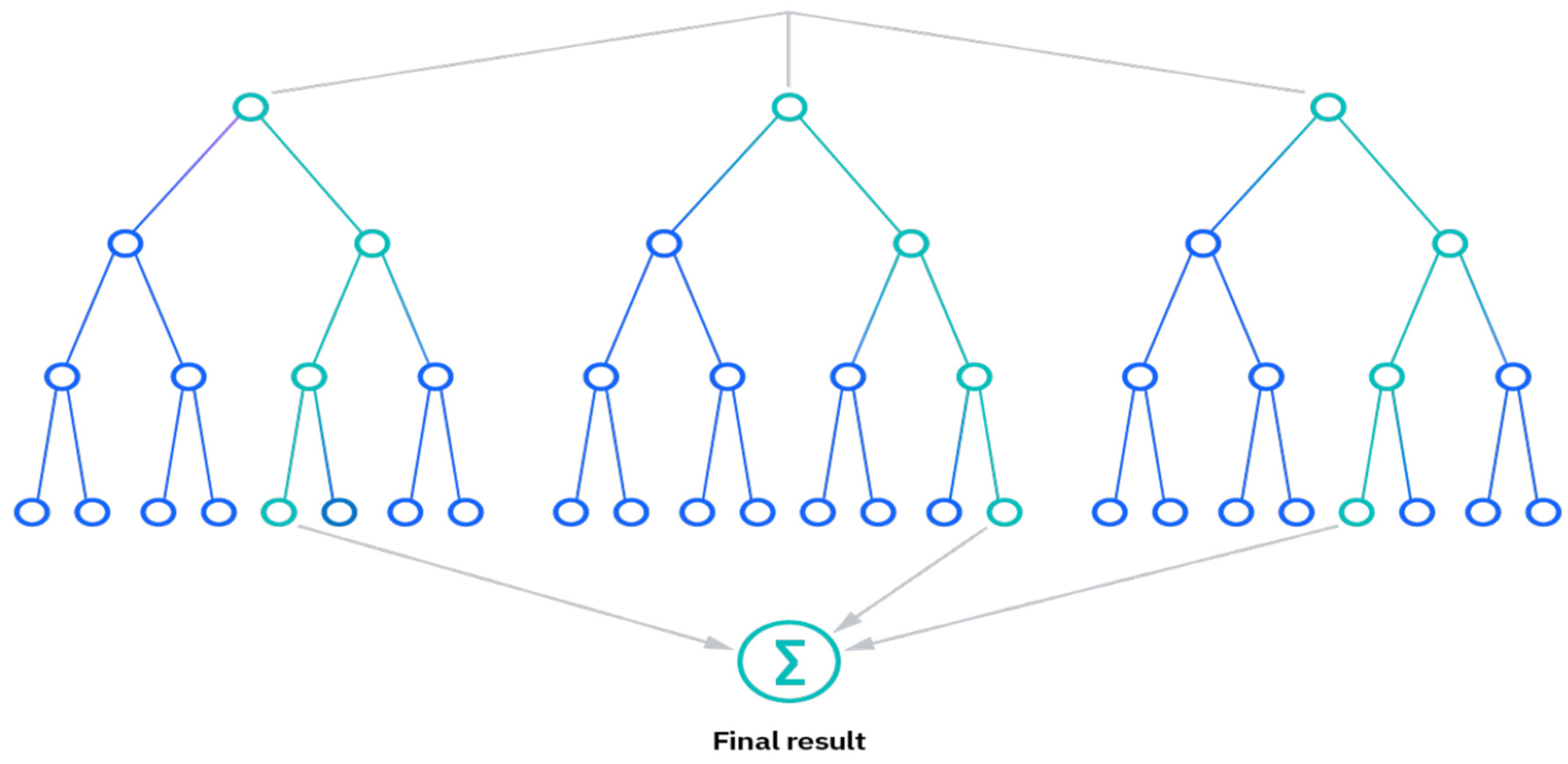
2.4. Random Forest Classifier
- Reduced risk of overfitting, as the presence of robust numbers of decision trees reduces the overall variance and prediction error.
- Provides flexibility, as it can perform both regression and classification tasks with a high level of accuracy. The model is useful for estimating missing values due to the inclusion of feature bagging because it retains accuracy even when some of the data is missing.
- Easy to determine feature importance by evaluating the importance of a variable’s contribution to the model is simple with RF.
2.5. Model Preparation
2.6. Performance Evaluation
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancer in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Zhang, Y.; Shao, X.; Wu, C.; Wu, J. PET/CT for predicting occult lymph node metastasis in gastric cancer. Curr. Oncol. 2022, 29, 6523–6539. [Google Scholar] [CrossRef]
- He, X.; Wu, W.; Lin, Z.; Ding, Y.; Si, J.; Sun, L.-M. Validation of the American Joint Committee on Cancer (AJCC) stage system for gastric cancer patients: A population-based analysis. Gastric Cancer 2018, 21, 391–400. [Google Scholar] [CrossRef] [PubMed]
- In, H.; Solsky, I.; Palis, B.; Langdon-Embry, M.; Ajani, J.; Sano, T. Validation of the 8th edition of the AJCC TNM staging system for gastric cancer using the National Cancer Database. Ann. Surg. Oncol. 2017, 24, 3683–3691. [Google Scholar] [CrossRef] [PubMed]
- Son, T.; Sun, J.; Choi, S.; Cho, M.; Kwon, I.G.; Kim, H.-I.; Hyung, W.J. Multi-institutional validation of the 8th AJCC TNM staging system for gastric cancer: Analysis of survival data from high-volume eastern centers and the SEER database. J. Surg. Oncol. 2019, 120, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Grabsch, H.; Ivanova, T.; Tan, I.B.; Murray, J.; Ooi, C.H.; Tan, P. Comprehensive genomic meta-analysis identifies intra-tumoural stroma as a predictor of survival in patients with gastric cancer. Gut 2013, 62, 1100–1111. [Google Scholar] [CrossRef] [PubMed]
- Davenport, T.; Kalakota, R. The potential for artificial intelligence in healthcare. Future Healthc. J. 2019, 6, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Briganti, G.; Le Moine, O. Artificial intelligence in medicine: Today and tomorrow. Front. Med. 2020, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.; Li, Q.; Feng, Q.X.; Qi, L.; Liu, X.S.; Arefan, D.; Wu, S. SurvivalCNN: A deep learning-based method for gastric cancer survival prediction using radiological imaging data and clinicopathological variables. Artif. Intell. Med. 2022, 134, 102424. [Google Scholar] [CrossRef]
- Bębas, E.; Borowska, M.; Derlatka, M.; Oczeretko, E.; Hładuński, M.; Szumowski, P.; Mojsak, M. Machine-learning-based classification of the histological subtype of non-small-cell lung cancer using MRI texture analysis. Biomed. Signal Process. Control. 2021, 66, 102446. [Google Scholar] [CrossRef]
- Mintz, Y.; Brodie, R. Introduction to artificial intelligence in medicine. Minim. Invasive Ther. Allied Technol. 2019, 28, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Lambin, P. Decoding tumor phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Islam, W.; Danala, G.; Pham, H.; Zheng, B. Improving the performance of computer-aided classification of breast lesions using a new feature fusion method. Proc. SPIE 2022, 12033, 84–91. [Google Scholar]
- Emaminejad, N. Fusion of quantitative image features and genomic biomarkers to improve prognosis assessment of early stage lung cancer patients. IEEE Trans. Biomed. Eng. 2016, 63, 1034–1043. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Gong, J.; Huang, X.; Lin, G.; Zheng, B.; Chen, J.; Lin, W. CT-based radiomics nomogram for preoperative prediction of No. 10 lymph nodes metastasis in advanced proximal gastric cancer. Eur. J. Surg. Oncol. 2021, 47, 1458–1465. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Lim, J.S.; Huh, Y.M.; Kim, J.H.; Hyung, W.J.; Chung, J.J.; Kim, S. A radiomics-based model for predicting prognosis of locally advanced gastric cancer in the preoperative setting. Sci. Rep. 2021, 11, 1879. [Google Scholar] [CrossRef]
- Pham, H.; Jones, M.A.; Gai, T.; Islam, W.; Danala, G.; Jo, J.; Zheng, B. Identifying an optimal machine learning generated image marker to predict survival of gastric cancer patients. In Proceedings of the Medical Imaging 2022: Computer-Aided Diagnosis, San Diego, CA, USA, 4 April 2022; Volume 12033, pp. 408–414. [Google Scholar]
- Ge, G.; Zhang, J. Feature selection methods and predictive models in CT lung cancer radiomics. J. Appl. Clin. Med. Phys. 2023, 24, e13869. [Google Scholar] [CrossRef] [PubMed]
- Hoi, H.; Jin, R.; Zhu, J.; Lyu, R. Semisupervised SVM batch mode active learning with applications to image retrieval. ACM Trans. Inf. Syst. 2009, 27, 1–16. [Google Scholar] [CrossRef]
- Désir, C.; Bernard, S.; Petitjean, C.; Heutte, L. A random forest based approach for one class classification in medical imaging. In Proceedings of the Machine Learning in Medical Imaging: Third International Workshop, MLMI 2012, Held in Conjunction with MICCAI 2012, Nice, France, 1 October 2012; Revised Selected Papers 3. Springer: Berlin/Heidelberg, Germany, 2012; pp. 250–257. [Google Scholar]
- Aydadenta, H. On the classification techniques in data mining for microarray data classification. J. Phys. Conf. Ser. 2018, 971, 012004. [Google Scholar] [CrossRef]
- Zheng, B.; Chang, Y.; Gur, D. Computerized detection of masses in digitized mammograms using single- image segmentation and a multilayer topographic feature analysis. Acad. Radiol. 1995, 2, 959–966. [Google Scholar] [CrossRef]
- Danala, G.; Patel, B.; Aghaei, F.; Heidari, M.; Li, J.; Wu, T.; Zheng, B. Classification of breast masses using a computer-aided diagnosis scheme of contrast enhanced digital mammograms. Ann. Biomed. Eng. 2018, 46, 1419–1431. [Google Scholar] [CrossRef] [PubMed]
- Mirniaharikandehei, S.; Heidari, M.; Danala, G.; Lakshmivarahan, S.; Zheng, B. Applying a random projection algorithm to optimize machine learning model for predicting peritoneal metastasis in gastric cancer patients using CT images. Comput. Methods Programs Biomed. 2021, 200, 105937. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.; Andrearczyk, V.; Apte, A.; Löck, S. The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Van Griethuysen, J.J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Aerts, H.J. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Park, Y.; Guldmann, J.M. Measuring continuous landscape patterns with Gray-Level Co-Occurrence Matrix (GLCM) indices: An alternative to patch metrics? Ecol. Indic. 2020, 109, 105802. [Google Scholar] [CrossRef]
- Zhou, J.; Yang, M. Bone region segmentation in medical images based on improved watershed algorithm. Comput. Intell. Neurosci. 2022, 2022, 3975853. [Google Scholar] [CrossRef]
- Venkatesh, U.; Balachander, B. Analysis of Textural Variations in Cerebellum in Brain to Identify Alzheimers by using Haralicks in Comparison with Gray Level Co-occurrence Matrix (GLRLM). In Proceedings of the 2022 2nd International Conference on Innovative Practices in Technology and Management (ICIPTM), Gautam Buddha Nagar, India, 23–25 February 2022; Volume 2, pp. 549–556. [Google Scholar]
- Chandraprabha, K.; Akila, S. Texture feature extraction for batik images using glcm and glrlm with neural network classification. Int. J. Sci. Res. Comput. Sci. Eng. Inf. Technol. 2019, 5, 6–15. [Google Scholar] [CrossRef]
- Rashed, B.M.; Popescu, N. Performance Investigation for Medical Image Evaluation and Diagnosis Using Machine-Learning and Deep-Learning Techniques. Computation 2023, 11, 63. [Google Scholar] [CrossRef]
- Khan, S.; Kaklis, P.; Serani, A.; Diez, M.; Kostas, K. Shape-supervised dimension reduction: Extracting geometry and physics associated features with geometric moments. Comput. -Aided Des. 2022, 150, 103327. [Google Scholar] [CrossRef]
- Bharti, R.; Khamparia, A.; Shabaz, M.; Dhiman, G.; Pande, S.; Singh, P. Prediction of heart disease using a combination of machine learning and deep learning. Comput. Intell. Neurosci. 2021, 2021, 8387680. [Google Scholar] [CrossRef]
- Zhang, H.; Hung, C.L.; Min, G.; Guo, J.P.; Liu, M.; Hu, X. GPU-accelerated GLRLM algorithm for feature extraction of MRI. Sci. Rep. 2019, 9, 10883. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Park, H.W. Statistical textural features for detection of microcalcifications in digitized mammograms. IEEE Trans. Med. Imaging 1999, 18, 231–238. [Google Scholar] [PubMed]
- Strano, G.; Hao, L.; Everson, R.M.; Evans, K.E. Surface roughness analysis, modelling and prediction in selective laser melting. J. Mater. Process. Technol. 2013, 213, 589–597. [Google Scholar] [CrossRef]
- Zhang, J.; Hu, J. Image segmentation based on 2D Otsu method with histogram analysis. In Proceedings of the 2008 International Conference on Computer Science and Software Engineering, Wuhan, China, 12–14 December 2008; Volume 6, pp. 105–108. [Google Scholar]
- Haralick, R.M.; Sternberg, S.R.; Zhuang, X. Image analysis using mathematical morphology. IEEE Trans. Pattern Anal. Mach. Intell. 1987, PAMI-9, 532–550. [Google Scholar]
- Dierking, W. Quantitative roughness characterization of geological surfaces and implications for radar signature analysis. IEEE Trans. Geosci. Remote Sens. 1999, 37, 2397–2412. [Google Scholar] [CrossRef]
- Singh, A.; Gaurav, K.; Rai, A.K.; Beg, Z. Machine learning to estimate surface roughness from satellite images. Remote Sens. 2021, 13, 3794. [Google Scholar] [CrossRef]
- Celik, T. Multiscale change detection in multitemporal satellite images. IEEE Geosci. Remote Sens. Lett. 2009, 6, 820–824. [Google Scholar] [CrossRef]
- Lubner, M.G.; Smith, A.D.; Sandrasegaran, K.; Sahani, D.V.; Pickhardt, P.J. CT texture analysis: Definitions, applications, biologic correlates, and challenges. Radiographics 2017, 37, 1483–1503. [Google Scholar] [CrossRef]
- Sánchez, C.I.; Niemeijer, M.; Išgum, I.; Dumitrescu, A.; Suttorp-Schulten, M.S.; Abràmoff, M.D.; van Ginneken, B. Contextual computer-aided detection: Improving bright lesion detection in retinal images and coronary calcification identification in CT scans. Med. Image Anal. 2012, 16, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.W.; Malhi, H.; Varghese, B.; Cen, S.; Hwang, D.; Aron, M.; Duddalwar, V. Computed tomography-based texture analysis of bladder cancer: Differentiating urothelial carcinoma from micropapillary carcinoma. Abdom. Radiol. 2019, 44, 201–208. [Google Scholar] [CrossRef]
- He, Z.; Chen, J.; Yang, F.; Pan, X.; Liu, C. Computed tomography-based texture assessment for the differentiation of benign, borderline, and early-stage malignant ovarian neoplasms. J. Int. Med. Res. 2023, 51, 03000605221150139. [Google Scholar] [CrossRef]
- Ba-Ssalamah, A.; Muin, D.; Schernthaner, R.; Kulinna-Cosentini, C.; Bastati, N.; Stift, J.; Mayerhoefer, M.E. Texture-based classification of different gastric tumors at contrast-enhanced CT. Eur. J. Radiol. 2013, 82, e537–e543. [Google Scholar] [CrossRef] [PubMed]
- Raja, J.V.; Khan, M.; Ramachandra, V.K.; Al-Kadi, O. Texture analysis of CT images in the characterization of oral cancers involving buccal mucosa. Dentomaxillofacial Radiol. 2012, 41, 475–480. [Google Scholar] [CrossRef]
- Bonnin, A.; Durot, C.; Barat, M.; Djelouah, M.; Grange, F.; Mulé, S.; Hoeffel, C. CT texture analysis as a predictor of favorable response to anti-PD1 monoclonal antibodies in metastatic skin melanoma. Diagn. Interv. Imaging 2022, 103, 97–102. [Google Scholar] [CrossRef] [PubMed]
- IBM. What Is Random Forest? Available online: https://www.ibm.com/topics/random-forest (accessed on 9 January 2024).
- Khandelwal, R. (2018, November 2). K Fold and Other Cross-Validation Techniques. Available online: https://medium.datadriveninvestor.com/k-fold-and-other-cross-validation-techniques-6c03a2563f1e (accessed on 17 July 2023).
- Liu, M.M.; Wen, L.; Liu, Y.J.; Cai, Q.; Li, L.T.; Cai, Y.M. Application of data mining methods to improve screening for the risk of early gastric cancer. BMC Med. Inform. Decis. Mak. 2018, 18, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Cai, Q.; Zhu, C.; Yuan, Y.; Feng, Q.; Feng, Y.; Hao, Y.; Li, Z. Development and validation of a prediction rule for estimating gastric cancer risk in the Chinese high-risk population: A nationwide multicentre study. Gut 2019, 68, 1576–1587. [Google Scholar] [CrossRef] [PubMed]
- Safdari, R.; Arpanahi, H.K.; Langarizadeh, M.; Ghazisaiedi, M.; Dargahi, H.; Zendehdel, K. Design a fuzzy rule-based expert system to aid earlier diagnosis of gastric cancer. Acta Inform. Medica 2018, 26, 19. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Shen, J.; Qian, H.; Ma, H.; Ji, J.; Ma, H.; Shou, C. Diagnosis of gastric cancer using decision tree classification of mass spectral data. Cancer Sci. 2007, 98, 37–43. [Google Scholar] [CrossRef]
- Brindha, S.K.; Kumar, N.S.; Chenkual, S.; Lalruatfela, S.T.; Zomuana, T.; Ralte, Z.; Nath, P. Data mining for early gastric cancer etiological factors from diet-lifestyle characteristics. In Proceedings of the 2020 4th International Conference on Intelligent Computing and Control Systems (ICICCS), Madurai, India, 13–15 May 2020; pp. 65–73. [Google Scholar]
- Mortezagholi, A.; Khosravizadeh, O.; Menhaj, M.B.; Shafigh, Y.; Kalhor, R. Make intelligent of gastric cancer diagnosis error in Qazvin’s medical centers: Using data mining method. Asian Pac. J. Cancer Prev. APJCP 2019, 20, 2607. [Google Scholar] [CrossRef]
- Taninaga, J.; Nishiyama, Y.; Fujibayashi, K.; Gunji, T.; Sasabe, N.; Iijima, K.; Naito, T. Prediction of future gastric cancer risk using a machine learning algorithm and comprehensive medical check-up data: A case-control study. Sci. Rep. 2019, 9, 12384. [Google Scholar] [CrossRef]
- Islam, W.; Jones, M.; Faiz, R.; Sadeghipour, N.; Qiu, Y.; Zheng, B. Improving performance of breast lesion classification using a ResNet50 model optimized with a novel attention mechanism. Tomography 2022, 8, 2411–2425. [Google Scholar] [CrossRef]
- Armi, L.; Fekri-Ershad, S. Texture image analysis and texture classification methods-A review. arXiv 2019, arXiv:1904.06554. [Google Scholar]
- Zhao, X.; Xia, X.; Wang, X.; Bai, M.; Zhan, D.; Shu, K. Deep learning-based protein features predict overall survival and chemotherapy benefit in gastric cancer. Front. Oncol. 2022, 12, 847706. [Google Scholar] [CrossRef] [PubMed]
- Yeganeh, A.; Johannssen, A.; Chukhrova, N.; Erfanian, M.; Azarpazhooh, M.R.; Morovatdar, N. A monitoring framework for health care processes using Generalized Additive Models and Auto-Encoders. Artif. Intell. Med. 2023, 146, 102689. [Google Scholar] [CrossRef] [PubMed]
- Aslam, M.A.; Xue, C.; Chen, Y.; Zhang, A.; Liu, M.; Wang, K.; Cui, D. Breath analysis based early gastric cancer classification from deep stacked sparse autoencoder neural network. Sci. Rep. 2021, 11, 4014. [Google Scholar] [CrossRef]


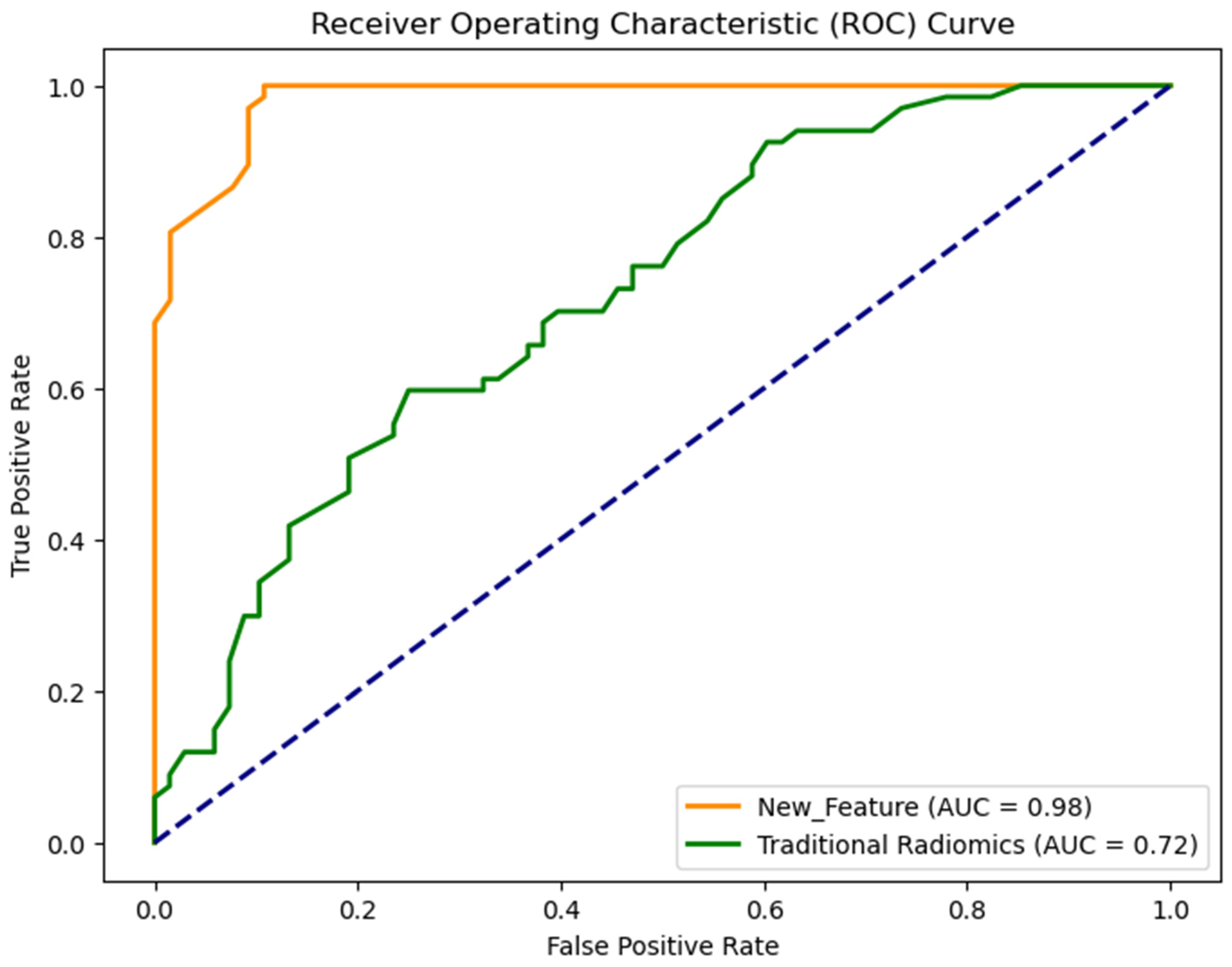

| Feature Set | AUC | Accuracy | Sensitivity | Specificity | F-1 Score |
|---|---|---|---|---|---|
| Radiomics | 0.72 ± 0.02 | 0.64 ± 0.04 | 0.64 ± 0.04 | 0.63 ± 0.02 | 0.64 ± 0.03 |
| New | 0.98 ± 0.01 | 0.92 ± 0.02 | 0.94 ± 0.03 | 0.90 ± 0.03 | 0.92 ± 0.04 |
| Machine Learning Model | Accuracy | AUC |
|---|---|---|
| SVM | 0.90 | 0.93 |
| Random Forest Classifier | 0.92 | 0.98 |
| KNN | 0.91 | 0.93 |
| Naïve Bayes | 0.91 | 0.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, W.; Abdoli, N.; Alam, T.E.; Jones, M.; Mutembei, B.M.; Yan, F.; Tang, Q. A Neoteric Feature Extraction Technique to Predict the Survival of Gastric Cancer Patients. Diagnostics 2024, 14, 954. https://doi.org/10.3390/diagnostics14090954
Islam W, Abdoli N, Alam TE, Jones M, Mutembei BM, Yan F, Tang Q. A Neoteric Feature Extraction Technique to Predict the Survival of Gastric Cancer Patients. Diagnostics. 2024; 14(9):954. https://doi.org/10.3390/diagnostics14090954
Chicago/Turabian StyleIslam, Warid, Neman Abdoli, Tasfiq E. Alam, Meredith Jones, Bornface M. Mutembei, Feng Yan, and Qinggong Tang. 2024. "A Neoteric Feature Extraction Technique to Predict the Survival of Gastric Cancer Patients" Diagnostics 14, no. 9: 954. https://doi.org/10.3390/diagnostics14090954
APA StyleIslam, W., Abdoli, N., Alam, T. E., Jones, M., Mutembei, B. M., Yan, F., & Tang, Q. (2024). A Neoteric Feature Extraction Technique to Predict the Survival of Gastric Cancer Patients. Diagnostics, 14(9), 954. https://doi.org/10.3390/diagnostics14090954









