Novel Mutations in GPR68 and SLC24A4 Cause Hypomaturation Amelogenesis Imperfecta
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subject Enrollment
2.2. DNA Isolation and Whole Exome Sequencing
2.3. Bioinformatics
2.4. Sanger Sequencing
3. Results
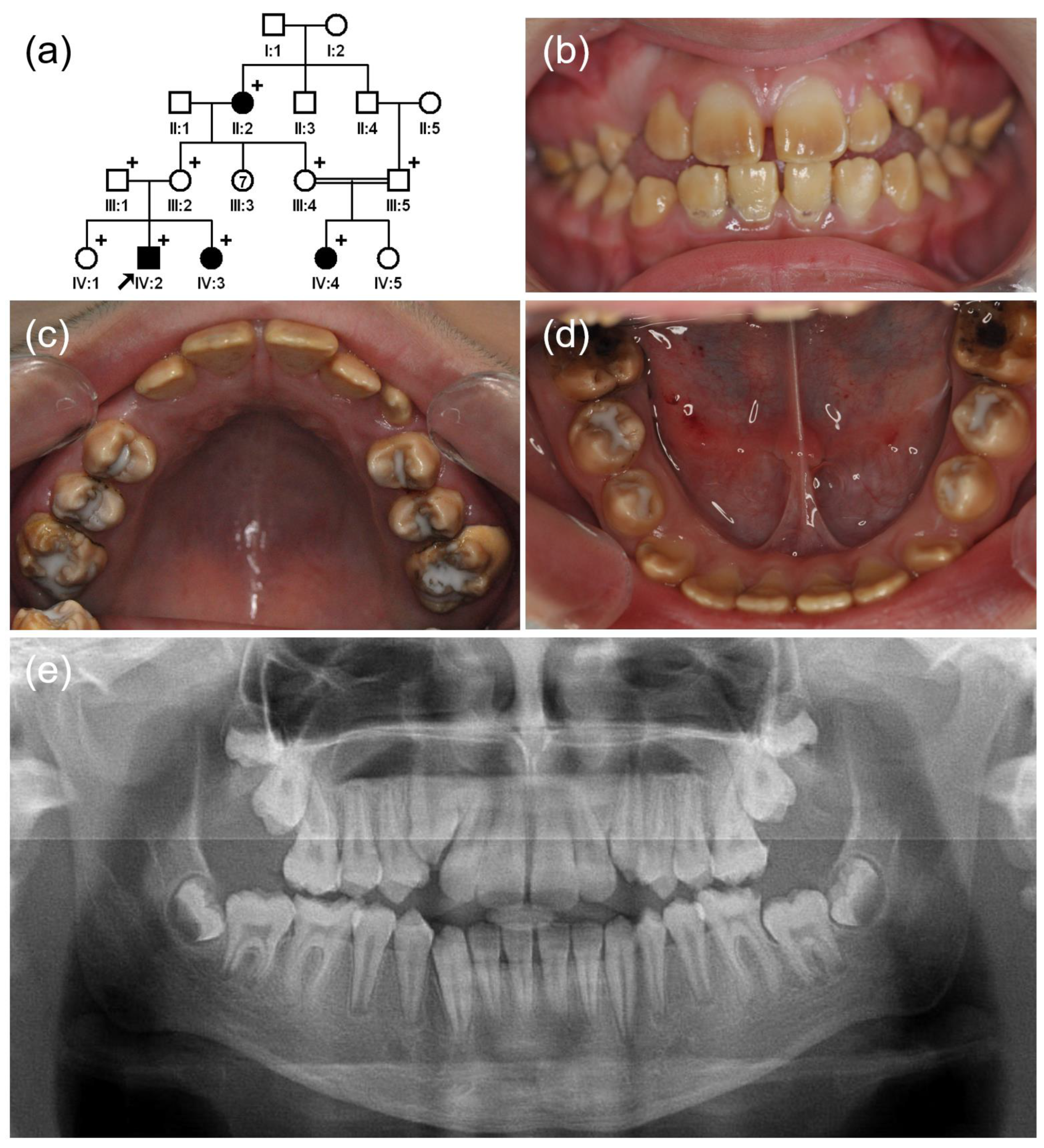
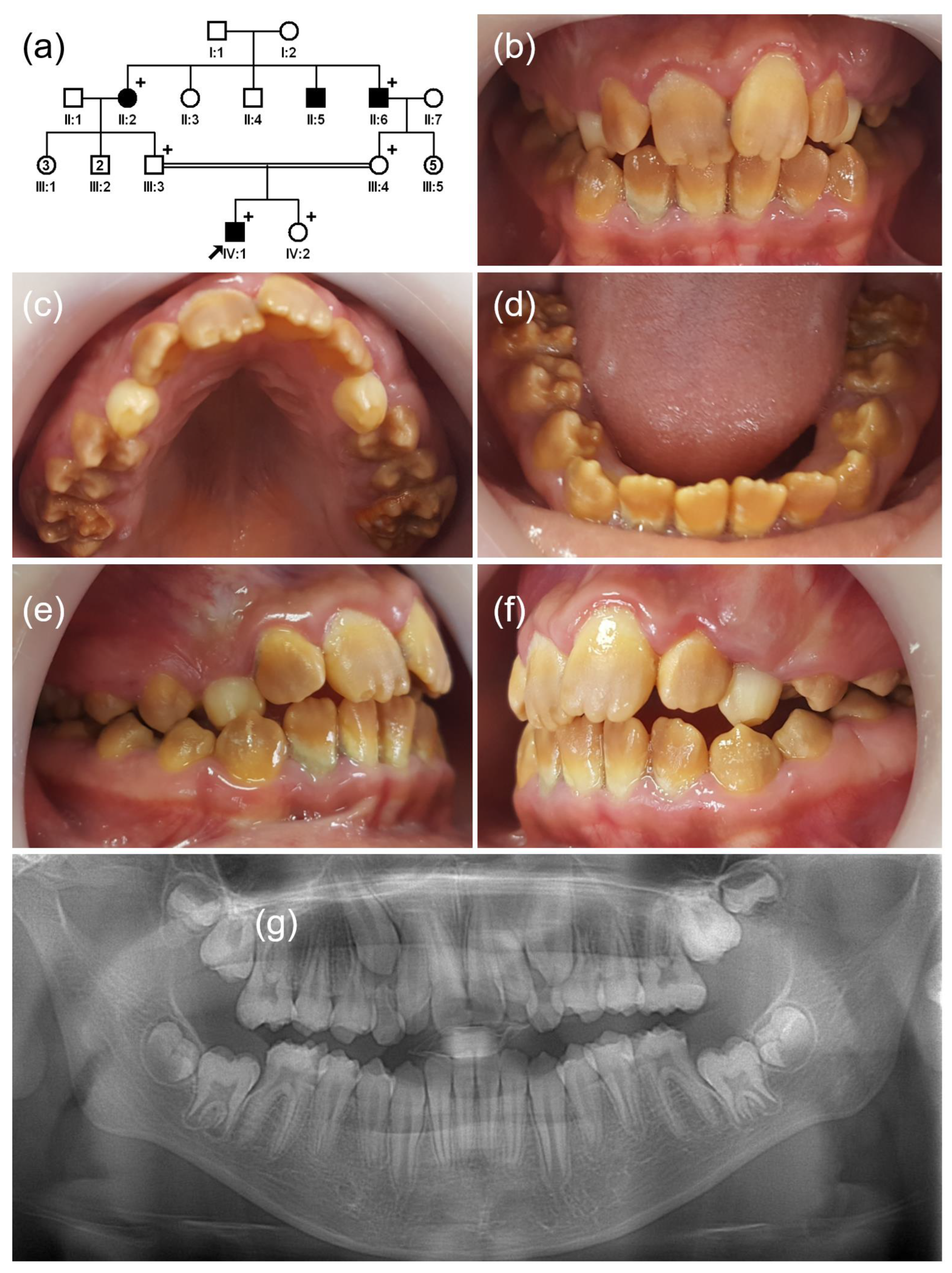
3.1. Family 1
3.2. Family 2
3.3. Family 3
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simmer, J.P.; Papagerakis, P.; Smith, C.E.; Fisher, D.C.; Rountrey, A.; Zheng, L.; Hu, J.C.-C. Regulation of Dental Enamel Shape and Hardness. J. Dent. Res. 2010, 89, 1024–1038. [Google Scholar] [CrossRef]
- Smith, C. Cellular and Chemical Events during Enamel Maturation. Crit. Rev. Oral Biol. Med. 1998, 9, 128–161. [Google Scholar] [CrossRef]
- Lacruz, R.S.; Nanci, A.; Kurtz, I.; Wright, J.T.; Paine, M.L. Regulation of pH During Amelogenesis. Calcif. Tissue Int. 2009, 86, 91–103. [Google Scholar] [CrossRef]
- Wright, J.T.; Carrion, I.A.; Morris, C. The Molecular Basis of Hereditary Enamel Defects in Humans. J. Dent. Res. 2015, 94, 52–61. [Google Scholar] [CrossRef]
- Witkop, C.J., Jr. Amelogenesis imperfecta, dentinogenesis imperfecta and dentin dysplasia revisited: Problems in classification. J. Oral Pathol. Med. 1988, 17, 547–553. [Google Scholar] [CrossRef]
- Coffield, K.D.; Phillips, C.; Brady, M.; Roberts, M.W.; Strauss, R.P.; Wright, J.T. The psychosocial impact of developmental dental defects in people with hereditary amelogenesis imperfecta. J. Am. Dent. Assoc. 2005, 136, 620–630. [Google Scholar] [CrossRef]
- Sujak, S.L.; Kadir, R.A.; Dom, T.N.M. Esthetic perception and psychosocial impact of developmental enamel defects among Malaysian adolescents. J. Oral Sci. 2004, 46, 221–226. [Google Scholar] [CrossRef]
- Fincham, A.G.; Belcourt, A.B.; Termine, J.D.; Butler, W.T.; Cothran, W.C. Dental enamel matrix: Sequences of two amelogenin polypeptides. Biosci. Rep. 1981, 1, 771–778. [Google Scholar] [CrossRef]
- Fincham, A.G.; Simmer, J.P. Amelogenin proteins of developing dental enamel. In Ciba Foundation Symposium 205-Dental Enamel: Dental Enamel: Ciba Foundation Symposium 205; Wiley & Sons, Ltd.: Chichester, UK, 2007; Volume 205, pp. 118–134. [Google Scholar] [CrossRef]
- Lagerström, M.; Dahl, N.; Nakahori, Y.; Nakagome, Y.; Bäckman, B.; Landegren, U.; Pettersson, U. A deletion in the amelogenin gene (AMG) causes X-linked amelogenesis imperfecta (AIH1). Genomics 1991, 10, 971–975. [Google Scholar] [CrossRef]
- Rajpar, M.H.; Harley, K.; Laing, C.; Davies, R.M.; Dixon, M.J. Mutation of the gene encoding the enamel-specific protein, enamelin, causes autosomal-dominant amelogenesis imperfecta. Hum. Mol. Genet. 2001, 10, 1673–1677. [Google Scholar] [CrossRef]
- Poulter, J.A.; Murillo, G.; Brookes, S.J.; Smith, C.E.L.; Parry, D.A.; Silva, S.; Kirkham, J.; Inglehearn, C.F.; Mighell, A.J. Deletion of ameloblastin exon 6 is associated with amelogenesis imperfecta. Hum. Mol. Genet. 2014, 23, 5317–5324. [Google Scholar] [CrossRef]
- Kim, J.W.; Zhang, H.; Seymen, F.; Koruyucu, M.; Hu, Y.; Kang, J.; Kim, Y.J.; Ikeda, A.; Kasimoglu, Y.; Bayram, M.; et al. Mutations in RELT cause autosomal recessive amelogenesis imperfecta. Clin. Genet. 2019, 95, 375–383. [Google Scholar] [CrossRef]
- Seymen, F.; Kim, Y.J.; Lee, Y.J.; Kang, J.; Kim, T.-H.; Choi, H.; Koruyucu, M.; Kasimoglu, Y.; Tuna, E.B.; Gencay, K.; et al. Recessive Mutations in ACPT, Encoding Testicular Acid Phosphatase, Cause Hypoplastic Amelogenesis Imperfecta. Am. J. Hum. Genet. 2016, 99, 1199–1205. [Google Scholar] [CrossRef][Green Version]
- Kim, J.-W.; Lee, S.-K.; Lee, Z.H.; Park, J.-C.; Lee, K.-E.; Lee, M.-H.; Park, J.-T.; Seo, B.-M.; Hu, J.C.-C.; Simmer, J.P. FAM83H Mutations in Families with Autosomal-Dominant Hypocalcified Amelogenesis Imperfecta. Am. J. Hum. Genet. 2008, 82, 489–494. [Google Scholar] [CrossRef]
- Parry, D.; Brookes, S.; Logan, C.; Poulter, J.; El-Sayed, W.; Al-Bahlani, S.; Al Harasi, S.; Sayed, J.; Raïf, E.M.; Shore, R.C.; et al. Mutations in C4orf26, Encoding a Peptide with In Vitro Hydroxyapatite Crystal Nucleation and Growth Activity, Cause Amelogenesis Imperfecta. Am. J. Hum. Genet. 2012, 91, 565–571. [Google Scholar] [CrossRef]
- Kim, J.-W.; Simmer, J.P.; Hart, T.C.; Hart, P.S.; Ramaswami, M.D.; Bartlett, J.D.; Hu, J.C.-C. MMP-20 mutation in autosomal recessive pigmented hypomaturation amelogenesis imperfecta. J. Med. Genet. 2005, 42, 271–275. [Google Scholar] [CrossRef]
- Hart, P.S.; Hart, T.C.; Michalec, M.D.; Ryu, O.H.; Simmons, D.; Hong, S.; Wright, J.T. Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta. J. Med. Genet. 2004, 41, 545–549. [Google Scholar] [CrossRef]
- El-Sayed, W.; Parry, D.A.; Shore, R.C.; Ahmed, M.; Jafri, H.; Rashid, Y.; Al-Bahlani, S.; Al Harasi, S.; Kirkham, J.; Inglehearn, C.F.; et al. Mutations in the Beta Propeller WDR72 Cause Autosomal-Recessive Hypomaturation Amelogenesis Imperfecta. Am. J. Hum. Genet. 2009, 85, 699–705. [Google Scholar] [CrossRef]
- Parry, D.; Poulter, J.; Logan, C.; Brookes, S.J.; Jafri, H.; Ferguson, C.H.; Anwari, B.M.; Rashid, Y.; Zhao, H.; Johnson, C.A.; et al. Identification of Mutations in SLC24A4, Encoding a Potassium-Dependent Sodium/Calcium Exchanger, as a Cause of Amelogenesis Imperfecta. Am. J. Hum. Genet. 2013, 92, 307–312. [Google Scholar] [CrossRef]
- Parry, D.A.; Smith, C.E.; El-Sayed, W.; Poulter, J.A.; Shore, R.C.; Logan, C.V.; Mogi, C.; Sato, K.; Okajima, F.; Harada, A.; et al. Mutations in the pH-Sensing G-protein-Coupled Receptor GPR68 Cause Amelogenesis Imperfecta. Am. J. Hum. Genet. 2016, 99, 984–990. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Van Der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar] [CrossRef]
- Miller, J.; Pearce, D.A. Nonsense-mediated decay in genetic disease: Friend or foe? Mutat. Res. Mutat. Res. 2014, 762, 52–64. [Google Scholar] [CrossRef]
- Brogna, S.; Wen, J. Nonsense-mediated mRNA decay (NMD) mechanisms. Nat. Struct. Mol. Biol. 2009, 16, 107–113. [Google Scholar] [CrossRef]
- Sisignano, M.; Fischer, M.; Geisslinger, G. Proton-Sensing GPCRs in Health and Disease. Cells 2021, 10, 2050. [Google Scholar] [CrossRef]
- Rowe, J.B.; Kapolka, N.J.; Taghon, G.J.; Morgan, W.M.; Isom, D.G. The evolution and mechanism of GPCR proton sensing. J. Biol. Chem. 2021, 296, 100167. [Google Scholar] [CrossRef]
- Sato, K.; Mogi, C.; Mighell, A.J.; Okajima, F. A missense mutation of Leu74Pro of OGR1 found in familial amelogenesis imperfecta actually causes the loss of the pH-sensing mechanism. Biochem. Biophys. Res. Commun. 2020, 526, 920–926. [Google Scholar] [CrossRef]
- Sulem, P.; Gudbjartsson, D.; Stacey, S.N.; Helgason, A.; Rafnar, T.; Magnusson, K.P.; Manolescu, A.; Karason, A.; Palsson, A.; Thorleifsson, G.; et al. Genetic determinants of hair, eye and skin pigmentation in Europeans. Nat. Genet. 2007, 39, 1443–1452. [Google Scholar] [CrossRef]
- Wang, S.-K.; Choi, M.; Richardson, A.; Reid, B.; Seymen, F.; Yildirim, M.; Tuna, E.; Gençay, K.; Simmer, J.; Hu, J. STIM1 and SLC24A4 Are Critical for Enamel Maturation. J. Dent. Res. 2014, 93, 94S–100S. [Google Scholar] [CrossRef]
- Li, X.-F.; Kraev, A.S.; Lytton, J. Molecular Cloning of a Fourth Member of the Potassium-dependent Sodium-Calcium Exchanger Gene Family, NCKX4. J. Biol. Chem. 2002, 277, 48410–48417. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, T.; Uehara, A.; Imanaga, I.; Shigekawa, M. The Na+/Ca2+ Exchanger NCX1 Has Oppositely Oriented Reentrant Loop Domains That Contain Conserved Aspartic Acids Whose Mutation Alters Its Apparent Ca2+Affinity. J. Biol. Chem. 2000, 275, 38571–38580. [Google Scholar] [CrossRef] [PubMed]
- Alam Khan, S.; Khan, M.A.; Muhammad, N.; Bashir, H.; Khan, N.; Muhammad, N.; Yilmaz, R.; Khan, S.; Wasif, N. A novel nonsense variant in SLC24A4 causing a rare form of amelogenesis imperfecta in a Pakistani family. BMC Med. Genet. 2020, 21, 97–99. [Google Scholar] [CrossRef] [PubMed]
- Herzog, C.R.; Reid, B.M.; Seymen, F.; Koruyucu, M.; Tuna, E.B.; Simmer, J.P.; Hu, J.C.-C. Hypomaturation amelogenesis imperfecta caused by a novel SLC24A4 mutation. Oral Surgery Oral Med. Oral Pathol. Oral Radiol. 2014, 119, e77–e81. [Google Scholar] [CrossRef] [PubMed]
- Seymen, F.; Gencay, K.; Lee, K.-E.; Le, C.T.; Lee, Z.; Kim, J.-W.; Yildirim, M. Exonal Deletion of SLC24A4 Causes Hypomaturation Amelogenesis Imperfecta. J. Dent. Res. 2014, 93, 366–370. [Google Scholar] [CrossRef]
- Lepperdinger, U.; Maurer, E.; Witsch-Baumgartner, M.; Stigler, R.; Zschocke, J.; Lussi, A.; Kapferer-Seebacher, I. Expanding the phenotype of hypomaturation amelogenesis imperfecta due to a novel SLC24A4 variant. Clin. Oral Investig. 2020, 24, 3519–3525. [Google Scholar] [CrossRef] [PubMed]

| Location | cDNA | Protein | Mode of Inheritance | References |
|---|---|---|---|---|
| Exon 2 | c.83_84del | p.(Tyr28Cysfs*146) | AR homo | This report |
| Exon 2 | c.221T>C | p.(Leu74Pro) | AR homo | Parry et al. (2016) [21] |
| Exon 2 | c.386_835del | p.(Phe129_Asn278del) | AR homo | Parry et al. (2016) [21] |
| Exon 2 | c.667_668delAA | p.(Lys223Glyfs*113) | AR homo | Parry et al. (2016) [21] |
| Location | cDNA | Protein | Mode of Inheritance | References |
|---|---|---|---|---|
| Exon 5 | c.437C>T | p.(Ala146Val) | AR homo | Wang et al. (2014) [31] This report |
| Exon 7 | c.613C>T | p.(Arg205*) | AR homo | This report |
| Exon 11 | c.1015C>T | p.(Arg339*) | AR homo | Parry et al. (2013) [20] |
| Exon 12 | c.1192C>T | p.(Gln398*) | AR homo | Khan et al. (2020) [34] |
| Exon 13 | c.1307T>G | p.(Leu436Arg) | AR homo | Herzog et al. (2015) [35] |
| Intron 14 | deletion of 10042 bp | (loss of exons 15–17) | AR homo | Seymen et al. (2014) [36] |
| Exon 14 | c.1495A>T | p.(Ser499Cys) | AR homo | Parry et al. (2013) [20] |
| Exon 15 | c.1604G>A | p.(Gly535Asp) | AR homo | Lepperdinger et al. (2020) [37] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seymen, F.; Zhang, H.; Kasimoglu, Y.; Koruyucu, M.; Simmer, J.P.; Hu, J.C.-C.; Kim, J.-W. Novel Mutations in GPR68 and SLC24A4 Cause Hypomaturation Amelogenesis Imperfecta. J. Pers. Med. 2022, 12, 13. https://doi.org/10.3390/jpm12010013
Seymen F, Zhang H, Kasimoglu Y, Koruyucu M, Simmer JP, Hu JC-C, Kim J-W. Novel Mutations in GPR68 and SLC24A4 Cause Hypomaturation Amelogenesis Imperfecta. Journal of Personalized Medicine. 2022; 12(1):13. https://doi.org/10.3390/jpm12010013
Chicago/Turabian StyleSeymen, Figen, Hong Zhang, Yelda Kasimoglu, Mine Koruyucu, James P. Simmer, Jan C.-C. Hu, and Jung-Wook Kim. 2022. "Novel Mutations in GPR68 and SLC24A4 Cause Hypomaturation Amelogenesis Imperfecta" Journal of Personalized Medicine 12, no. 1: 13. https://doi.org/10.3390/jpm12010013
APA StyleSeymen, F., Zhang, H., Kasimoglu, Y., Koruyucu, M., Simmer, J. P., Hu, J. C.-C., & Kim, J.-W. (2022). Novel Mutations in GPR68 and SLC24A4 Cause Hypomaturation Amelogenesis Imperfecta. Journal of Personalized Medicine, 12(1), 13. https://doi.org/10.3390/jpm12010013







