Association between Gut Microbiota and Emotional-Behavioral Symptoms in Children with Attention-Deficit/Hyperactivity Disorder
Abstract
:1. Introduction
2. Materials and Methods
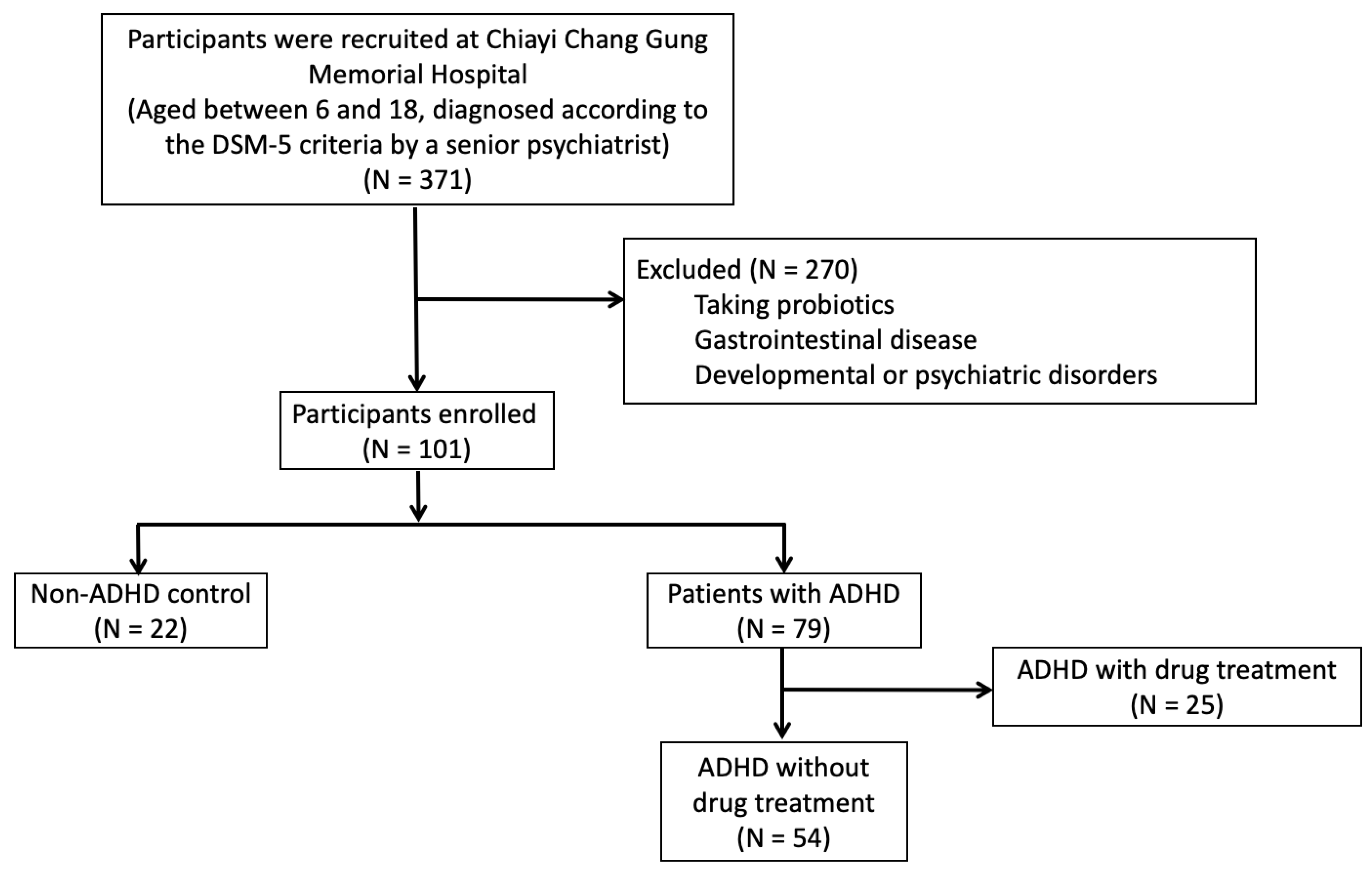
2.1. Participants
2.2. Sample Collection and DNA Extraction
2.3. Sequencing
2.4. Data Processing and Analysis
2.5. Measurements
2.6. Statistical and Bioinformatic Analysis
3. Results
3.1. Demographic Data
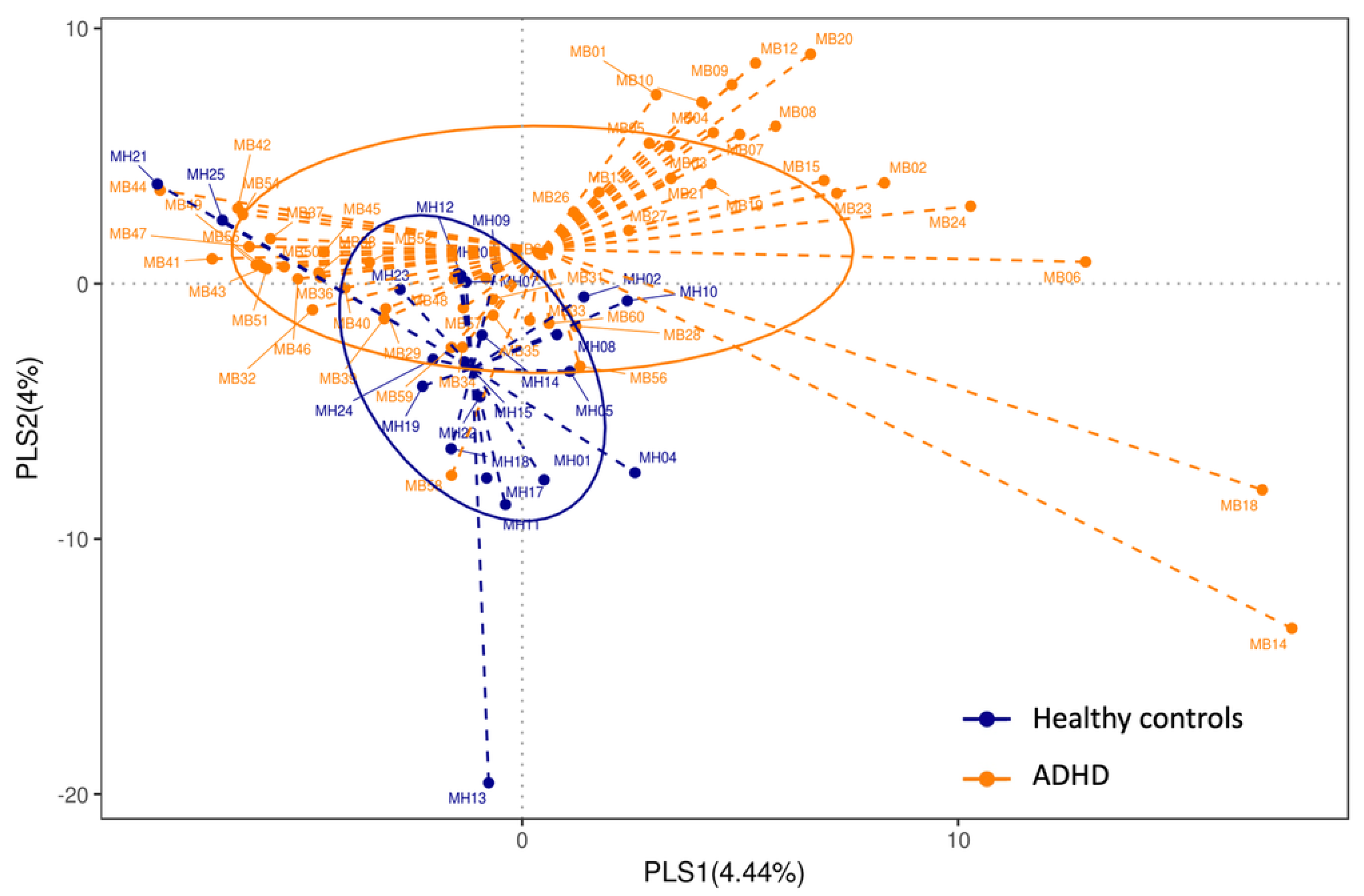
3.2. Alpha Diversity and Beta Diversity
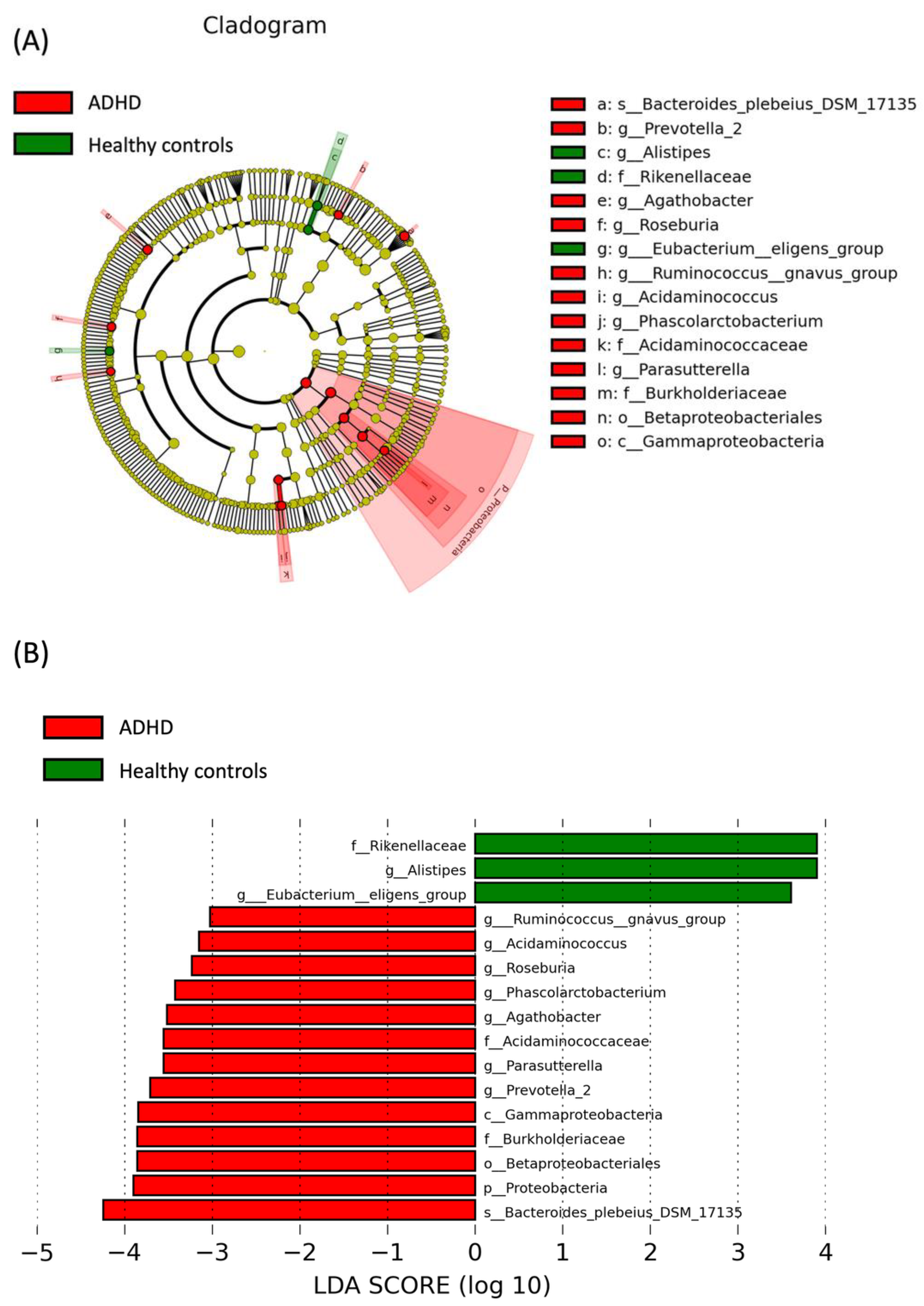
3.3. LEfSe Analysis
3.4. Association between Clinical Symptoms and Relative Abundance of Bacteria
4. Adjusted for Sex, Age and Score of SNAP-Iv Discussion
Strengths and Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Polanczyk, G.V.; Willcutt, E.G.; Salum, G.A.; Kieling, C.; Rohde, L.A. ADHD prevalence estimates across three decades: An updated systematic review and meta-regression analysis. Int. J. Epidemiol. 2014, 43, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.J.; Yang, C.Y.; Chou, W.J.; Lee, M.J.; Chou, M.C.; Kuo, H.C.; Yeh, Y.M.; Lee, S.Y.; Huang, L.H.; Li, S.C. Gut microbiota and dietary patterns in children with attention-deficit/hyperactivity disorder. Eur. Child Adolesc. Psychiatry 2020, 29, 287–297. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.C.; Yang, Y.H.; Yu Kuo, T.; Lu, M.L.; Tseng, W.T.; Hou, T.Y.; Yeh, J.Y.; Lee, C.T.; Chen, Y.L.; Lee, M.J.; et al. Methylphenidate and the risk of burn injury among children with attention-deficit/hyperactivity disorder. Epidemiol. Psychiatr. Sci. 2020, 29, e146. [Google Scholar] [CrossRef]
- Faraone, S.V.; Rostain, A.L.; Blader, J.; Busch, B.; Childress, A.C.; Connor, D.F.; Newcorn, J.H. Practitioner Review: Emotional dysregulation in attention-deficit/hyperactivity disorder—Implications for clinical recognition and intervention. J. Child Psychol. Psychiatry 2019, 60, 133–150. [Google Scholar] [CrossRef]
- Surman, C.B.; Biederman, J.; Spencer, T.; Miller, C.A.; McDermott, K.M.; Faraone, S.V. Understanding deficient emotional self-regulation in adults with attention deficit hyperactivity disorder: A controlled study. Atten. Deficit Hyperact. Disord. 2013, 5, 273–281. [Google Scholar] [CrossRef] [Green Version]
- Gallo, E.F.; Posner, J. Moving towards causality in attention-deficit hyperactivity disorder: Overview of neural and genetic mechanisms. Lancet Psychiatry 2016, 3, 555–567. [Google Scholar] [CrossRef] [Green Version]
- Richarte, V.; Rosales, K.; Corrales, M.; Bellina, M.; Fadeuilhe, C.; Calvo, E.; Ibanez, P.; Sanchez-Mora, C.; Ribases, M.; Ramos-Quiroga, J.A. The gut-brain axis in attention deficit hyperactivity disorder: The role of the microbiota. Rev. Neurol. 2018, 66, S109–S114. [Google Scholar]
- Sandgren, A.M.; Brummer, R.J.M. ADHD-originating in the gut? The emergence of a new explanatory model. Med. Hypotheses 2018, 120, 135–145. [Google Scholar] [CrossRef]
- Lima-Ojeda, J.M.; Rupprecht, R.; Baghai, T.C. “I Am I and My Bacterial Circumstances”: Linking Gut Microbiome, Neurodevelopment, and Depression. Front. Psychiatry 2017, 8, 153. [Google Scholar] [CrossRef] [Green Version]
- Cenit, M.C.; Sanz, Y.; Codoñer-Franch, P. Influence of gut microbiota on neuropsychiatric disorders. World J. Gastroenterol. 2017, 23, 5486–5498. [Google Scholar] [CrossRef] [Green Version]
- Malan-Muller, S.; Valles-Colomer, M.; Raes, J.; Lowry, C.A.; Seedat, S.; Hemmings, S.M.J. The Gut Microbiome and Mental Health: Implications for Anxiety- and Trauma-Related Disorders. Omics 2018, 22, 90–107. [Google Scholar] [CrossRef] [PubMed]
- Cenit, M.C.; Nuevo, I.C.; Codoñer-Franch, P.; Dinan, T.G.; Sanz, Y. Gut microbiota and attention deficit hyperactivity disorder: New perspectives for a challenging condition. Eur. Child Adolesc. Psychiatry 2017, 26, 1081–1092. [Google Scholar] [CrossRef] [PubMed]
- Srikantha, P.; Mohajeri, M.H. The Possible Role of the Microbiota-Gut-Brain-Axis in Autism Spectrum Disorder. Int. J. Mol. Sci. 2019, 20, 2115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, G.; Grenham, S.; Scully, P.; Fitzgerald, P.; Moloney, R.D.; Shanahan, F.; Dinan, T.G.; Cryan, J.F. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol. Psychiatry 2013, 18, 666–673. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Ling, Z.; Zhang, Y.; Mao, H.; Ma, Z.; Yin, Y.; Wang, W.; Tang, W.; Tan, Z.; Shi, J.; et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 2015, 48, 186–194. [Google Scholar] [CrossRef] [Green Version]
- Aarts, E.; Ederveen, T.H.A.; Naaijen, J.; Zwiers, M.P.; Boekhorst, J.; Timmerman, H.M.; Smeekens, S.P.; Netea, M.G.; Buitelaar, J.K.; Franke, B.; et al. Gut microbiome in ADHD and its relation to neural reward anticipation. PLoS ONE 2017, 12, e0183509. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.Y.; Zhou, Y.Y.; Zhou, G.L.; Li, Y.C.; Yuan, J.; Li, X.H.; Ruan, B. Gut microbiota profiles in treatment-naïve children with attention deficit hyperactivity disorder. Behav. Brain Res. 2018, 347, 408–413. [Google Scholar] [CrossRef]
- Wan, L.; Ge, W.R.; Zhang, S.; Sun, Y.L.; Wang, B.; Yang, G. Case-Control Study of the Effects of Gut Microbiota Composition on Neurotransmitter Metabolic Pathways in Children With Attention Deficit Hyperactivity Disorder. Front. Neurosci. 2020, 14, 127. [Google Scholar] [CrossRef] [Green Version]
- Prehn-Kristensen, A.; Zimmermann, A.; Tittmann, L.; Lieb, W.; Schreiber, S.; Baving, L.; Fischer, A. Reduced microbiome alpha diversity in young patients with ADHD. PLoS ONE 2018, 13, e0200728. [Google Scholar] [CrossRef] [Green Version]
- Szopinska-Tokov, J.; Dam, S.; Naaijen, J.; Konstanti, P.; Rommelse, N.; Belzer, C.; Buitelaar, J.; Franke, B.; Aarts, E.; Arias Vasquez, A. Investigating the Gut Microbiota Composition of Individuals with Attention-Deficit/Hyperactivity Disorder and Association with Symptoms. Microorganisms 2020, 8, 406. [Google Scholar] [CrossRef] [Green Version]
- Reale, L.; Bartoli, B.; Cartabia, M.; Zanetti, M.; Costantino, M.A.; Canevini, M.P.; Termine, C.; Bonati, M. Comorbidity prevalence and treatment outcome in children and adolescents with ADHD. Eur. Child Adolesc. Psychiatry 2017, 26, 1443–1457. [Google Scholar] [CrossRef] [PubMed]
- Bussing, R.; Fernandez, M.; Harwood, M.; Wei, H.; Garvan, C.W.; Eyberg, S.M.; Swanson, J.M. Parent and teacher SNAP-IV ratings of attention deficit hyperactivity disorder symptoms: Psychometric properties and normative ratings from a school district sample. Assessment 2008, 15, 317–328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef]
- Chen, T.; Yu, W.H.; Izard, J.; Baranova, O.V.; Lakshmanan, A.; Dewhirst, F.E. The Human Oral Microbiome Database: A web accessible resource for investigating oral microbe taxonomic and genomic information. Database 2010, 2010, baq013. [Google Scholar] [CrossRef] [Green Version]
- Escapa, I.F.; Chen, T.; Huang, Y.; Gajare, P.; Dewhirst, F.E.; Lemon, K.P. New Insights into Human Nostril Microbiome from the Expanded Human Oral Microbiome Database (eHOMD): A Resource for the Microbiome of the Human Aerodigestive Tract. mSystems 2018, 3, e00187-18. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2014, 42, D643–D648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caporaso, J.G.; Bittinger, K.; Bushman, F.D.; DeSantis, T.Z.; Andersen, G.L.; Knight, R. PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics 2010, 26, 266–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Gau, S.S.; Shang, C.Y.; Liu, S.K.; Lin, C.H.; Swanson, J.M.; Liu, Y.C.; Tu, C.L. Psychometric properties of the Chinese version of the Swanson, Nolan, and Pelham, version IV scale–parent form. Int. J. Methods Psychiatr. Res. 2008, 17, 35–44. [Google Scholar] [CrossRef]
- Leung, P.W.; Kwong, S.L.; Tang, C.P.; Ho, T.P.; Hung, S.F.; Lee, C.C.; Hong, S.L.; Chiu, C.M.; Liu, W.S. Test-retest reliability and criterion validity of the Chinese version of CBCL, TRF, and YSR. J. Child Psychol. Psychiatry 2006, 47, 970–973. [Google Scholar] [CrossRef]
- Yang, H.J.; Soong, W.T.; Chiang, C.N.; Chen, W.J. Competence and behavioral/emotional problems among Taiwanese adolescents as reported by parents and teachers. J. Am. Acad. Child Adolesc. Psychiatry 2000, 39, 232–239. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [Green Version]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2011, 5, 169–172. [Google Scholar] [CrossRef] [Green Version]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breban, M.; Tap, J.; Leboime, A.; Said-Nahal, R.; Langella, P.; Chiocchia, G.; Furet, J.P.; Sokol, H. Faecal microbiota study reveals specific dysbiosis in spondyloarthritis. Ann. Rheum. Dis. 2017, 76, 1614–1622. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Liang, H.; Wang, Y.; Miao, M.; Shi, T.; Yang, F.; Liu, E.; Yuan, W.; Ji, Z.S.; Li, D.K. Altered Gut Microbiota Composition Associated with Eczema in Infants. PLoS ONE 2016, 11, e0166026. [Google Scholar] [CrossRef] [Green Version]
- Henke, M.T.; Kenny, D.J.; Cassilly, C.D.; Vlamakis, H.; Xavier, R.J.; Clardy, J. Ruminococcus gnavus, a member of the human gut microbiome associated with Crohn’s disease, produces an inflammatory polysaccharide. Proc. Natl. Acad. Sci. USA 2019, 116, 12672–12677. [Google Scholar] [CrossRef] [Green Version]
- Willing, B.P.; Dicksved, J.; Halfvarson, J.; Andersson, A.F.; Lucio, M.; Zheng, Z.; Järnerot, G.; Tysk, C.; Jansson, J.K.; Engstrand, L. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 2010, 139, 1844–1854.e1841. [Google Scholar] [CrossRef]
- Joossens, M.; Huys, G.; Cnockaert, M.; De Preter, V.; Verbeke, K.; Rutgeerts, P.; Vandamme, P.; Vermeire, S. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 2011, 60, 631–637. [Google Scholar] [CrossRef] [Green Version]
- Hall, A.B.; Yassour, M.; Sauk, J.; Garner, A.; Jiang, X.; Arthur, T.; Lagoudas, G.K.; Vatanen, T.; Fornelos, N.; Wilson, R.; et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 2017, 9, 103. [Google Scholar] [CrossRef]
- Painold, A.; Mörkl, S.; Kashofer, K.; Halwachs, B.; Dalkner, N.; Bengesser, S.; Birner, A.; Fellendorf, F.; Platzer, M.; Queissner, R.; et al. A step ahead: Exploring the gut microbiota in inpatients with bipolar disorder during a depressive episode. Bipolar Disord. 2019, 21, 40–49. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.T.; Kosciolek, T.; Maldonado, Y.; Daly, R.E.; Martin, A.S.; McDonald, D.; Knight, R.; Jeste, D.V. Differences in gut microbiome composition between persons with chronic schizophrenia and healthy comparison subjects. Schizophr. Res. 2019, 204, 23–29. [Google Scholar] [CrossRef]
- Rose, D.R.; Yang, H.; Serena, G.; Sturgeon, C.; Ma, B.; Careaga, M.; Hughes, H.K.; Angkustsiri, K.; Rose, M.; Hertz-Picciotto, I.; et al. Differential immune responses and microbiota profiles in children with autism spectrum disorders and co-morbid gastrointestinal symptoms. Brain Behav. Immun. 2018, 70, 354–368. [Google Scholar] [CrossRef] [PubMed]
- Tengeler, A.C.; Dam, S.A.; Wiesmann, M.; Naaijen, J.; van Bodegom, M.; Belzer, C.; Dederen, P.J.; Verweij, V.; Franke, B.; Kozicz, T.; et al. Gut microbiota from persons with attention-deficit/hyperactivity disorder affects the brain in mice. Microbiome 2020, 8, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cervera-Tison, M.; Tailford, L.E.; Fuell, C.; Bruel, L.; Sulzenbacher, G.; Henrissat, B.; Berrin, J.G.; Fons, M.; Giardina, T.; Juge, N. Functional analysis of family GH36 α-galactosidases from Ruminococcus gnavus E1: Insights into the metabolism of a plant oligosaccharide by a human gut symbiont. Appl. Environ. Microbiol. 2012, 78, 7720–7732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crost, E.H.; Tailford, L.E.; Le Gall, G.; Fons, M.; Henrissat, B.; Juge, N. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS ONE 2013, 8, e76341. [Google Scholar] [CrossRef] [Green Version]
- Dam, S.A.; Mostert, J.C.; Szopinska-Tokov, J.W.; Bloemendaal, M.; Amato, M.; Arias-Vasquez, A. The Role of the Gut-Brain Axis in Attention-Deficit/Hyperactivity Disorder. Gastroenterol. Clin. N. Am. 2019, 48, 407–431. [Google Scholar] [CrossRef]
- Liu, S.; Li, E.; Sun, Z.; Fu, D.; Duan, G.; Jiang, M.; Yu, Y.; Mei, L.; Yang, P.; Tang, Y.; et al. Altered gut microbiota and short chain fatty acids in Chinese children with autism spectrum disorder. Sci. Rep. 2019, 9, 287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosero, J.A.; Killer, J.; Sechovcová, H.; Mrázek, J.; Benada, O.; Fliegerová, K.; Havlík, J.; Kopečný, J. Reclassification of Eubacterium rectale (Hauduroy et al. 1937) Prévot 1938 in a new genus Agathobacter gen. nov. as Agathobacter rectalis comb. nov., and description of Agathobacter ruminis sp. nov., isolated from the rumen contents of sheep and cows. Int. J. Syst. Evol. Microbiol. 2016, 66, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, G.T.; Macfarlane, S. Fermentation in the human large intestine: Its physiologic consequences and the potential contribution of prebiotics. J. Clin. Gastroenterol. 2011, 45, S120–S127. [Google Scholar] [CrossRef] [Green Version]
- Cheung, S.G.; Goldenthal, A.R.; Uhlemann, A.C.; Mann, J.J.; Miller, J.M.; Sublette, M.E. Systematic Review of Gut Microbiota and Major Depression. Front. Psychiatry 2019, 10, 34. [Google Scholar] [CrossRef] [Green Version]
- Bhandari, S.; Larson, M.E.; Kumar, N.; Stein, D. Association of Inflammatory Bowel Disease (IBD) with Depressive Symptoms in the United States Population and Independent Predictors of Depressive Symptoms in an IBD Population: A NHANES Study. Gut Liver 2017, 11, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.J.; Li, S.C.; Li, S.W.; Kuo, H.C.; Lee, S.Y.; Huang, L.H.; Chin, C.Y.; Yang, C.Y. Gut microbiota and plasma cytokine levels in patients with attention-deficit/hyperactivity disorder. Transl. Psychiatry 2022, 12, 76. [Google Scholar] [CrossRef] [PubMed]
- Hua, X.; Zhu, J.; Yang, T.; Guo, M.; Li, Q.; Chen, J.; Li, T. The Gut Microbiota and Associated Metabolites Are Altered in Sleep Disorder of Children With Autism Spectrum Disorders. Front. Psychiatry 2020, 11, 855. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Liu, G.; Ding, M.; Zong, G.; Hu, F.B.; Willett, W.C.; Rimm, E.B.; Manson, J.E.; Sun, Q. Isoflavone Intake and the Risk of Coronary Heart Disease in US Men and Women: Results from 3 Prospective Cohort Studies. Circulation 2020, 141, 1127–1137. [Google Scholar] [CrossRef]
- Curtis, P.J.; Sampson, M.; Potter, J.; Dhatariya, K.; Kroon, P.A.; Cassidy, A. Chronic ingestion of flavan-3-ols and isoflavones improves insulin sensitivity and lipoprotein status and attenuates estimated 10-year CVD risk in medicated postmenopausal women with type 2 diabetes: A 1-year, double-blind, randomized, controlled trial. Diabetes Care 2012, 35, 226–232. [Google Scholar] [CrossRef]

| ADHD Children with Treatment-Naïve (N = 54) | Healthy Controls (N = 22) | p-Value | Effect Size a | |
|---|---|---|---|---|
| Sex, n (%) | 0.042 * | 3.046 | ||
| Male | 44 (81.5%) | 13 (59.1%) | ||
| Female | 10 (18.5%) | 9 (40.9%) | ||
| Age (mean ± SD year) | 8.39 (1.75) | 9.73 (2.23) | 0.007 * | 0.669 |
| BMI (mean ± SD) | 18.32 (3.91) | 19.87 (3.72) | 0.112 | 0.406 |
| Preterm birth, n (%) | 5 (9.3%) | 2 (9.1%) | 0.675 | 1.020 |
| Allergic rhinitis, n (%) | 19 (35.2%) | 7 (31.8%) | 0.499 | 1.163 |
| SNAP-IV | ||||
| Inattention (SD) | 15.15 (5.7) | 4.5 (4.53) | <0.001 * | 2.069 |
| Hyperactivity/Impulsivity (SD) | 11.65 (6.37) | 3.41 (4.67) | <0.001 * | 1.475 |
| Opposition/defiance (SD) | 9.83 (6.26) | 5.64 (5.28) | 0.007 * | 0.724 |
| Total Score (SD) | 26.8 (10.58) | 7.91 (8.19) | <0.001 * | 1.997 |
| CBCL syndromes, median (SD) | ||||
| Anxious/Depressed | 59.15 (8.72) | 54.91 (7.09) | 0.047 * | 0.534 |
| Withdrawn/Depressed | 60.41 (9.26) | 55.82 (5.35) | 0.033 * | 0.607 |
| Somatic Complaints | 56.7 (5.79) | 53.95 (5.75) | 0.064 | 0.477 |
| Social Problems | 63.72 (8.33) | 55.5 (8.76) | <0.001 * | 0.962 |
| Thought Problems | 59.52 (7.91) | 53.45 (5.35) | 0.002 * | 0.899 |
| Attention Problems | 68.93 (9.8) | 55.45 (6.06) | <0.001 * | 1.654 |
| Rule-Breaking Behavior | 62.2 (7.94) | 53.86 (5.33) | <0.001 * | 1.233 |
| Aggressive Behavior | 62.59 (8.56) | 53.91 (5.42) | <0.001 * | 1.212 |
| Internalising | 58.48 (10.54) | 51.36 (10.09) | 0.009 * | 0.690 |
| Externalising | 62.41 (9.36) | 49.77 (9.05) | <0.001 * | 1.373 |
| Total Score | 64.22 (8.63) | 50.09 (11.21) | <0.001 * | 1.413 |
| Genera | Inattention | Hyperactivity/Impulsivity | Total Score | |||
|---|---|---|---|---|---|---|
| r | p | r | p | r | p | |
| Proteobacteria | 0.018 | 0.899 | 0.012 | 0.932 | 0.041 | 0.768 |
| Gammaproteobacteria | −0.065 | 0.641 | −0.030 | 0.829 | −0.022 | 0.874 |
| Betaproteobacteriales | 0.078 | 0.575 | −0.082 | 0.558 | 0.007 | 0.959 |
| Burkholderiaceae | 0.078 | 0.575 | −0.082 | 0.558 | 0.007 | 0.959 |
| Acidaminococcaceae | −0.107 | 0.443 | −0.125 | 0.368 | −0.120 | 0.389 |
| Agathobacter | 0.236 | 0.086 | 0.176 | 0.204 | 0.236 | 0.086 |
| Phascolarctobacterium | −0.070 | 0.617 | −0.109 | 0.433 | −0.097 | 0.485 |
| Prevotella_2 | 0.134 | 0.336 | −0.071 | 0.612 | 0.021 | 0.881 |
| Parasutterella | 0.108 | 0.437 | −0.042 | 0.762 | 0.037 | 0.793 |
| Acidaminococcus | 0.116 | 0.402 | 0.082 | 0.558 | 0.124 | 0.372 |
| Roseburia | 0.024 | 0.864 | 0.134 | 0.333 | 0.090 | 0.518 |
| Ruminococcus_gnavus_group | −0.054 | 0.700 | 0.070 | 0.615 | 0.016 | 0.906 |
| Bacteroides_plebeius_DSM_17135 | 0.157 | 0.256 | 0.039 | 0.779 | 0.094 | 0.498 |
| Rikenellaceae | −0.074 | 0.743 | −0.022 | 0.921 | −0.032 | 0.888 |
| Alistipes | −0.074 | 0.743 | −0.022 | 0.921 | −0.032 | 0.888 |
| Eubacterium_eligens_group | 0.289 | 0.191 | −0.057 | 0.801 | 0.124 | 0.582 |
| Genera | Withdrawn /Depressed | Somatic Complaints | Social Problems | Thought Problems | Attention Problems | Rule-Breaking Behavior | Aggressive Behavior | Internalizing | Externalizing | Total Score | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r | p | r | p | r | p | r | p | r | p | r | p | r | p | r | p | r | p | r | p | |
| Proteobacteria | 0.210 | 0.127 | 0.225 | 0.102 | 0.153 | 0.268 | 0.119 | 0.393 | 0.152 | 0.273 | 0.106 | 0.446 | 0.162 | 0.242 | 0.222 | 0.107 | 0.148 | 0.284 | 0.192 | 0.165 |
| Gammaproteobacteria | 0.197 | 0.154 | 0.190 | 0.168 | 0.180 | 0.194 | 0.081 | 0.560 | 0.126 | 0.364 | 0.109 | 0.433 | 0.155 | 0.263 | 0.210 | 0.127 | 0.150 | 0.280 | 0.180 | 0.193 |
| Betaproteobacteriales | 0.057 | 0.683 | 0.023 | 0.870 | −0.071 | 0.610 | −0.053 | 0.705 | −0.142 | 0.305 | −0.043 | 0.757 | −0.067 | 0.630 | −0.025 | 0.860 | −0.076 | 0.586 | −0.095 | 0.495 |
| Burkholderiaceae | 0.057 | 0.683 | 0.023 | 0.870 | −0.071 | 0.610 | −0.053 | 0.705 | −0.142 | 0.305 | −0.043 | 0.757 | −0.067 | 0.630 | −0.025 | 0.860 | −0.076 | 0.586 | −0.095 | 0.495 |
| Acidaminococcaceae | −0.039 | 0.779 | 0.073 | 0.602 | −0.077 | 0.582 | 0.122 | 0.380 | −0.026 | 0.853 | −0.049 | 0.726 | −0.051 | 0.716 | 0.040 | 0.774 | −0.057 | 0.680 | −0.030 | 0.832 |
| Agathobacter | 0.317 | 0.020 * | 0.119 | 0.391 | 0.241 | 0.079 | 0.250 | 0.069 | 0.120 | 0.389 | 0.162 | 0.241 | 0.083 | 0.551 | 0.146 | 0.292 | 0.109 | 0.431 | 0.184 | 0.184 |
| Phascolarctobacterium | −0.139 | 0.318 | 0.104 | 0.454 | −0.161 | 0.244 | 0.042 | 0.762 | −0.049 | 0.727 | −0.132 | 0.341 | −0.135 | 0.331 | −0.032 | 0.816 | −0.144 | 0.299 | −0.107 | 0.442 |
| Prevotella_2 | 0.000 | 0.999 | −0.019 | 0.894 | 0.095 | 0.494 | −0.019 | 0.890 | −0.097 | 0.485 | 0.081 | 0.562 | 0.061 | 0.661 | −0.038 | 0.788 | 0.097 | 0.486 | 0.029 | 0.838 |
| Parasutterella | −0.044 | 0.750 | 0.006 | 0.963 | −0.155 | 0.264 | −0.017 | 0.901 | −0.156 | 0.259 | −0.160 | 0.248 | −0.134 | 0.335 | −0.060 | 0.668 | −0.161 | 0.246 | −0.148 | 0.287 |
| Acidaminococcus | 0.288 | 0.035 * | 0.082 | 0.556 | 0.223 | 0.105 | 0.313 | 0.021 * | 0.202 | 0.143 | 0.224 | 0.104 | 0.204 | 0.140 | 0.154 | 0.265 | 0.221 | 0.108 | 0.244 | 0.076 |
| Roseburia | −0.058 | 0.678 | −0.103 | 0.461 | 0.104 | 0.456 | 0.003 | 0.986 | −0.073 | 0.599 | 0.058 | 0.678 | 0.030 | 0.828 | −0.095 | 0.493 | 0.056 | 0.685 | 0.010 | 0.943 |
| Ruminococcus_gnavus_group | 0.195 | 0.157 | 0.151 | 0.277 | 0.175 | 0.205 | 0.264 | 0.054 | 0.242 | 0.077 | 0.272 | 0.046 * | 0.295 | 0.031 * | 0.251 | 0.068 | 0.285 | 0.036 * | 0.291 | 0.033 * |
| Bacteroides_plebeius_DSM_17135 | −0.033 | 0.814 | 0.027 | 0.845 | −0.080 | 0.567 | 0.008 | 0.953 | 0.058 | 0.678 | −0.104 | 0.454 | −0.113 | 0.416 | −0.060 | 0.664 | −0.090 | 0.518 | −0.035 | 0.799 |
| Rikenellaceae | −0.131 | 0.562 | 0.505 | 0.016 * | 0.119 | 0.598 | 0.088 | 0.697 | 0.058 | 0.799 | −0.138 | 0.540 | 0.162 | 0.472 | 0.341 | 0.121 | 0.038 | 0.868 | 0.225 | 0.315 |
| Alistipes | −0.131 | 0.562 | 0.505 | 0.016 * | 0.119 | 0.598 | 0.088 | 0.697 | 0.058 | 0.799 | −0.138 | 0.540 | 0.162 | 0.472 | 0.341 | 0.121 | 0.038 | 0.868 | 0.225 | 0.315 |
| Eubacterium_eligens_group | −0.224 | 0.317 | −0.112 | 0.620 | 0.048 | 0.832 | 0.108 | 0.634 | −0.022 | 0.923 | −0.112 | 0.620 | −0.210 | 0.348 | −0.113 | 0.615 | −0.217 | 0.332 | −0.100 | 0.658 |
| Genera | Withdrawn/Depressed | Thought Problems | Rule-Breaking Behavior | Aggressive Behavior | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | SE | p Value | B | SE | p Value | B | SE | p Value | B | SE | p Value | |
| Agathobacter | 0.002 | 0.001 | 0.044 * | |||||||||
| Acidaminococcus | 0.003 | 0.002 | 0.124 | 0.002 | 0.001 | 0.079 | ||||||
| Ruminococcus_gnavus_group | 0.005 | 0.003 | 0.046 * | 0.004 | 0.003 | 0.111 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, M.-J.; Lai, H.-C.; Kuo, Y.-L.; Chen, V.C.-H. Association between Gut Microbiota and Emotional-Behavioral Symptoms in Children with Attention-Deficit/Hyperactivity Disorder. J. Pers. Med. 2022, 12, 1634. https://doi.org/10.3390/jpm12101634
Lee M-J, Lai H-C, Kuo Y-L, Chen VC-H. Association between Gut Microbiota and Emotional-Behavioral Symptoms in Children with Attention-Deficit/Hyperactivity Disorder. Journal of Personalized Medicine. 2022; 12(10):1634. https://doi.org/10.3390/jpm12101634
Chicago/Turabian StyleLee, Min-Jing, Hsin-Chih Lai, Yu-Lun Kuo, and Vincent Chin-Hung Chen. 2022. "Association between Gut Microbiota and Emotional-Behavioral Symptoms in Children with Attention-Deficit/Hyperactivity Disorder" Journal of Personalized Medicine 12, no. 10: 1634. https://doi.org/10.3390/jpm12101634
APA StyleLee, M.-J., Lai, H.-C., Kuo, Y.-L., & Chen, V. C.-H. (2022). Association between Gut Microbiota and Emotional-Behavioral Symptoms in Children with Attention-Deficit/Hyperactivity Disorder. Journal of Personalized Medicine, 12(10), 1634. https://doi.org/10.3390/jpm12101634






