Oral Biofluid Biomarker Research: Current Status and Emerging Frontiers
Abstract
:1. Introduction
- Saliva collection is undemanding: procurement of saliva does not require highly trained personnel, and can be performed easily and readily, in contrast with blood sampling. To obtain sample saliva, expensive tools are not necessary.
- Saliva collection is noninvasive: Individual patients are usually more comfortable with saliva sampling, and are more likely to participate.
- Saliva samples are easier to handle and store: secretions in saliva that are not present in serum or plasma help decrease the risk of HIV transmission, and saliva does not clot.
2. Properties of Saliva as a Diagnostic Fluid
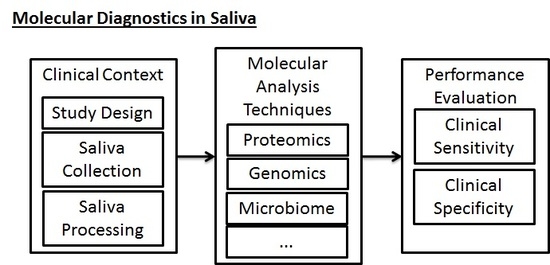
3. Biomarker Development and Clinical Reality
Biomarker Background
- Sample Collection and Processing: Following the selection of the clinical scenario, the sample collection and processing phase is the next important phase of the study that must be well-regulated. In traditional collection of blood samples for testing of biomarkers, trained personnel must perform venipuncture, collect blood samples in vacuum tubes, and then process the samples to remove red blood cells. Saliva collection, inasmuch as it does not require venipuncture, can be done more conveniently and efficiently by laypeople and physicians, increasing the probability of a study’s success. However, in a similar fashion to blood-based testing, saliva-based testing still requires some specific parameters to be set by the study designer and to be uniformly applied during the collection phase of experimental work. Table 1 includes a preliminary assessment of some clinical considerations that must be made when collection saliva.
- Laboratory Analysis of Biomarkers: Regardless of the specific molecular constituents that are being targeted (i.e., the test could be in relation to the proteome, transcriptome, genome, microbiome, metabolome, epigenome, etc.), the portion of study following study design and sample collection must take into account a proper laboratory workflow for processing the samples and ensuring that their sample quality is adequate for performing discovery or validation work. In this phase, it is necessary for the study to have well-designed quality control steps, thorough sample inventorying and storing (for future reference), and thorough documentation of the workflow for future reference and reproducibility. These steps must be taken in addition to the parameters that must be optimized for the specific technical procedures themselves.
4. Varieties of Biomarkers for Diagnostics in Saliva
4.1. Proteomics
4.2. Immunomics
4.3. The Salivary Microbiome
4.4. Genomics—Transcriptomics and Epigenomics
4.5. Metabolomics
5. Electric Field-Induced Release and Measurement (EFIRM)
- Basic Science: Complementing the diagnostic evaluation of saliva in a clinical setting is the need for rigorous scientific understanding of saliva’s relation to distal diseases. Specifically, this involves examining model systems (whether cell-based or animal-model based) in a rigorous and systematic fashion, which allows us to thoroughly understand the nature of salivary biomarkers and why biomarkers can often be found in the oral cavity. At present, there is intense interest in evaluating exosomes—microvesicular structures 30–100 nm in diameter found in saliva and other biofluids. They have been found to contain proteins, DNA, mRNA, and noncoding RNAs. Thus, some hypothesize that these exosomes may be the pathway where information is being carried from one portion of the body to another. Already, exosomes have been examined as prognostic markers for diseases such as lung cancer, squamous cell carcinoma, and breast cancer [42,88,89,90,91]. In examining exosomal entities as a possible transmitter of biomarkers to the oral cavity, EFIRM was used in conjunction with magnetic beads to extract exosomes from saliva, rapidly use electric fields to cause cargo unloading, and capture exosomal reference markers [88]. This method was used by Lau et al. for examining tumor-derived exosomes in a pancreatic cancer mouse model [42].
- Translational Research: In regards to the clinical utility of the EFIRM method, EFIRM has been deployed on a number of clinical contexts. EFIRM was first deployed for successfully performing multiplexed targeting of the IL-8 protein and IL-8 mRNA markers for oral cancer [92]. More recently, EFIRM has taken an exciting direction forward by being able to detect nonsquamous cell lung cancer (NSCLC) oncogenic mutations, which determine the susceptibility of NSCLC to treatment by tyrosine kinase inhibitors. This examination of the ability to detect oncogenic mutations also showed high correlation. Most notably, EFIRM was able to successfully identify mutations in the endothelial growth factor receptor (EGFR) within saliva samples with a clinical sensitivity and specificity above 95% in two blinded cohort groups [93,94].
6. Conclusions and Future Direction
Acknowledgments
Conflicts of Interest
References
- Lee, Y.-H.; Wong, D.T. Saliva: An emerging biofluid for early detection of diseases. Am. J. Dent. 2009, 22, 241–248. [Google Scholar] [PubMed]
- Spielmann, N.; Wong, D.T. Saliva: Diagnostics and therapeutic perspectives. Oral Dis. 2011, 17, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, J.M.; Schafer, C.A.; Schafer, J.J.; Farrell, J.J.; Paster, B.J.; Wong, D.T.W. Salivary biomarkers: Toward future clinical and diagnostic utilities. Clin. Microbiol. Rev. 2013, 26, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Schafer, C.A.; Schafer, J.J.; Yakob, M.; Lima, P.; Camargo, P.; Wong, D.T.W. Saliva Diagnostics: Utilizing Oral Fluids to Determine Health Status. In Monographs in Oral Science; Ligtenberg, A.J.M., Veerman, E.C.I., Eds.; S. KARGER AG: Basel, Switzerland, 2014; Volume 24, pp. 88–98. [Google Scholar]
- Park, N.J. Characterization of RNA in Saliva. Clin. Chem. 2006, 52, 988–994. [Google Scholar] [CrossRef] [PubMed]
- Bonne, N.J.; Wong, D.T. Salivary biomarker development using genomic, proteomic and metabolomic approaches. Genome Med. 2012, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Ploussard, G.; de la Taille, A. Urine biomarkers in prostate cancer. Nat. Rev. Urol. 2010, 7, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Varghese, S.A.; Powell, T.B.; Budisavljevic, M.N.; Oates, J.C.; Raymond, J.R.; Almeida, J.S.; Arthur, J.M. Urine Biomarkers Predict the Cause of Glomerular Disease. J. Am. Soc. Nephrol. 2007, 18, 913–922. [Google Scholar] [CrossRef] [PubMed]
- Majem, B.; Rigau, M.; Reventós, J.; Wong, D. Non-Coding RNAs in Saliva: Emerging Biomarkers for Molecular Diagnostics. Int. J. Mol. Sci. 2015, 16, 8676–8698. [Google Scholar] [CrossRef] [PubMed]
- Zelles, T.; Purushotham, K.R.; Macauley, S.P.; Oxford, G.E.; Humphreys-Beher, M.G. Saliva and growth factors: The fountain of youth resides in us all. J. Dent. Res. 1995, 74, 1826–1832. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, M. Science behind human saliva. J. Nat. Sci. Biol. Med. 2011, 2, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, S.P.; Williamson, R.T. A review of saliva: Normal composition, flow, and function. J. Prosthet. Dent. 2001, 85, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Zalewska, A.; Zwierz, K.; Zółkowski, K.; Gindzieński, A. Structure and biosynthesis of human salivary mucins. Acta Biochim. Pol. 2000, 47, 1067–1079. [Google Scholar] [PubMed]
- Edgar, W.M. Saliva and dental health. Clinical implications of saliva: Report of a consensus meeting. Br. Dent. J. 1990, 169, 96–98. [Google Scholar] [CrossRef] [PubMed]
- Haeckel, R.; Hänecke, P. Application of saliva for drug monitoring. An in vivo model for transmembrane transport. Eur. J. Clin. Chem. Clin. Biochem. J. Forum Eur. Clin. Chem. Soc. 1996, 34, 171–191. [Google Scholar]
- Jusko, W.J.; Milsap, R.L. Pharmacokinetic principles of drug distribution in saliva. Ann. N. Y. Acad. Sci. 1993, 694, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Silberring, J.; Ciborowski, P. Biomarker discovery and clinical proteomics. Trends Anal. Chem. TRAC 2010, 29, 128. [Google Scholar] [CrossRef] [PubMed]
- Ilyin, S.E.; Belkowski, S.M.; Plata-Salamán, C.R. Biomarker discovery and validation: Technologies and integrative approaches. Trends Biotechnol. 2004, 22, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Pepe, M.S.; Feng, Z. Improving Biomarker Identification with Better Designs and Reporting. Clin. Chem. 2011, 57, 1093–1095. [Google Scholar] [CrossRef] [PubMed]
- Pepe, M.S.; Feng, Z.; Janes, H.; Bossuyt, P.M.; Potter, J.D. Pivotal Evaluation of the Accuracy of a Biomarker Used for Classification or Prediction: Standards for Study Design. JNCI J. Natl. Cancer Inst. 2008, 100, 1432–1438. [Google Scholar] [CrossRef] [PubMed]
- Johansson, I.; Ericson, T.; Steen, L. Studies of the Effect of Diet on Saliva Secretion and Caries Development: The Effect of Fasting on Saliva Composition of Female Subjects. J. Nutr. 1984, 114, 2010–2020. [Google Scholar] [PubMed]
- Chiang, S.H.; Thomas, G.A.; Liao, W.; Grogan, T.; Buck, R.L.; Fuentes, L.; Yakob, M.; Laughlin, M.J.; Schafer, C.; Nazmul-Hossain, A.; et al. RNAPro•SAL: A device for rapid and standardized collection of saliva RNA and proteins. BioTechniques 2014, 58, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Binnie, V.; McHugh, S.; Macpherson, L.; Borland, B.; Moir, K.; Malik, K. The validation of self-reported smoking status by analysing cotinine levels in stimulated and unstimulated saliva, serum and urine. Oral Dis. 2004, 10, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Loo, J.A.; Wong, D.T. Human saliva proteome analysis. Ann. N. Y. Acad. Sci. 2007, 1098, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Al Kawas, S.; Rahim, Z.H.A.; Ferguson, D.B. Potential uses of human salivary protein and peptide analysis in the diagnosis of disease. Arch. Oral Biol. 2012, 57, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Xie, Y.; Ramachandran, P.; Ogorzalek Loo, R.R.; Li, Y.; Loo, J.A.; Wong, D.T. Large-scale identification of proteins in human salivary proteome by liquid chromatography/mass spectrometry and two-dimensional gel electrophoresis-mass spectrometry. Proteomics 2005, 5, 1714–1728. [Google Scholar] [CrossRef] [PubMed]
- Schulz, B.L.; Cooper-White, J.; Punyadeera, C.K. Saliva proteome research: Current status and future outlook. Crit. Rev. Biotechnol. 2013, 33, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.-M. Comparative proteomic analysis of human whole saliva. Arch. Oral Biol. 2004, 49, 951–962. [Google Scholar] [CrossRef] [PubMed]
- Genco, R.J.; Grossi, S.G.; Ho, A.; Nishimura, F.; Murayama, Y. A proposed model linking inflammation to obesity, diabetes, and periodontal infections. J. Periodontol. 2005, 76, 2075–2084. [Google Scholar] [CrossRef] [PubMed]
- Rao, P.V.; Reddy, A.P.; Lu, X.; Dasari, S.; Krishnaprasad, A.; Biggs, E.; Roberts, C.T.; Nagalla, S.R. Proteomic identification of salivary biomarkers of type-2 diabetes. J. Proteome Res. 2009, 8, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.L.; Wilson, C.H.; Perri, L.P.; Hannibal, S.L.; O’Connell, R.J. Usefulness of a rapid human immunodeficiency virus-1 antibody test for the management of occupational exposure to blood and body fluid. Infect. Control Hosp. Epidemiol. 2005, 26, 768–774. [Google Scholar] [CrossRef] [PubMed]
- St John, M.A.R.; Li, Y.; Zhou, X.; Denny, P.; Ho, C.-M.; Montemagno, C.; Shi, W.; Qi, F.; Wu, B.; Sinha, U.; et al. Interleukin 6 and interleukin 8 as potential biomarkers for oral cavity and oropharyngeal squamous cell carcinoma. Arch. Otolaryngol. Head Neck Surg. 2004, 130, 929–935. [Google Scholar] [CrossRef] [PubMed]
- Moore, H.D.; Ivey, R.G.; Voytovich, U.J.; Lin, C.; Stirewalt, D.L.; Pogosova-Agadjanyan, E.L.; Paulovich, A.G. The Human Salivary Proteome is Radiation Responsive. Radiat. Res. 2014, 181, 521–530. [Google Scholar] [CrossRef] [PubMed]
- Pernot, E.; Cardis, E.; Badie, C. Usefulness of Saliva Samples for Biomarker Studies in Radiation Research. Cancer Epidemiol. Biomarkers Prev. 2014, 23, 2673–2680. [Google Scholar] [CrossRef] [PubMed]
- Soni, S.; Agrawal, P.; Kumar, N.; Mittal, G.; Nishad, D.; Chaudhury, N.; Bhatnagar, A.; Basu, M.; Chhillar, N. Salivary biochemical markers as potential acute toxicity parameters for acute radiation injury: A study on small experimental animals. Hum. Exp. Toxicol. 2016, 35, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.-S.L.; Rees, T.; Wright, J. A review of research on salivary biomarkers for oral cancer detection. Clin. Transl. Med. 2014, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Yakob, M.; Fuentes, L.; Wang, M.B.; Abemayor, E.; Wong, D.T.W. Salivary biomarkers for detection of oral squamous cell carcinoma—Current state and recent advances. Curr. Oral Health Rep. 2014, 1, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Gleber-Netto, F.O.; Yakob, M.; Li, F.; Feng, Z.; Dai, J.; Kao, H.-K.; Chang, Y.-L.; Chang, K.-P.; Wong, D.T.W. Salivary Biomarkers for Detection of Oral Squamous Cell Carcinoma in a Taiwanese Population. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 3340–3347. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Arellano, M.; Boontheung, P.; Wang, J.; Zhou, H.; Jiang, J.; Elashoff, D.; Wei, R.; Loo, J.A.; Wong, D.T. Salivary Proteomics for Oral Cancer Biomarker Discovery. Clin. Cancer Res. 2008, 14, 6246–6252. [Google Scholar] [CrossRef] [PubMed]
- Bigler, L.R.; Streckfus, C.F.; Copeland, L.; Burns, R.; Dai, X.; Kuhn, M.; Martin, P.; Bigler, S.A. The potential use of saliva to detect recurrence of disease in women with breast carcinoma. J. Oral Pathol. Med. 2002, 31, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xiao, H.; Karlan, S.; Zhou, H.; Gross, J.; Elashoff, D.; Akin, D.; Yan, X.; Chia, D.; Karlan, B.; et al. Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non-invasive detection of breast cancer. PLoS ONE 2010, 5, e15573. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.; Kim, Y.; Chia, D.; Spielmann, N.; Eibl, G.; Elashoff, D.; Wei, F.; Lin, Y.-L.; Moro, A.; Grogan, T.; et al. Role of pancreatic cancer-derived exosomes in salivary biomarker development. J. Biol. Chem. 2013, 288, 26888–26897. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Gao, K.; Pollard, R.; Arellano-Garcia, M.; Zhou, H.; Zhang, L.; Elashoff, D.; Kallenberg, C.G.M.; Vissink, A.; Wong, D.T. Preclinical validation of salivary biomarkers for primary Sjögren’s syndrome. Arthritis Care Res. 2010, 62, 1633–1638. [Google Scholar] [CrossRef] [PubMed]
- Katsiougiannis, S.; Wong, D.T.W. The Proteomics of Saliva in Sjögren’s Syndrome. Rheum. Dis. Clin. N. Am. 2016, 42, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Vissink, A.; Arellano, M.; Roozendaal, C.; Zhou, H.; Kallenberg, C.G.M.; Wong, D.T. Identification of autoantibody biomarkers for primary Sjögren’s syndrome using protein microarrays. Proteomics 2011, 11, 1499–1507. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.; Zhang, L.; Zhou, H.; Lee, J.M.; Garon, E.B.; Wong, D.T.W. Proteomic analysis of human saliva from lung cancer patients using two-dimensional difference gel electrophoresis and mass spectrometry. Mol. Cell. Proteomics 2012, 11, M111.012112. [Google Scholar] [CrossRef] [PubMed]
- Ching, K.H.; Burbelo, P.D.; Gonzalez-Begne, M.; Roberts, M.E.P.; Coca, A.; Sanz, I.; Iadarola, M.J. Salivary anti-Ro60 and anti-Ro52 Antibody Profiles to Diagnose Sjögren’s Syndrome. J. Dent. Res. 2011, 90, 445–449. [Google Scholar] [CrossRef] [PubMed]
- Ben-Chetrit, E.; Fischel, R.; Rubinow, A. Anti-SSA/Ro and anti-SSB/La antibodies in serum and saliva of patients with Sjogren’s syndrome. Clin. Rheumatol. 1993, 12, 471–474. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Qiang, L.; Ding, Y.; Wei, P.; Li, Y.N.; Hua, H.; Li, Z.G. The role of muscarinic acetylcholine receptor type 3 polypeptide (M3RP205-220) antibody in the saliva of patients with primary Sjogren’s syndrome. Clin. Exp. Rheumatol. 2011, 30, 322–326. [Google Scholar]
- Berra, A.; Sterin-Borda, L.; Bacman, S.; Borda, E. Role of salivary IgA in the pathogenesis of Sjögren syndrome. Clin. Immunol. 2002, 104, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Moody, M.; Zipp, M.; Al-Hashimi, I. Salivary anti-spectrin autoantibodies in Sjögren’s syndrome. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 2001, 91, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Nasidze, I.; Quinque, D.; Li, J.; Li, M.; Tang, K.; Stoneking, M. Comparative analysis of human saliva microbiome diversity by barcoded pyrosequencing and cloning approaches. Anal. Biochem. 2009, 391, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, M.; Umeda, M.; Benno, Y. Molecular analysis of human oral microbiota. J. Periodontal Res. 2005, 40, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, F.; Fan, X.; Gao, J.; Chen, N.; Wong, T.; Wu, J.; Wen, S.W. Detection of hepatitis B surface antigen, hepatitis B core antigen, and hepatitis B virus DNA in parotid tissues. Int. J. Infect. Dis. 2009, 13, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Keijser, B.J.F.; Zaura, E.; Huse, S.M.; van der Vossen, J.M.B.M.; Schuren, F.H.J.; Montijn, R.C.; ten Cate, J.M.; Crielaard, W. Pyrosequencing analysis of the oral microflora of healthy adults. J. Dent. Res. 2008, 87, 1016–1020. [Google Scholar] [CrossRef] [PubMed]
- Paster, B.J.; Boches, S.K.; Galvin, J.L.; Ericson, R.E.; Lau, C.N.; Levanos, V.A.; Sahasrabudhe, A.; Dewhirst, F.E. Bacterial diversity in human subgingival plaque. J. Bacteriol. 2001, 183, 3770–3783. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, M.; Umeda, M.; Ishikawa, I.; Benno, Y. Comparison of the oral bacterial flora in saliva from a healthy subject and two periodontitis patients by sequence analysis of 16S rDNA libraries. Microbiol. Immunol. 2000, 44, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Faller, L.L.; Klitgord, N.; Mazumdar, V.; Ghodsi, M.; Sommer, D.D.; Gibbons, T.R.; Treangen, T.J.; Chang, Y.-C.; Li, S.; et al. Deep Sequencing of the Oral Microbiome Reveals Signatures of Periodontal Disease. PLoS ONE 2012, 7, e37919. [Google Scholar] [CrossRef] [PubMed]
- Delaney, K.P.; Branson, B.M.; Uniyal, A.; Kerndt, P.R.; Keenan, P.A.; Jafa, K.; Gardner, A.D.; Jamieson, D.J.; Bulterys, M. Performance of an oral fluid rapid HIV-1/2 test: Experience from four CDC studies. AIDS Lond. Engl. 2006, 20, 1655–1660. [Google Scholar] [CrossRef] [PubMed]
- Munshi, S.; Andalib, S.; Tabassum, S.; Islam, M.; Ahmed, M. Testing Hepatitis A virus antibody in oral fluid among the prospective vaccinees foster the need of new oral HAV rapid test. Indian J. Med. Microbiol. 2011, 29, 72. [Google Scholar] [CrossRef] [PubMed]
- Oba, I.T.; Spina, A.M.; Saraceni, C.P.; Lemos, M.F.; Senhoras, R.; Moreira, R.C.; Granato, C.F. Detection of hepatitis A antibodies by ELISA using saliva as clinical samples. Rev. Inst. Med. Trop. São Paulo 2000, 42, 197–200. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, P.L.; Cunha, C.B.; Busek, S.C.U.; Oliveira, G.C.; Ribeiro-Rodrigues, R.; Pereira, F.E. Detection of hepatitis C virus RNA in saliva samples from patients with seric anti-HCV antibodies. Braz. J. Infect. Dis. 2005, 9, 28–34. [Google Scholar] [CrossRef] [PubMed]
- Estévez, P.T.; Satoguina, J.; Nwakanma, D.C.; West, S.; Conway, D.J.; Drakeley, C.J. Human saliva as a source of anti-malarial antibodies to examine population exposure to Plasmodium falciparum. Malar. J. 2011, 10, 104. [Google Scholar] [CrossRef] [PubMed]
- Yap, G.; Sil, B.K.; Ng, L.-C. Use of Saliva for Early Dengue Diagnosis. PLoS Negl. Trop. Dis. 2011, 5, e1046. [Google Scholar] [CrossRef] [PubMed]
- Eguchi, J.; Ishihara, K.; Watanabe, A.; Fukumoto, Y.; Okuda, K. PCR method is essential for detecting Mycobacterium tuberculosis in oral cavity samples. Oral Microbiol. Immunol. 2003, 18, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Formenty, P.; Leroy, E.M.; Epelboin, A.; Libama, F.; Lenzi, M.; Sudeck, H.; Yaba, P.; Allarangar, Y.; Boumandouki, P.; Nkounkou, V.B.; et al. Detection of Ebola virus in oral fluid specimens during outbreaks of Ebola virus hemorrhagic fever in the Republic of Congo. Clin. Infect. Dis. 2006, 42, 1521–1526. [Google Scholar] [CrossRef] [PubMed]
- Furuta, Y.; Fukuda, S.; Chida, E.; Takasu, T.; Ohtani, F.; Inuyama, Y.; Nagashima, K. Reactivation of herpes simplex virus type 1 in patients with Bell’s palsy. J. Med. Virol. 1998, 54, 162–166. [Google Scholar] [CrossRef]
- Blackbourn, D.J.; Lennette, E.T.; Ambroziak, J.; Mourich, D.V.; Levy, J.A. Human herpesvirus 8 detection in nasal secretions and saliva. J. Infect. Dis. 1998, 177, 213–216. [Google Scholar] [CrossRef] [PubMed]
- Gautheret, A.; Aubin, J.T.; Fauveau, V.; Rozenbaum, W.; Huraux, J.M.; Agut, H. Rate of detection of human herpesvirus-6 at different stages of HIV infection. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 14, 820–824. [Google Scholar] [CrossRef]
- Boppana, S.B.; Ross, S.A.; Shimamura, M.; Palmer, A.L.; Ahmed, A.; Michaels, M.G.; Sánchez, P.J.; Bernstein, D.I.; Tolan, R.W.; Novak, Z.; et al. Saliva polymerase-chain-reaction assay for cytomegalovirus screening in newborns. N. Engl. J. Med. 2011, 364, 2111–2118. [Google Scholar] [CrossRef] [PubMed]
- Docktor, M.J.; Paster, B.J.; Abramowicz, S.; Ingram, J.; Wang, Y.E.; Correll, M.; Jiang, H.; Cotton, S.L.; Kokaras, A.S.; Bousvaros, A. Alterations in diversity of the oral microbiome in pediatric inflammatory bowel disease. Inflamm. Bowel Dis. 2012, 18, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Farrell, J.J.; Zhang, L.; Zhou, H.; Chia, D.; Elashoff, D.; Akin, D.; Paster, B.J.; Joshipura, K.; Wong, D.T.W. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 2012, 61, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Zimmermann, B.G.; Zhou, H.; Wang, J.; Henson, B.S.; Yu, W.; Elashoff, D.; Krupp, G.; Wong, D.T. Exon-level expression profiling: A comprehensive transcriptome analysis of oral fluids. Clin. Chem. 2008, 54, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Li, Y. Salivary Transcriptome Diagnostics for Oral Cancer Detection. Clin. Cancer Res. 2004, 10, 8442–8450. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Wang, J.; Meijer, J.; Ieong, S.; Xie, Y.; Yu, T.; Zhou, H.; Henry, S.; Vissink, A.; Pijpe, J.; et al. Salivary proteomic and genomic biomarkers for primary Sjögren’s syndrome. Arthritis Rheum. 2007, 56, 3588–3600. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Zhou, H.; Zhang, L.; Lee, J.W.; Zhou, Q.; Hu, S.; Wolinsky, L.E.; Farrell, J.; Eibl, G.; Wong, D.T. Systemic Disease-Induced Salivary Biomarker Profiles in Mouse Models of Melanoma and Non-Small Cell Lung Cancer. PLoS ONE 2009, 4, e5875. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The Majority of MicroRNAs Detectable in Serum and Saliva Is Concentrated in Exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef] [PubMed]
- Park, N.J.; Zhou, H.; Elashoff, D.; Henson, B.S.; Kastratovic, D.A.; Abemayor, E.; Wong, D.T. Salivary microRNA: Discovery, Characterization, and Clinical Utility for Oral Cancer Detection. Clin. Cancer Res. 2009, 15, 5473–5477. [Google Scholar] [CrossRef] [PubMed]
- Bahn, J.H.; Zhang, Q.; Li, F.; Chan, T.-M.; Lin, X.; Kim, Y.; Wong, D.T.W.; Xiao, X. The Landscape of MicroRNA, Piwi-Interacting RNA, and Circular RNA in Human Saliva. Clin. Chem. 2015, 61, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Metabolomics and Systems Biology in Human Health and Medicine; Jones, O.A.H. (Ed.) CABI: Wallingford, UK, 2014.
- Hoerr, V.; Vogel, H.J. Metabolomics. In Encyclopedia of Biophysics; Roberts, G.C.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; p. 1510. [Google Scholar]
- Dame, Z.T.; Aziat, F.; Mandal, R.; Krishnamurthy, R.; Bouatra, S.; Borzouie, S.; Guo, A.C.; Sajed, T.; Deng, L.; Lin, H.; et al. The human saliva metabolome. Metabolomics 2015, 11, 1864–1883. [Google Scholar] [CrossRef]
- Sugimoto, M.; Wong, D.T.; Hirayama, A.; Soga, T.; Tomita, M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 2010, 6, 78–95. [Google Scholar] [CrossRef] [PubMed]
- Aimetti, M.; Cacciatore, S.; Graziano, A.; Tenori, L. Metabonomic analysis of saliva reveals generalized chronic periodontitis signature. Metabolomics 2012, 8, 465–474. [Google Scholar] [CrossRef]
- Kageyama, G.; Saegusa, J.; Irino, Y.; Tanaka, S.; Tsuda, K.; Takahashi, S.; Sendo, S.; Morinobu, A. Metabolomics analysis of saliva from patients with primary Sjögren’s syndrome: Salivary metabolomics among Sjögren’s syndrome. Clin. Exp. Immunol. 2015, 182, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Mikkonen, J.J. Metabolic Profiling of Saliva in Patients with Primary Sjögren’s syndrome. J. Postgenomics Drug Biomark. Dev. 2012, 3, 128. [Google Scholar] [CrossRef]
- Laiakis, E.C.; Strawn, S.J.; Brenner, D.J.; Fornace, A.J. Assessment of Saliva as a Potential Biofluid for Biodosimetry: A Pilot Metabolomics Study in Mice. Radiat. Res. 2016, 186, 92–97. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Yang, J.; Wong, D.T.W. Detection of exosomal biomarker by electric field-induced release and measurement (EFIRM). Biosens. Bioelectron. 2013, 44, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yuan, X.; Shi, H.; Wu, L.; Qian, H.; Xu, W. Exosomes in cancer: Small particle, big player. J. Hematol. Oncol. 2015, 8, 83. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.S.; Wong, D.T.W. Breast Cancer Exosome-like Microvesicles and Salivary Gland Cells Interplay Alters Salivary Gland Cell-Derived Exosome-like Microvesicles In Vitro. PLoS ONE 2012, 7, e33037. [Google Scholar] [CrossRef] [PubMed]
- Rosell, R.; Wei, J.; Taron, M. Circulating MicroRNA Signatures of Tumor-Derived Exosomes for Early Diagnosis of Non–Small-Cell Lung Cancer. Clin. Lung Cancer 2009, 10, 8–9. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Patel, P.; Liao, W.; Chaudhry, K.; Zhang, L.; Arellano-Garcia, M.; Hu, S.; Elashoff, D.; Zhou, H.; Shukla, S.; et al. Electrochemical Sensor for Multiplex Biomarkers Detection. Clin. Cancer Res. 2009, 15, 4446–4452. [Google Scholar] [CrossRef] [PubMed]
- Pu, D.; Liang, H.; Wei, F.; Akin, D.; Feng, Z.; Yan, Q.; Li, Y.; Zhen, Y.; Xu, L.; Dong, G.; et al. Evaluation of a novel saliva-based epidermal growth factor receptor mutation detection for lung cancer: A pilot study: Saliva-based EGFR mutation detection. Thorac. Cancer 2016, 7, 428–436. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Lin, C.-C.; Joon, A.; Feng, Z.; Troche, G.; Lira, M.E.; Chia, D.; Mao, M.; Ho, C.-L.; Su, W.-C.; et al. Noninvasive Saliva-based EGFR Gene Mutation Detection in Patients with Lung Cancer. Am. J. Respir. Crit. Care Med. 2014, 190, 1117–1126. [Google Scholar] [CrossRef] [PubMed]
- Sholl, L.M.; Aisner, D.L.; Allen, T.C.; Beasley, M.B.; Cagle, P.T.; Capelozzi, V.L.; Dacic, S.; Hariri, L.P.; Kerr, K.M.; Lantuejoul, S.; et al. Liquid Biopsy in Lung Cancer: A Perspective From Members of the Pulmonary Pathology Society. Arch. Pathol. Lab. Med. 2016, 140, 825–829. [Google Scholar] [CrossRef] [PubMed]
| Parameter | Description |
|---|---|
| Subject Status | Prior to collection of samples, study researchers should prescribe either fasted or unfasted states to patient cohorts. It has been observed that saliva in a fasted state may lead to differences in composition of saliva [21]. |
| Sample Collection Time | When instructing patients on sample collection, it is necessary to specify a window of time that the patient may be allowed to contribute their saliva to a sample collection instance. These windows are important precautions against sample degradation if the time is long, and also allow adequate time for saliva to be collected with biomarker content. |
| Sample Collection Volume Requirement | Typically, running biomarker identifications or bioassays on a salivary sample will require a specific volume that must be collected for running tests. If the subject has a pathology that severely limits the flow of saliva to the oral cavity, it may be necessary for the study to have modifications made to account for the reduced volume that may be achievable. |
| Sample Collection Method | A multitude of different saliva collection methods can be used for testing. Typical collection protocol used at facilities such as UCLA involves the usage of traditional falcon tubes on ice, but saliva collectors have also been explored for collection [22]. This method can be designated as “unstimulated” since it uses saliva that has naturally pooled in the mouth. This is differs from the class of “stimulated” collection, where samples of saliva are attained through methods such as absorbent pads or chewing on parafilm [23]. The methods used must be appropriately identified, as results of analysis may differ depending on the saliva collection method. |
| Sample Processing and Storage | Collections of saliva must be properly optimized based on desired targets to be tested for. The inclusion of constituents in the saliva such as epithelial cells may contribute background that may hinder assessments of whether molecular targets are truly in the saliva. For this reason, centrifugation may be considered for removing cells and creating cell-free saliva. Stabilizing agents may be necessary for preservation of samples, depending on the target. |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, A.; Wang, C.P.; Tu, M.; Wong, D.T.W. Oral Biofluid Biomarker Research: Current Status and Emerging Frontiers. Diagnostics 2016, 6, 45. https://doi.org/10.3390/diagnostics6040045
Wang A, Wang CP, Tu M, Wong DTW. Oral Biofluid Biomarker Research: Current Status and Emerging Frontiers. Diagnostics. 2016; 6(4):45. https://doi.org/10.3390/diagnostics6040045
Chicago/Turabian StyleWang, Austin, Chris P. Wang, Michael Tu, and David T.W. Wong. 2016. "Oral Biofluid Biomarker Research: Current Status and Emerging Frontiers" Diagnostics 6, no. 4: 45. https://doi.org/10.3390/diagnostics6040045
APA StyleWang, A., Wang, C. P., Tu, M., & Wong, D. T. W. (2016). Oral Biofluid Biomarker Research: Current Status and Emerging Frontiers. Diagnostics, 6(4), 45. https://doi.org/10.3390/diagnostics6040045