Development and Validation of a Machine Learning Model for Early Prediction of Sepsis Onset in Hospital Inpatients from All Departments
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design and Setting
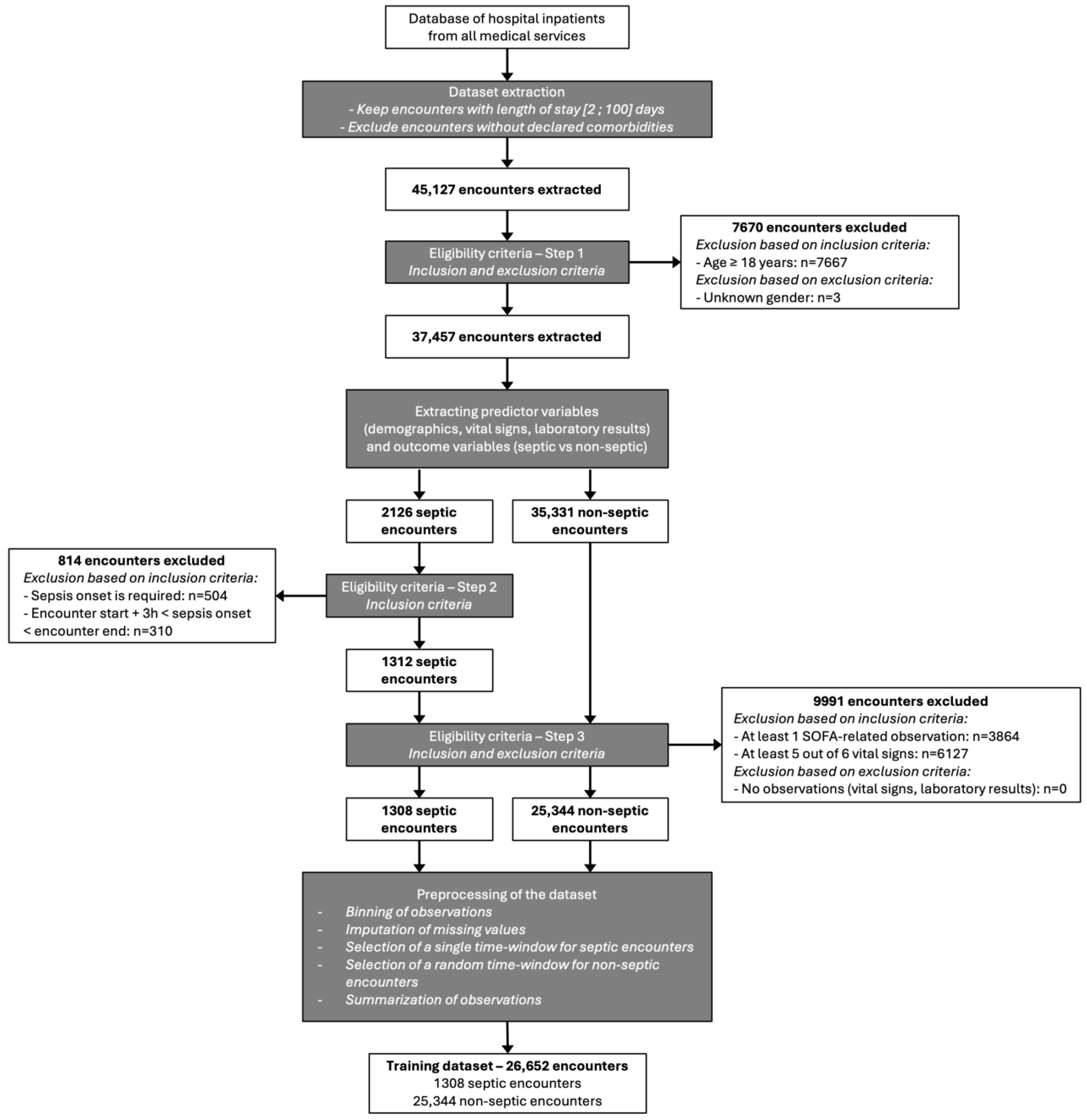
2.1.1. Study Population
2.1.2. Data Source
2.2. Procedures
2.2.1. Predictors and Other Variables
- Vital signs (heart rate—HR, respiratory rate—RR, diastolic blood pressure—DBP, systolic blood pressure—SBP, oxygen saturation—SpO2, and body temperature);
- Laboratory values (creatinine, lactate, white blood cells—WBC, platelets, bilirubin, diuresis, partial pressure of oxygen—PaO2, and fraction of inspired oxygen—FiO2);
- Assessment of level of consciousness (Glasgow Coma Scale—GCS).
2.2.2. Outcome Variables
2.2.3. Preprocessing of the Datasets
2.2.4. Training Dataset
2.2.5. Study Dataset
2.2.6. Implementation Details on SEPSI Score
2.3. Evaluation of the Model
2.3.1. Performance Metrics
2.3.2. Comparison with Existing Scores
2.3.3. Detection of Early Sepsis Onset
2.3.4. Statistical Analysis
2.4. SEPSI Score Among Other Advanced ML Models
3. Results
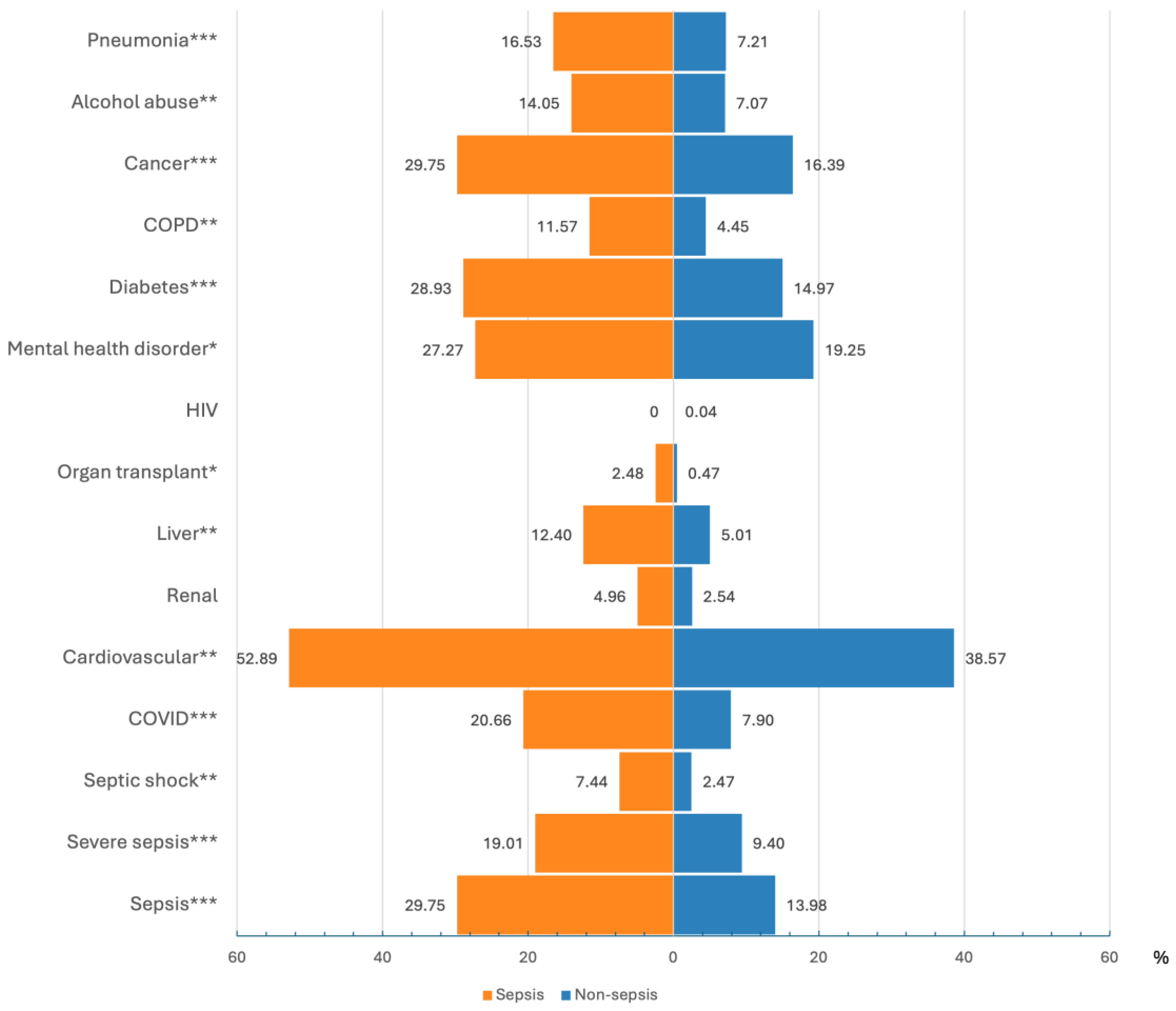
3.1. Study Cohort Characteristics
3.2. Performance Metrics of the Model
3.3. Detection of Early Sepsis Onset
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dupuis, C.; Bouadma, L.; Ruckly, S.; Perozziello, A.; Van-Gysel, D.; Mageau, A.; Mourvillier, B.; de Montmollin, E.; Bailly, S.; Papin, G.; et al. Sepsis and Septic Shock in France: Incidences, Outcomes and Costs of Care. Ann. Intensive Care 2020, 10, 145. [Google Scholar] [CrossRef]
- Global Report on the Epidemiology and Burden of Sepsis. Available online: https://www.who.int/publications/i/item/9789240010789 (accessed on 20 November 2024).
- Sepsis/Septicémie. Available online: https://www.pasteur.fr/fr/centre-medical/fiches-maladies/sepsis-septicemie (accessed on 20 November 2024).
- Berg, D.; Gerlach, H. Recent Advances in Understanding and Managing Sepsis. F1000Research 2018, 7, 1570. [Google Scholar] [CrossRef] [PubMed]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.-D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- WHA Adopts Resolution on Sepsis. Available online: https://globalsepsisalliance.org/news/2017/5/26/wha-adopts-resolution-on-sepsis (accessed on 20 November 2024).
- Kumar, A.; Roberts, D.; Wood, K.E.; Light, B.; Parrillo, J.E.; Sharma, S.; Suppes, R.; Feinstein, D.; Zanotti, S.; Taiberg, L.; et al. Duration of Hypotension before Initiation of Effective Antimicrobial Therapy Is the Critical Determinant of Survival in Human Septic Shock. Crit. Care Med. 2006, 34, 1589–1596. [Google Scholar] [CrossRef]
- Sartelli, M.; Kluger, Y.; Ansaloni, L.; Hardcastle, T.C.; Rello, J.; Watkins, R.R.; Bassetti, M.; Giamarellou, E.; Coccolini, F.; Abu-Zidan, F.M.; et al. Raising Concerns about the Sepsis-3 Definitions. World J. Emerg. Surg. 2018, 13, 6. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.-L.; Moreno, R.; Takala, J.; Willatts, S.; De Mendonça, A.; Bruining, H.; Reinhart, C.K.; Suter, P.M.; Thijs, L.G. The SOFA (Sepsis-Related Organ Failure Assessment) Score to Describe Organ Dysfunction/Failure. Intensive Care Med. 1996, 22, 707–710. [Google Scholar] [CrossRef] [PubMed]
- Lambden, S.; Laterre, P.F.; Levy, M.M.; Francois, B. The SOFA Score—Development, Utility and Challenges of Accurate Assessment in Clinical Trials. Crit. Care 2019, 23, 374. [Google Scholar] [CrossRef]
- Subbe, C.P.; Kruger, M.; Rutherford, P.; Gemmel, L. Validation of a Modified Early Warning Score in Medical Admissions. QJM 2001, 94, 521–526. [Google Scholar] [CrossRef] [PubMed]
- Sabir, L.; Ramlakhan, S.; Goodacre, S. Comparison of qSOFA and Hospital Early Warning Scores for Prognosis in Suspected Sepsis in Emergency Department Patients: A Systematic Review. Emerg. Med. J. 2022, 39, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xu, R.; Zeng, Y.; Zhao, Y.; Hu, X. A Comparison of qSOFA, SIRS and NEWS in Predicting the Accuracy of Mortality in Patients with Suspected Sepsis: A Meta-Analysis. PLoS ONE 2022, 17, e0266755. [Google Scholar] [CrossRef] [PubMed]
- Evans, L.; Rhodes, A.; Alhazzani, W.; Antonelli, M.; Coopersmith, C.M.; French, C.; Machado, F.R.; Mcintyre, L.; Ostermann, M.; Prescott, H.C.; et al. Surviving Sepsis Campaign: International Guidelines for Management of Sepsis and Septic Shock 2021. Intensive Care Med. 2021, 47, 1181–1247. [Google Scholar] [CrossRef]
- Chua, W.L.; Rusli, K.D.B.; Aitken, L.M. Early Warning Scores for Sepsis Identification and Prediction of In-Hospital Mortality in Adults with Sepsis: A Systematic Review and Meta-Analysis. J. Clin. Nurs. 2024, 33, 2005–2018. [Google Scholar] [CrossRef]
- Hassan, N.; Slight, R.; Weiand, D.; Vellinga, A.; Morgan, G.; Aboushareb, F.; Slight, S.P. Preventing Sepsis; How Can Artificial Intelligence Inform the Clinical Decision-Making Process? A Systematic Review. Int. J. Med. Inf. 2021, 150, 104457. [Google Scholar] [CrossRef] [PubMed]
- Fleuren, L.M.; Klausch, T.L.T.; Zwager, C.L.; Schoonmade, L.J.; Guo, T.; Roggeveen, L.F.; Swart, E.L.; Girbes, A.R.J.; Thoral, P.; Ercole, A.; et al. Machine Learning for the Prediction of Sepsis: A Systematic Review and Meta-Analysis of Diagnostic Test Accuracy. Intensive Care Med. 2020, 46, 383–400. [Google Scholar] [CrossRef]
- Persson, I.; Östling, A.; Arlbrandt, M.; Söderberg, J.; Becedas, D. A Machine Learning Sepsis Prediction Algorithm for Intended Intensive Care Unit Use (NAVOY Sepsis): Proof-of-Concept Study. JMIR Form. Res. 2021, 5, e28000. [Google Scholar] [CrossRef] [PubMed]
- Persson, I.; Macura, A.; Becedas, D.; Sjövall, F. Early Prediction of Sepsis in Intensive Care Patients Using the Machine Learning Algorithm NAVOY® Sepsis, a Prospective Randomized Clinical Validation Study. J. Crit. Care 2024, 80, 154400. [Google Scholar] [CrossRef] [PubMed]
- Shimabukuro, D.W.; Barton, C.W.; Feldman, M.D.; Mataraso, S.J.; Das, R. Effect of a Machine Learning-Based Severe Sepsis Prediction Algorithm on Patient Survival and Hospital Length of Stay: A Randomised Clinical Trial. BMJ Open Respir. Res. 2017, 4, e000234. [Google Scholar] [CrossRef]
- Desautels, T.; Calvert, J.; Hoffman, J.; Jay, M.; Kerem, Y.; Shieh, L.; Shimabukuro, D.; Chettipally, U.; Feldman, M.D.; Barton, C.; et al. Prediction of Sepsis in the Intensive Care Unit with Minimal Electronic Health Record Data: A Machine Learning Approach. JMIR Med. Inform. 2016, 4, e5909. [Google Scholar] [CrossRef] [PubMed]
- Burdick, H.; Pino, E.; Gabel-Comeau, D.; McCoy, A.; Gu, C.; Roberts, J.; Le, S.; Slote, J.; Pellegrini, E.; Green-Saxena, A.; et al. Effect of a Sepsis Prediction Algorithm on Patient Mortality, Length of Stay and Readmission: A Prospective Multicentre Clinical Outcomes Evaluation of Real-World Patient Data from US Hospitals. BMJ Health Care Inf. 2020, 27, e100109. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.; Das, R. Reducing Patient Mortality, Length of Stay and Readmissions through Machine Learning-Based Sepsis Prediction in the Emergency Department, Intensive Care Unit and Hospital Floor Units. BMJ Open Qual. 2017, 6, e000158. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Otles, E.; Donnelly, J.P.; Krumm, A.; McCullough, J.; DeTroyer-Cooley, O.; Pestrue, J.; Phillips, M.; Konye, J.; Penoza, C.; et al. External Validation of a Widely Implemented Proprietary Sepsis Prediction Model in Hospitalized Patients. JAMA Intern. Med. 2021, 181, 1065–1070. [Google Scholar] [CrossRef] [PubMed]
- Ramspek, C.L.; Jager, K.J.; Dekker, F.W.; Zoccali, C.; van Diepen, M. External Validation of Prognostic Models: What, Why, How, When and Where? Clin. Kidney J. 2021, 14, 49–58. [Google Scholar] [CrossRef] [PubMed]
- International Classification of Diseases (ICD). Available online: https://www.who.int/standards/classifications/classification-of-diseases (accessed on 25 March 2024).
- Python. Available online: https://www.python.org/ (accessed on 25 March 2024).
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: New York, NY, USA, 2016; pp. 785–794. [Google Scholar]
- Grinsztajn, L.; Oyallon, E.; Varoquaux, G. Why Do Tree-Based Models Still Outperform Deep Learning on Typical Tabular Data? arXiv 2022, arXiv:2207.08815. [Google Scholar]
- Saito, T.; Rehmsmeier, M. The Precision-Recall Plot Is More Informative than the ROC Plot When Evaluating Binary Classifiers on Imbalanced Datasets. PLoS ONE 2015, 10, e0118432. [Google Scholar] [CrossRef] [PubMed]
- Pandas. Available online: https://pandas.pydata.org/ (accessed on 25 March 2024).
- SciPy. Available online: https://scipy.org/ (accessed on 29 March 2024).
- Taneja, I.; Damhorst, G.L.; Lopez-Espina, C.; Zhao, S.D.; Zhu, R.; Khan, S.; White, K.; Kumar, J.; Vincent, A.; Yeh, L.; et al. Diagnostic and Prognostic Capabilities of a Biomarker and EMR-Based Machine Learning Algorithm for Sepsis. Clin. Transl. Sci. 2021, 14, 1578–1589. [Google Scholar] [CrossRef]
- VFusionTM Sepsis. Available online: https://www.vivacehealthsolutionsinc.com/vfusion/ (accessed on 20 November 2024).
- Bhattacharjee, P.; Edelson, D.P.; Churpek, M.M. Identifying Patients With Sepsis on the Hospital Wards. CHEST 2017, 151, 898–907. [Google Scholar] [CrossRef]
- Islam, M.M.; Nasrin, T.; Walther, B.A.; Wu, C.-C.; Yang, H.-C.; Li, Y.-C. Prediction of Sepsis Patients Using Machine Learning Approach: A Meta-Analysis. Comput. Methods Programs Biomed. 2019, 170, 1–9. [Google Scholar] [CrossRef]
- Mao, Q.; Jay, M.; Hoffman, J.L.; Calvert, J.; Barton, C.; Shimabukuro, D.; Shieh, L.; Chettipally, U.; Fletcher, G.; Kerem, Y.; et al. Multicentre Validation of a Sepsis Prediction Algorithm Using Only Vital Sign Data in the Emergency Department, General Ward and ICU. BMJ Open 2018, 8, e017833. [Google Scholar] [CrossRef] [PubMed]
- Theodosiou, A.A.; Read, R.C. Artificial Intelligence, Machine Learning and Deep Learning: Potential Resources for the Infection Clinician. J. Infect. 2023, 87, 287–294. [Google Scholar] [CrossRef]
- Schinkel, M.; Paranjape, K.; Nannan Panday, R.S.; Skyttberg, N.; Nanayakkara, P.W.B. Clinical Applications of Artificial Intelligence in Sepsis: A Narrative Review. Comput. Biol. Med. 2019, 115, 103488. [Google Scholar] [CrossRef] [PubMed]
- Wulff, A.; Montag, S.; Marschollek, M.; Jack, T. Clinical Decision-Support Systems for Detection of Systemic Inflammatory Response Syndrome, Sepsis, and Septic Shock in Critically Ill Patients: A Systematic Review. Methods Inf. Med. 2019, 58, e43–e57. [Google Scholar] [CrossRef]




| Sepsis Class | Corresponding ICD-10 Codes |
|---|---|
| Septic shock | R572 |
| Severe sepsis | R651 |
| Sepsis | A021, A227, A267, A327, A427, B377, O85, A400, A401, A402, A403, A408, A409, A410, A411, A412, A413, A414, A415, A418, A419, P3600, P3610, P3620, P3630, P3640, P3650, P3680, P3690 |
| Sepsis Encounters (N = 121) | Non-Sepsis Encounters (N = 5149) | ||
|---|---|---|---|
| Demographics | |||
| Age | Mean ± SD | 69.8 ± 12.7 | 65.6 ± 17.5 |
| Male | N (%) | 72 (59.5) | 2575 (50.0) |
| Female | N (%) | 49 (40.5) | 2574 (50.0) |
| Vital signs | |||
| HR (beats per minute) | Mean ± SD | 85.6 ± 19.7 | 81.0 ± 17.6 |
| Hours between measurements | Mean | 5 | 7 |
| Body temperature (°Celsius) | Mean ± SD | 37.1 ± 0.8 | 36.9 ± 0.7 |
| Hours between measurements | Mean | 6 | 8 |
| RR (breaths per minute) | Mean ± SD | 21.7 ± 5.7 | 21.2 ± 6.1 |
| Hours between measurements | Mean | 13 | 56 |
| DBP (mmHg) | Mean ± SD | 67.2 ± 14.9 | 70.9 ± 15.1 |
| Hours between measurements | Mean | 5 | 7 |
| SBP (mmHg) | Mean ± SD | 126.1 ± 22.1 | 128.6 ± 22.2 |
| Hours between measurements | Mean | 5 | 7 |
| SpO2 (%) | Mean ± SD | 96.5 ± 2.5 | 96.6 ± 2.6 |
| Hours between measurements | Mean | 6 | 9 |
| Level of consciousness | |||
| GCS score | Mean ± SD | 10.1 ± 5.2 | 11.4 ± 5.2 |
| Hours between measurements | Mean | 16 | 96 |
| Laboratory results | |||
| Creatinine (mg/L) | Mean ± SD | 18.3 ± 15.6 | 12.3 ± 12.0 |
| Hours between measurements | Mean | 37 | 48 |
| Lactate (mg/L) | Mean ± SD | 213.6 ± 255.5 | 180.0 ± 172.4 |
| Hours between measurements | Mean | 1655 | 3134 |
| WBC (mg/dL) | Mean ± SD | 11.6 ± 9.7 | 9.8 ± 7.6 |
| Hours between measurements | Mean | 40 | 54 |
| Platelet count (×109/L) | Mean ± SD | 218.2 ± 146.7 | 258.7 ± 119.2 |
| Hours between measurements | Mean | 40 | 54 |
| Bilirubin (μmol/L) | Mean ± SD | 22.2 ± 36.7 | 11.5 ± 26.5 |
| Hours between measurements | Mean | 72 | 105 |
| Diuresis (mL) | Mean ± SD | 409.3 ± 670.3 | 466.7 ± 563.5 |
| Hours between measurements | Mean | 9 | 22 |
| PaO2 (mmHg) | Mean ± SD | 90.5 ± 34.9 | 86.7 ± 39.0 |
| Hours between measurements | Mean | 54 | 247 |
| FiO2 (%) | Mean ± SD | 42.5 ± 21.0 | 45.9 ± 22.6 |
| Hours between measurements | Mean | 69 | 525 |
| Length of hospital stay | |||
| Day(s) | Median | 16.7 | 5.9 |
| Q1; Q3 | 10.1; 24.9 | 3.7; 9.5 | |
| Study Dataset N = 5270 121 Sepsis/5149 Non-Sepsis | ||||||
|---|---|---|---|---|---|---|
| SEPSI Score | SOFA | qSOFA | SIRS | MEWS | ||
| AUROC | Mean ± SD | 0.992 ± 0.001 | 0.944 | 0.537 | 0.628 | 0.617 |
| Median | 0.992 | - | - | - | - | |
| Min; Max | 0.992; 0.993 | - | - | - | - | |
| AUPR | Mean ± SD | 0.738 ± 0.029 | 0.174 | 0.036 | 0.041 | 0.048 |
| Median | 0.728 | - | - | - | - | |
| Min; Max | 0.711; 0.775 | - | - | - | - | |
| Positive predictive value | Mean ± SD | 0.610 ± 0.018 | 0.174 | 0.175 | 0.070 | 0.108 |
| Median | 0.606 | - | - | - | - | |
| Min; Max | 0.593; 0.640 | - | - | - | - | |
| Sensitivity | Mean ± SD | 0.845 ± 0.018 | 1.000 | 0.083 | 0.372 | 0.289 |
| Median | 0.851 | - | - | - | - | |
| Min; Max | 0.826; 0.868 | - | - | - | - | |
| Specificity | Mean ± SD | 0.987 ± 0.001 | 0.889 | 0.991 | 0.884 | 0.944 |
| Median | 0.987 | - | - | - | - | |
| Min; Max | 0.986; 0.989 | - | - | - | - | |
| F1 score | Mean ± SD | 0.708 ± 0.013 | 0.297 | 0.112 | 0.118 | 0.158 |
| Median | 0.705 | - | - | - | - | |
| Min; Max | 0.699; 0.730 | - | - | - | - | |
| Accuracy | Mean ± SD | 0.984 ± 0.001 | 0.891 | 0.970 | 0.872 | 0.929 |
| Median | 0.984 | - | - | - | - | |
| Min; Max | 0.983; 0.986 | - | - | - | - | |
| Negative predictive value | Mean ± SD | 0.996 ± 0.000 | 1.000 | 0.979 | 0.984 | 0.983 |
| Median | 0.996 | - | - | - | - | |
| Min; Max | 0.996; 0.997 | - | - | - | - | |
| Miss rate (false negative rate) | Mean ± SD | 0.155 ± 0.018 | 0.000 | 0.917 | 0.628 | 0.711 |
| Median | 0.149 | - | - | - | - | |
| Min; Max | 0.132; 0.174 | - | - | - | - | |
| Fall-out (false positive rate) | Mean ± SD | 0.013 ± 0.001 | 0.111 | 0.009 | 0.116 | 0.056 |
| Median | 0.013 | - | - | - | - | |
| Min; Max | 0.011; 0.014 | - | - | - | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thiboud, P.-E.; François, Q.; Faure, C.; Chaufferin, G.; Arribe, B.; Ettahar, N. Development and Validation of a Machine Learning Model for Early Prediction of Sepsis Onset in Hospital Inpatients from All Departments. Diagnostics 2025, 15, 302. https://doi.org/10.3390/diagnostics15030302
Thiboud P-E, François Q, Faure C, Chaufferin G, Arribe B, Ettahar N. Development and Validation of a Machine Learning Model for Early Prediction of Sepsis Onset in Hospital Inpatients from All Departments. Diagnostics. 2025; 15(3):302. https://doi.org/10.3390/diagnostics15030302
Chicago/Turabian StyleThiboud, Pierre-Elliott, Quentin François, Cécile Faure, Gilles Chaufferin, Barthélémy Arribe, and Nicolas Ettahar. 2025. "Development and Validation of a Machine Learning Model for Early Prediction of Sepsis Onset in Hospital Inpatients from All Departments" Diagnostics 15, no. 3: 302. https://doi.org/10.3390/diagnostics15030302
APA StyleThiboud, P.-E., François, Q., Faure, C., Chaufferin, G., Arribe, B., & Ettahar, N. (2025). Development and Validation of a Machine Learning Model for Early Prediction of Sepsis Onset in Hospital Inpatients from All Departments. Diagnostics, 15(3), 302. https://doi.org/10.3390/diagnostics15030302






