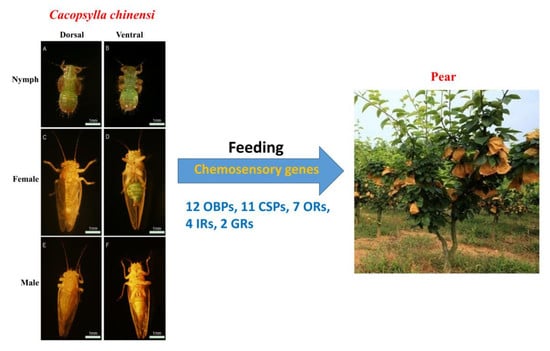
Chemosensory Gene Families in the Oligophagous Pear Pest Cacopsylla chinensis (Hemiptera: Psyllidae)
Abstract
1. Introduction
2. Material and Methods
2.1. Tissue Sample Collection
2.2. Total RNA Extraction and cDNA Synthesis
2.3. Library Preparation for Transcriptome Sequencing
2.4. De Novo Assembly and Unigene Annotation
2.5. Phylogenetic Analysis
2.6. Quantitative Real-Time PCR
2.7. Statistical Analysis
3. Results
3.1. Transcriptome Sequencing and Assembly
3.2. Homology Analysis
3.3. Non-Receptor Chemosensory Gene Families
Odorant Binding Proteins (OBPs)
3.4. Chemosensory Proteins (CSPs)
3.5. Chemosensory Receptor Gene Families
Odorant receptors (ORs), ionotropic receptors (IRs), and gustatory receptors (GRs)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Field, L.M.; Pickett, J.A.; Wadhams, L.J. Molecular studies in insect olfaction. Insect Mol. Biol. 2000, 9, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Leal, W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Francis, F.; Liu, Y.; Chen, J.L.; Cheng, D.F. An overview of odorant-binding protein functions in insect peripheral olfactory reception. Genet. Mol. Res. 2011, 10, 3056. [Google Scholar] [CrossRef] [PubMed]
- Montagné, N.; Fouchier, A.D.; Newcomb, R.D.; Jacquin-Joly, E. Advances in the Identification and Characterization of Olfactory Receptors in Insects. Prog. Mol. Biol. Transl. 2014, 130, 55–80. [Google Scholar] [CrossRef]
- Paula, D.P.; Togawa, R.C.; Costa, M.M.; Grynberg, P.; Martins, N.F.; Andow, D.A. Identification and expression profile of odorant-binding proteins in Halyomorpha halys (Hemiptera: Pentatomidae). Insect Mol. Biol. 2016, 25, 580–594. [Google Scholar] [CrossRef]
- Tunstall, N.E.; Warr, C.G. Chemical communication in insects: The peripheral odour coding system of Drosophila melanogaster. In Sensing in Nature; Springer: Berlin, Germany, 2012; pp. 59–77. [Google Scholar]
- Zhang, J.; Walker, W.B.; Wang, G. Pheromone reception in moths: From molecules to behaviors. Prog. Mol. Biol. Transl. Sci. 2015, 130, 109–128. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Calvello, M.; Ban, L. Diversity of Odorant-binding Proteins and Chemosensory Proteins in Insects. Chem. Senses 2005, 30 (Suppl. 1), i291–i292. [Google Scholar] [CrossRef]
- Xu, Y.-L.; He, P.; Zhang, L.; Fang, S.-Q.; Dong, S.-L.; Zhang, Y.-J.; Li, F. Large-scale identification of odorant-binding proteins and chemosensory proteins from expressed sequence tags in insects. BMC. Genom. 2009, 10, 632. [Google Scholar] [CrossRef]
- Mckenna, M.P.; Hekmat-Scafe, D.S.; Gaines, P.; Carlson, J.R. Putative Drosophila pheromone-binding proteins expressed in a subregion of the olfactory system. J. Biol. Chem. 1994, 269, 16340–16347. [Google Scholar] [CrossRef]
- Vogt, R.G.; Riddiford, L.M. Pheromone binding and inactivation by moth antennae. Nature 1981, 293, 161–163. [Google Scholar] [CrossRef]
- Vogt, R.G. Biochemical diversity of odor detection: OBPs, ODEs and SNMPs. In Insect Pheromone Biochemistry and Molecular Biology; Blomquist, G., Vogt, R., Eds.; Academic Press: San Diego, CA, USA, 2003; pp. 391–445. [Google Scholar]
- Sanchez-Gracia, A.; Vieira, F.G.; Rozas, J. Molecular evolution of the major chemosensory gene families in insects. Heredity 2009, 103, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Maleszka, J.; Forêt, S.; Saint, R.; Maleszka, R. RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera). Dev. Genes Evol. 2007, 217, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Christensen, T.A.; Hildebrand, J.G. Pheromonal and host-odor processing in the insect antennal lobe: How different? Curr. Opin. Neurobiol. 2002, 12, 393–399. [Google Scholar] [CrossRef]
- Cunningham, J.P.; Moore, C.J.; Zalucki, M.P.; Cribb, B.W. Insect odour perception: Recognition of odour components by flower foraging moths. Proc. R. Soc. B Biol. Sci. 2006, 273, 2035–2040. [Google Scholar] [CrossRef] [PubMed]
- Carey, A.F.; Wang, G.; Su, C.Y.; Zwiebel, L.J.; Carlson, J.R. Odorant reception in the malaria mosquito Anopheles gambiae. Nature 2010, 464, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Hopf, T.A.; Morinaga, S.; Ihara, S.; Touhara, K.; Marks, D.S.; Benton, R. Amino acid coevolution reveals three-dimensional structure and functional domains of insect odorant receptors. Nat. Commun. 2015, 6, 6077. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.P. Odor and pheromone detection in Drosophila melanogaster. Pflüg. Arch. Eur. J. Physiol. 2007, 454, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Pitts, R.J.; Fox, A.N.; Zwiebel, L.J. A highly conserved candidate chemoreceptor expressed in both olfactory and gustatory tissues in the malaria vector Anopheles gambiae. Proc. Natl. Acad. Sci. USA 2004, 101, 5058–5063. [Google Scholar] [CrossRef]
- Haverkamp, A.; Hansson, B.S.; Knaden, M. Combinatorial codes and labeled lines: How insects use olfactory cues to find and judge food, mates, and oviposition sites in complex environments. Front. Physiol. 2018, 9, 49. [Google Scholar] [CrossRef]
- Knaden, M.; Strutz, A.; Ahsan, J.; Sachse, S.; Hansson, B.S. Spatial representation of odorant valence in an insect brain. Cell Rep. 2012, 1, 392–399. [Google Scholar] [CrossRef]
- Dweck, H.M.; Ebrahim, S.M.; Kromann, S.; Bown, D.; Hillbur, Y.; Sachse, S.; Hansson, B.; Stensmyr, M. Olfactory preference for egg laying on citrus substrates in Drosophila. Curr. Biol. 2013, 23, 2472–2480. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.T.; Si, F.L.; He, Z.B.; Chen, B. Genome-wide identification, characterization and classification of ionotropic glutamate receptor genes (iGluRs) in the malaria vector Anopheles sinensis (Diptera: Culicidae). Parasites Vectors 2018, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Benton, R.; Vannice, K.S.; Gomez-Diaz, C.; Vosshall, L.B. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 2009, 136, 149–162. [Google Scholar] [CrossRef] [PubMed]
- Abuin, L.; Bargeton, B.; Ulbrich, M.H.; Isacoff, E.Y.; Kellenberger, S.; Benton, R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron 2011, 69, 44–60. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.Y.; Xu, W.; Dong, S.L.; Zhu, J.Y.; Xu, Y.X.; Anderson, A. Genome-wide analysis of ionotropic receptor gene repertoire in Lepidoptera with an emphasis on its functions of Helicoverpa armigera. Insect Biochem. Mol. Biol. 2018, 99, 37–53. [Google Scholar] [CrossRef]
- Enjin, A.; Zaharieva, E.E.; Frank, D.D.; Mansourian, S.; Suh, G.S.; Gallio, M.; Stensmyr, M.C. Humidity Sensing in Drosophila. Curr. Biol. 2016, 26, 1352–1358. [Google Scholar] [CrossRef]
- Chen, C.; Buhl, E.; Xu, M.; Croset, V.; Rees, J.S.; Lilley, K.S.; Benton, R.; Hodge, J.J.; Stanewsky, R.J.N. Drosophila Ionotropic Receptor 25a mediates circadian clock resetting by temperature. Nature 2015, 527, 516. [Google Scholar] [CrossRef]
- Liman, E.; Zhang, Y.; Montell, C. Peripheral coding of taste. Neuron 2014, 81, 984–1000. [Google Scholar] [CrossRef]
- Robertson, H.M. Molecular evolution of the major arthropod chemoreceptor gene families. Annu. Rev. Entomol. 2019, 64, 227–242. [Google Scholar] [CrossRef]
- Pentzold, S.; Burse, A.; Boland, W. Contact chemosensation of phytochemicals by insect herbivores. Nat. Prod. Rep. 2017, 34, 478–483. [Google Scholar] [CrossRef]
- Wright, G.A. To feed or not to feed: Circuits involved in the control of feeding in insects. Curr. Opin. Neurobiol. 2016, 41, 87–91. [Google Scholar] [CrossRef]
- Cui, H.H.; Gu, S.H.; Zhu, X.Q.; Wei, Y.; Liu, H.W.; Khalid, H.D.; Guo, Y.Y.; Zhang, Y.J. Odorant-binding and chemosensory proteins identified in the antennal transcriptome of Adelphocoris suturalis Jakovlev. Comp. Biochem. Physiol. 2017, 24, 139–145. [Google Scholar] [CrossRef] [PubMed]
- Bin, S.Y.; Qu, M.Q.; Pu, X.H.; Wu, Z.Z.; Lin, J.T. Antennal transcriptome and expression analyses of olfactory genes in the sweetpotato weevil Cylas formicarius. Sci. Rep. 2017, 7, 11073. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.L.; Yang, J.; Boykin, L.M.; Zhao, Q.Y.; Li, Q.; Wang, X.W.; Liu, S.S. The characteristics and expression profiles of the mitochondrial genome for the Mediterranean species of the Bemisia tabaci complex. BMC. Genom. 2013, 14, 401. [Google Scholar] [CrossRef] [PubMed]
- Hekmat-Scafe, D.S.; Scafe, C.R.; Mckinney, A.J.; Tanouye, M.A. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 2002, 12, 1357–1369. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Li, F. The Pear psylla (Homoptera) of China with descriptions of seven new species. Entomotaxonomia 1981, 3, 35–47. [Google Scholar]
- Sun, J.R.; Yan, L.I.; Yan, S.; Zhang, Q.W.; Xu, H.L. Microsatellite marker analysis of genetic diversity of Cacopsylla chinensis (Yang et Li) (Hemiptera: Psyllidae) populations in China. Acta Entomol. Sin. 2011, 54, 820–827. [Google Scholar] [CrossRef]
- Blomquist, C.L.; Kirkpatrick, B.C. Frequency and Seasonal Distribution of Pear Psylla Infected with the Pear Decline Phytoplasma in California Pear Orchards. Plant. Pathol. 2002, 92, 1218–1226. [Google Scholar] [CrossRef] [PubMed]
- Kang, A.R.; Baek, J.Y.; Lee, S.H.; Cho, Y.S.; Kim, W.S.; Han, Y.S.; Kim, I. Geographic homogeneity and high gene flow of the pear psylla, Cacopsylla pyricola (Hemiptera: Psyllidae), detected by mitochondrial COI gene and nuclear ribosomal internal transcribed spacer 2. Anim. Cells Syst. 2012, 16, 145–153. [Google Scholar] [CrossRef]
- Yang, C.L.; Zhu, H.Y.; Zhang, F. Comparative proteomics analysis between the short-term stress and long-term adaptation of the Blattella germanica (Blattodea: Blattellidae) in response to beta-cypermethrin. J. Econ. Entomol. 2019. [Google Scholar] [CrossRef]
- Zhang, X.C.; Zhang, F. The potential control strategies based on the interaction between indoor cockroaches and their symbionts in China. Adv. Insect. Physiol. 2018, 55, 55–122. [Google Scholar]
- Guo, S.J.; Yan, X.Y.; Shi, F.F.; Ma, K.; Chen, Z.J.; Zhang, C. Expression and distribution of the zinc finger protein, SNAI3, in mouse ovaries and pre-implantation embryos. J. Reprod. Dev. 2018, 64, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, T.; Guo, Y.J.; Li, Y.J.; Zhang, Y.; Teng, N.; Zhang, F.M.; Yang, G.W. Molecular characterization and expression patterns of a non-mammalian toll-like receptor gene (TLR21) in larvae ontogeny of common carp (Cyprinus carpio L.) and upon immune stimulation. BMC. Vet. Res 2018, 14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, X.J.; Huang, Y.H.; Zhao, Z.G.; Zhang, S.S.; Gong, X.S.; Xie, L.; Kang, D.M.; Jing, X. Differential expression of hemolymph proteins between susceptible and insecticide-resistant Blattella germanica (Blattodea: Blattellidae). Environ. Entomol. 2014, 43, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.G.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.Y.; Li, J.Q.; Wu, S.F. Integrated nr database in protein annotation system and its localization. Comput. Eng. 2006, 32, 71–74. [Google Scholar]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic. Acids. Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef]
- Jensen, L.J.; Julien, P.; Kuhn, M.; Mering, C.; Muller, J.; Doerks, T.; Bork, P. eggNOG: Automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008, 36, D250–D254. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic. Acids. Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Zhang, Z.; Wood, W. A profile hidden Markov model for signal peptides generated by HMMER. Bioinformatics 2003, 19, 307–308. [Google Scholar] [CrossRef] [PubMed]
- Bateman, A.; Coin, L.; Durbin, R.; Finn, R.; Hollich, V.; Griffiths-Jones, S.; Khanna, A.; Marshall, M.; Moxon, S.; Sonnhammer, E. The Pfam protein families database. Nucleic. Acids. Res. 2004, 32, D138–D141. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, H. Predicting Secretory Proteins with SignalP. Methods. Mol. Biol. 2017, 1611, 59–73. [Google Scholar] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Le, S.Q.; Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008, 25, 1307–1320. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.Y.; Janovjak, H.; Miserez, A.Z. Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 2002, 32, 1372–1374. [Google Scholar] [CrossRef]
- Hou, P.L.; Zhao, G.M.; He, C.Q.; Wang, H.M.; He, H.B. Biopanning of polypeptides binding to bovine ephemeral fever virus G(1) protein from phage display peptide library. BMC. Vet. Res. 2018, 14, 9. [Google Scholar] [CrossRef]
- He, M.; Zhang, Y.N.; He, P. Molecular characterization and differential expression of an olfactory receptor gene family in the white-backed planthopper Sogatella furcifera based on transcriptome analysis. PLoS ONE 2015, 10, e0140605. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Zhao, N.; Yang, B. Global transcriptional analysis of olfactory genes in the head of pine shoot beetle, Tomicus yunnanensis. Comp. Funct. Genom. 2012, 2012. [Google Scholar] [CrossRef][Green Version]
- Liu, Z.; Wang, X.; Lei, C.; Zhu, F. Sensory genes identification with head transcriptome of the migratory armyworm, Mythimna separata. Sci. Rep. 2017, 7, 46033. [Google Scholar] [CrossRef] [PubMed]
- Cao, D.; Liu, Y.; Walker, W.B.; Li, J.; Wang, G. Molecular characterization of the Aphis gossypii olfactory receptor gene families. PLoS ONE 2014, 9, e101187. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.H.; Wu, K.M.; Guo, Y.Y.; Field, L.M.; Pickett, J.A.; Zhang, Y.J.; Zhou, J.J. Identification and expression profiling of odorant binding proteins and chemosensory proteins between two wingless morphs and a winged morph of the cotton aphid Aphis gossypii Glover. PLoS ONE 2013, 8, e73524. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Zhang, H.; Bin, S.; Chen, L.; Han, Q.; Lin, J. Antennal and Abdominal Transcriptomes Reveal Chemosensory Genes in the Asian Citrus Psyllid, Diaphorina citri. PLoS ONE 2016, 11, e0159372. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Yuan, X.; Qian, P.; Cheng, J.; Zhang, C.; Gurr, G.; Zhu, Z.R. Identification and expression profiling of putative chemosensory protein genes in two rice planthoppers, Laodelphax striatellus (Fallén) and Sogatella furcifera (Horváth). J. Asia-Pac. Entomol. 2015, 18, 771–778. [Google Scholar] [CrossRef]
- He, M.; He, P. Molecular characterization, expression profiling, and binding properties of odorant binding protein genes in the whitebacked planthopper, Sogatella furcifera. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2014, 174, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.S.; Sun, Z.; Ma, W.; Chen, W.; Wang, M.Q. De novo analysis of the Nilaparvata lugens (Stål) antenna transcriptome and expression patterns of olfactory genes. Comp. Biochem. Physiol. D Genomics Proteomics 2014, 9, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; He, P.; Dong, S.L. Different expression profiles suggest functional differentiation among chemosensory proteins in Nilaparvata lugens (Hemiptera: Delphacidae). J. Insect Sci. 2014, 14, 270. [Google Scholar] [CrossRef]
- Mao, Y.; Xu, X.; Xu, W.; Ishida, Y.; Leal, W.S.; Ames, J.B.; Clardy, J. Crystal and solution structures of an odorant-binding protein from the southern house mosquito complexed with an oviposition pheromone. Proc. Natl. Acad. Sci. USA 2010, 107, 19102–19107. [Google Scholar] [CrossRef]
- Pelletier, J.; Guidolin, A.; Syed, Z.; Cornel, A.J.; Leal, W.S. Knockdown of a mosquito odorant-binding protein involved in the sensitive detection of oviposition attractants. J. Chem. Ecol. 2010, 36, 245–248. [Google Scholar] [CrossRef]
- Liu, N.Y.; Liu, C.C.; Dong, S.L. Functional differentiation of pheromone-binding proteins in the common cutworm Spodoptera litura. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2013, 165, 254–262. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Zhang, J.; Liu, N.Y.; Zhang, Y.N.; Yang, K.; Dong, S.L. Distinct expression profiles and different functions of odorant binding proteins in Nilaparvata lugens Stål. PLoS ONE 2011, 6, e28921. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.L.; Pan, Y.F.; Ma, Y.F.; Wang, J.; He, M.; He, P. Binding affinity characterization of an antennae-enriched chemosensory protein from the white-backed planthopper, Sogatella furcifera (Horvath), with host plant volatiles. Pestic. Biochem. Physiol. 2018, 152, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Gu, S.H.; Wang, S.Y.; Zhang, X.Y.; Ji, P.; Liu, J.T.; Wang, G.R.; Wu, K.M.; Guo, Y.Y.; Zhou, J.J.; Zhang, Y.J. Functional characterizations of chemosensory proteins of the Alfalfa Plant Bug Adelphocoris lineolatus indicate their involvement in host recognition. PLoS ONE 2012, 7, e42871. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.N.; Ye, Z.F.; Yang, K.; Dong, S.L. Antenna-predominant and male-biased CSP19 of Sesamia inferens is able to bind the female sex pheromones and host plant volatiles. Gene 2014, 536, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Calvello, M.; Brandazza, A.; Navarrini, A.; Dani, F.R.; Turillazzi, S.; Felicioli, A.; Pelosi, P. Expression of odorant-binding proteins and chemosensory proteins in some Hymenoptera. Insect Biochem. Mol. Biol. 2005, 35, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.J.; Kan, Y.; Antoniw, J.; Pickett, J.A.; Field, L.M. Genome and eST analyses and expression of a gene family with putative functions in insect chemoreception. Chem. Senses 2006, 31, 453. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-N.; Jin, J.-Y.; Jin, R.; Xia, Y.-H.; Zhou, J.-J.; Deng, J.-Y.; Dong, S.-L. Differential expression patterns in chemosensory and non-chemosensory tissues of putative chemosensory genes identified by transcriptome analysis of insect pest the purple stem borer Sesamia inferens (Walker). PLoS ONE 2013, 8, e69715. [Google Scholar] [CrossRef]
- Qiao, H.L.; Deng, P.Y.; Li, D.D.; Chen, M.; Jiao, Z.J.; Liu, Z.C.; Zhang, Y.Z.; Kan, Y.C. Expression analysis and binding experiments of chemosensory proteins indicate multiple roles in Bombyx mori. J. Insect Physiol. 2013, 59, 667–675. [Google Scholar] [CrossRef]
- Yi, X.; Qi, J.; Zhou, X.; Hu, M.Y.; Zhong, G.H. Differential expression of chemosensory-protein genes in midguts in response to diet of Spodoptera litura. Sci. Rep. 2017, 7, 296. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.N.; Zhu, X.Y.; Fang, L.P.; He, P.; Wang, Z.Q.; Chen, G.; Sun, L.; Ye, Z.F.; Deng, D.G.; Li, J.B. Identification and expression profiles of sex pheromone biosynthesis and transport related genes in Spodoptera litura. PLoS ONE 2015, 10, e0140019. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Luo, Q.; Rizwan-Ul-Haq, M.; Hu, M.Y. Cloning and characterization of three chemosensory proteins from Spodoptera exigua and effects of gene silencing on female survival and reproduction. Bull. Entomol. Res. 2012, 102, 600–609. [Google Scholar] [CrossRef] [PubMed]
- Foret, S.; Wanner, K.W.; Maleszka, R. Chemosensory proteins in the honey bee: Insights from the annotated genome, comparative analyses and expressional profiling. Insect Biochem. Mol. Biol. 2007, 37, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Crasto, C.J. Olfactory Receptors. In Methods in Molecular Biology; Walker, J.M., Ed.; Humana Press: New York, NY, USA, 2013; Volume 2013. [Google Scholar] [CrossRef]
- Rytz, R.; Croset, V.; Benton, R. Ionotropic receptors (IRs): Chemosensory ionotropic glutamate receptors in Drosophila and beyond. Insect Biochem. Mol. Biol. 2013, 43, 888–897. [Google Scholar] [CrossRef] [PubMed]
- Agnihotri, A.R.; Roy, A.A.; Joshi, R.S. Gustatory receptors in Lepidoptera: Chemosensation and beyond. Insect Mol. Biol. 2016, 25, 519–529. [Google Scholar] [CrossRef] [PubMed]
- Stengl, M.; Funk, N.W. The role of the coreceptor Orco in insect olfactory transduction. J. Comp. Physiol. A 2013, 199, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Butterwick, J.A.; Del Marmol, J.; Kim, K.H.; Kahlson, M.A.; Rogow, J.A.; Walz, T.; Ruta, V. Cryo-EM structure of the insect olfactory receptor Orco. Nature 2018, 560, 447–452. [Google Scholar] [CrossRef] [PubMed]
- DeGennaro, M.; McBride, C.S.; Seeholzer, L.; Nakagawa, T.; Dennis, E.J.; Goldman, C.; Jasinskiene, N.; James, A.A.; Vosshall, L.B. orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature 2013, 498, 487–491. [Google Scholar] [CrossRef]
- Chang, H.; Liu, Y.; Ai, D.; Jiang, X.; Dong, S.; Wang, G. A Pheromone antagonist regulates optimal mating time in the moth Helicoverpa armigera. Curr. Biol. 2017, 27, 1610–1615. [Google Scholar] [CrossRef]
- Zhang, Y.N.; Du, L.X.; Xu, J.W.; Wang, B.; Zhang, X.Q.; Yan, Q.; Wang, G. Functional characterization of four sex pheromone receptors in the newly discovered maize pest Athetis lepigone. J. Insect Physiol. 2019, 113, 59–66. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, M.; Yan, S.; Wang, G.; Liu, Y. A female-biased odorant receptor from Apolygus lucorum (Meyer-Dur) tuned to some plant odors. Int. J. Mol. Sci. 2016, 17, 1165. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.R.; Wanner, K.W.; Trowell, S.C.; Warr, C.G.; Jaquin-Joly, E.; Zagatti, P.; Robertson, H.; Newcomb, R.D. Molecular basis of female-specific odorant responses in Bombyx mori. Insect Biochem. Mol. Biol. 2009, 39, 189–197. [Google Scholar] [CrossRef]
- Widmayer, P.; Heifetz, Y.; Breer, H. Expression of a pheromone receptor in ovipositor sensilla of the female moth (Heliothis virescens). Insect Mol. Biol. 2009, 18, 541–547. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Uda, Y.; Ono, Y.; Nakagawa, T.; Suwa, M.; Yamaoka, R.; Touhara, K. Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr. Biol. 2009, 19, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Mang, D.; Shu, M.; Tanaka, S.; Nagata, S.; Takada, T.; Endo, H.; Kikuta, S.; Tabunoki, H.; Iwabuchi, K.; Sato, R. Expression of the fructose receptor BmGr9 and its involvement in the promotion of feeding, suggested by its co-expression with neuropeptide F1 in Bombyx mori. Insect Biochem. Mol. Biol. 2016, 75, 58–69. [Google Scholar] [CrossRef] [PubMed]
- Mang, D.; Shu, M.; Endo, H.; Yoshizawa, Y.; Nagata, S.; Kikuta, S.; Sato, R. Expression of a sugar clade gustatory receptor, BmGr6, in the oral sensory organs, midgut, and central nervous system of larvae of the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2016, 70, 85–98. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhang, H.J.; Anderson, A. A sugar gustatory receptor identified from the foregut of cotton bollworm Helicoverpa armigera. J. Chem. Ecol. 2012, 38, 1513–1520. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Xu, Z.W.; Zhang, X.M.; Liu, N.Y. Genome-based identification and analysis of ionotropic receptors in Spodoptera litura. Naturwissenschaften 2018, 105, 38. [Google Scholar] [CrossRef]
- Jiang, X.J.; Ning, C.; Guo, H.; Jia, Y.Y.; Huang, L.Q.; Qu, M.J.; Wang, C.Z. A gustatory receptor tuned to d-fructose in antennal sensilla chaetica of Helicovera armigera. Insect Biochem. Mol. Biol. 2015, 60, 39–46. [Google Scholar] [CrossRef]










| Statistics Project | Number |
|---|---|
| Total clean reads | 32,879,148 |
| GC percentage | 34.11% |
| Q20 percentage | 99% |
| Total Unigene nucleotides | 40,874,177 |
| Total Unigenes | 63,052 |
| N50 of Unigenes (nt) | 702 |
| Min length of Unigenes (nt) | 201 |
| Mean length of Unigenes (nt) | 648.26 |
| Max length of Unigenes (nt) | 20,509 |
| Unigenes with homolog in NR | 22,392 |
| Gene | ORF | Signal | Complete | Best Blastx Match | ||||
|---|---|---|---|---|---|---|---|---|
| Name | (aa) | Peptide | ORF | Name | Acc. No. | Species | E-value | Identity (%) |
| Odorant Binding Protein (OBP) | ||||||||
| OBP1 | 238 | 1–24 | Y | odorant-binding protein 5 | AHB59658.1 | Sogatella furcifera | 8.00 × 10−34 | 36 |
| OBP2 | 105 | -- | N | odorant-binding protein 1 | ARR95844.1 | Diaphorina citri | 4.00 × 10−26 | 52 |
| OBP3 | 65 | -- | N | general OBP 83a-like | XP_008470659.1 | Diaphorina citri | 1.00 × 10−226 | 65 |
| OBP4 | 122 | 1–25 | Y | odorant-binding protein 5 | ATO59032.1 | Schistocerca gregaria | 8.00 × 10−13 | 37 |
| OBP5 | 145 | 1–21 | Y | odorant-binding protein 1 | ARR95844.1 | Diaphorina citri | 1.00 × 10−27 | 38 |
| OBP6 | 75 | -- | N | odorant-binding protein 8 | AMD82868.1 | Bemisia tabaci | 8.00 × 10−18 | 56 |
| OBP7 | 61 | -- | N | odorant-binding protein 1 | ARR95844.1 | Diaphorina citri | 2.00 × 10−26 | 84 |
| OBP8 | 82 | -- | N | odorant-binding protein 1 | ARR95844.1 | Diaphorina citri | 7.00 × 10−19 | 54 |
| OBP9 | 135 | 1–21 | Y | general OBP 57c isoform X2 | XP_021924930.1 | Zootermopsis nevadensis | 5.00 × 10−25 | 39 |
| OBP10 | 135 | N | Y | odorant-binding protein 3 | AGE97633.1 | Aphis gossypii | 2.00 × 10−13 | 32 |
| OBP11 | 86 | -- | N | odorant-binding protein 1 | ARR95844.1 | Diaphorina citri | 6.00 × 10−25 | 54 |
| OBP12 | 104 | 1–24 | Y | putative odorant-binding protein A10 | XP_008473937.1 | Diaphorina citri | 5.00 × 10−46 | 55 |
| Chemosensory Protein (CSP) | ||||||||
| CSP1 | 80 | -- | N | chemosensory protein | AJP61962.1 | Phenacoccus solenopsis | 1.00 × 10−16 | 39 |
| CSP2 | 104 | 1–24 | Y | chemosensory protein | AVM86436.1 | Corythucha ciliata | 3.00 × 10−16 | 37 |
| CSP3 | 161 | 1–16 | Y | ejaculatory bulb-specific protein 3-like | XP_008478860.1 | Diaphorina citri | 5.00 × 10−52 | 66 |
| CSP4 | 103 | 1–19 | Y | ejaculatory bulb-specific protein 3-like | XP_008471453.1 | Diaphorina citri | 2.00 × 10−60 | 75 |
| CSP5 | 91 | -- | N | chemosensory protein 1 | ARR95843.1 | Diaphorina citri | 1.00 × 10−38 | 74 |
| CSP6 | 105 | -- | N | ejaculatory bulb-specific protein 3-like | P_008478140.1 | Diaphorina citri | 1.00 × 10−64 | 89 |
| CSP7 | 121 | 1–19 | Y | ejaculatory bulb-specific protein 3-like | XP_008473947.1 | Diaphorina citri | 3.00 × 10−52 | 79 |
| CSP8 | 141 | 1–21 | Y | ejaculatory bulb-specific protein 3-like | XP_018916603.1 | Bemisia tabaci | 1.00 × 10−38 | 79 |
| CSP9 | 91 | -- | N | ejaculatory bulb-specific protein 3-like | XP_017300318.1 | Diaphorina citri | 4.00 × 10−59 | 81 |
| CSP10 | 132 | 1–19 | Y | chemosensory protein | AJP61957.1 | Phenacoccus solenopsis | 1.00 × 10−22 | 41 |
| CSP11 | 103 | 1–19 | Y | ejaculatory bulb-specific protein 3-like | P_008478860.1 | Diaphorina citri | 2.00 × 10−74 | 89 |
| Odorant receptors (OR) | ||||||||
| OR1 | 242 | -- | N | odorant receptor 82a | XP_008477835.1 | Diaphorina citri | 3.00 × 10−25 | 46 |
| OR2 | 96 | -- | N | odorant receptor 83b | ADB82908.1 | Loxostege sticticalis | 8.00 × 10−50 | 83 |
| OR3 | 150 | -- | N | odorant receptor 85c-like | XP_008476350.1 | Diaphorina citri | 3.00 × 10−18 | 33 |
| OR4 | 123 | -- | N | odorant receptor 82a | XP_008477835.1 | Diaphorina citri | 1.00 × 10−14 | 32 |
| OR5 | 333 | -- | N | odorant receptor 82a | XP_008477835.1 | Diaphorina citri | 4.00 × 10−04 | 53 |
| OR6 | 352 | -- | N | odorant receptor 82a | XP_008477835.1 | Diaphorina citri | 1.00 × 10−11 | 24 |
| OR7 | 321 | -- | N | odorant receptor 82a | XP_008477835.1 | Diaphorina citri | 6.00 × 10−06 | 38 |
| Ionotropic receptors (IR) | ||||||||
| IR1 | 73 | -- | N | ionotropic receptor 21a isoform X2 | XP_024936966.1 | Cephus cinctus | 4.00 × 10−09 | 45 |
| IR2 | 205 | -- | N | ionotropic receptor | AUF73076.1 | Anoplophora chinensis | 7.00 × 10−78 | 79 |
| IR3 | 120 | -- | N | ionotropic receptor 24 | ALD51351.1 | Locusta migratoria | 2.00 × 10−22 | 50 |
| IR4 | 145 | -- | N | ionotropic receptor 6 | AVH87294.1 | Holotrichia parallela | 1.00 × 10−36 | 59 |
| Gustatory receptor (GR) | ||||||||
| GR1 | 207 | -- | N | GR for sugar taste 64f-like | XP_008467720.2 | Diaphorina citri | 2.00 × 10−84 | 78 |
| GR2 | 200 | -- | N | gustatory receptor | ABY40623.1 | Tribolium castaneum | 5.00 × 10−41 | 45 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.-W.; Zhu, X.-Y.; Chao, Q.-J.; Zhang, Y.-J.; Yang, Y.-X.; Wang, R.-R.; Zhang, Y.; Xie, M.-Z.; Ge, Y.-T.; Wu, X.-L.; et al. Chemosensory Gene Families in the Oligophagous Pear Pest Cacopsylla chinensis (Hemiptera: Psyllidae). Insects 2019, 10, 175. https://doi.org/10.3390/insects10060175
Xu J-W, Zhu X-Y, Chao Q-J, Zhang Y-J, Yang Y-X, Wang R-R, Zhang Y, Xie M-Z, Ge Y-T, Wu X-L, et al. Chemosensory Gene Families in the Oligophagous Pear Pest Cacopsylla chinensis (Hemiptera: Psyllidae). Insects. 2019; 10(6):175. https://doi.org/10.3390/insects10060175
Chicago/Turabian StyleXu, Ji-Wei, Xiu-Yun Zhu, Qiu-Jie Chao, Yong-Jie Zhang, Yu-Xia Yang, Ran-Ran Wang, Yu Zhang, Meng-Zhen Xie, Ya-Ting Ge, Xin-Lai Wu, and et al. 2019. "Chemosensory Gene Families in the Oligophagous Pear Pest Cacopsylla chinensis (Hemiptera: Psyllidae)" Insects 10, no. 6: 175. https://doi.org/10.3390/insects10060175
APA StyleXu, J.-W., Zhu, X.-Y., Chao, Q.-J., Zhang, Y.-J., Yang, Y.-X., Wang, R.-R., Zhang, Y., Xie, M.-Z., Ge, Y.-T., Wu, X.-L., Zhang, F., Zhang, Y.-N., Ji, L., & Xu, L. (2019). Chemosensory Gene Families in the Oligophagous Pear Pest Cacopsylla chinensis (Hemiptera: Psyllidae). Insects, 10(6), 175. https://doi.org/10.3390/insects10060175