Abstract
Lipases play essential roles in digestion, transport, and processing of dietary lipids in insects. For parasitoid wasps with a unique life cycle, lipase functions could be multitudinous in particular. Pteromalus puparum is a pupal endoparasitoid of butterflies. The female adult deposits eggs into its host, along with multifunctional venom, and the developing larvae consume host as its main nutrition source. Parasitoid lipases are known to participate in the food digestion process, but the mechanism remains unclear. P. puparum genome and transcriptome data were interrogated. Multiple alignments and phylogenetic trees were constructed. We annotated a total of 64 predicted lipase genes belonging to five lipase families and suggested that eight venom and four salivary lipases could determine host nutrition environment post-parasitization. Many putative venom lipases were found with incomplete catalytic triads, relatively long β9 loops, and short lids. Data analysis reveals the loss of catalytic activities and weak triacylglycerol (TAG) hydrolytic activities of lipases in venom. Phylogenetic trees indicate various predicted functions of lipases in P. puparum. Our information enriches the database of parasitoid lipases and the knowledge of their functional diversification, providing novel insight into how parasitoid wasps manipulate host lipid storage by using venom lipases.
1. Introduction
Lipases (EC 3.1.1.3), also defined as triacylglycerol (TAG) acylhydrolases, usually catalyze the hydrolysis of lipids at the lipid–water interface and thus play a crucial role in controlling lipid uptake, transport, and utilization in insects. Most lipases from all organisms can be grouped into six families according to the sequence relationship within the α/β hydrolase fold superfamily of proteins. They are neutral (PF00151), acid (PF04083), lipase2 (PF01674), lipase3 (PF01764), GDSL (named after the conserved amino acid motif of Gly, Asp, Ser, and Leu around the active site Ser; PF00657), and hormone-sensitive lipases (PF06350) [1,2]. With a catalytic triad (usually Ser–Asp/Glu–His), which is capable of generating a charge relay system and a highly nucleophilic serine, all lipases from those six families use a two-step reaction mechanism. Besides these six families, phospholipase A2 (EC 3.1.1.4) is another lipase family belonging to a separate superfamily but using a different catalytic mechanism [3]. Although it contains lots of phospholipases, it is not considered or analyzed in this study.
Each of these six lipase families has a functional responsibility. The neutral lipases can hydrolyze not only neutral lipids such as TAGs, diacylglycerides (DAGs), and monoacylglycerols (MAGs) but also the sn-1 position in phospholipids and galactolipids [1,4]. The acid or abhydrolipase family was named after the function of the gastric lipase of mammals at acidic pH. It mainly hydrolyzes TAGs and cholesterol esters [5]. Lipases from lipase2 and lipase3 families can hydrolyze TAGs or carboxylesters with differing fatty acid elements [2]. GDSL family members can hydrolyze fatty acids from TAGs, DAGs, MAGs, phospholipids with the sn-2 position, and carboxylester or thioester substrates [6]. The hormone-sensitive lipases can hydrolyze TAGs and cholesterol esters [7] but are restrained by hormonal and neuronal controls via reversible cAMP-dependent phosphorylation of a serine residue [8].
Pteromalus puparum (Hymenoptera: Pteromalidae) is a pupal endoparasitoid with a wide host range but preferably lays eggs in the pupae of certain papilionid and pieridid butterfly species. Benefitting from its prominent venom, which is co-injected into hosts during oviposition, P. puparum can suppress the population of the cabbage butterfly Pieris rapae (Lepidoptera: Pieridae) with a parasitism rate greater than 90% in the cruciferous crops fields in China [9]. It has been used as an endoparasitoid model to study the parasitoid–host interaction in recent years, and the compositions of its venom have also been investigated. Combining the proteomics and transcriptomics data of P. puparum venom, 70 putative venom proteins have been previously identified [10,11], including a group of venom lipases. Interestingly, the number of predicted lipases in P. puparum venom is higher than that in other parasitoid wasps (Supplementary Materials Table S1) [10,12,13,14,15,16,17,18,19,20,21,22].
We had previously assumed that P. puparum venom lipases might act on catalyzing the hydrolysis of TAG to free fatty acid and glycerol [23,24,25,26] as usual. However, this assumption was questioned by some results of lipidomics analysis, which used a liquid chromatography and mass spectrometry (LC–MS)-based approach to identify and quantify the lipids in the parasitized hosts. The lipidomic analysis revealed that hemolymph TAG levels in P. rapae increased while DAGs levels significantly decreased post-parasitization by P. puparum [27]. In the host cotton–melon aphid Aphis gossypii, which was parasitized by Lysiphlebia japonica, levels of TAG mostly increased as well [28]. The accumulation of TAGs in parasitized hosts is antagonistic to the typical function of lipases, implying that lipases participating in adjusting lipid availability in hosts to benefit wasp offspring likely lose TAG activity or have evolved novel functions. Therefore, in this paper, we interrogated the genome and transcriptome of P. puparum to annotate the predicted lipases and analyze their expression profiles. We were especially interested in the sequence characteristics of the putative venom lipases and larval salivary lipases since they are responsible for regulating the lipid metabolism in hosts in particular.
To investigate the enzyme activities and potential functions of P. puparum lipases, we first identified their catalytic triads. Subsequently, multiple sequence alignments of β9 loop and lid motifs were used to estimate the TAG hydrolytic activity of neutral lipases. Putative functions of P. puparum neutral and acid lipases were predicted by phylogeny analysis. Our information provides an overview of the potential lipase functions in P. puparum. It also gives novel insights into studying the diversity and evolution of venom/salivary lipases in parasitoid wasps.
2. Materials and Methods
2.1. Annotation of Encoding Lipase Genes
Seven lipase protein sequences from the thoroughly annotated Drosophila melanogaster genome (flybase.org) were used as probes in tBLASTn analysis to identify lipases in Hymenoptera species. The Pteromalus puparum genome was sequenced and assembled in our previous work [29]. Other nine genome assemblies of parasitoid wasps, including Ceratosolen solmsi, Copidosoma floridanum, Diachasma alloeum, Fopius arisanus, Nasonia vitripennis, Orussus abietinus, Trichogramma pretiosum, and Solenopsis invicta were downloaded from NCBI GenBank. The probe CG6283 was used to identify natural lipases; CG6753, CG11598, and LIP (CG7279) for acid lipases; CG11029 for the family GDSL; CG33174 for the family lipase3; and CG11055 for hormone-sensitive lipases. Since lipase2 sequence was not found in D. melanogaster and other insect genomes, a Caenorhabditis elegans lipase2 (NP_496693) was added and used as a probe of this family (Supplementary file 1). Using the tool HMMER v3.1b2 and Pfam31.0 database [30], we searched the domains of all candidate sequences and filtered sequences as follows: PF00151 (neutral), PF04083 (acid), PF01674 (lipase2), PF01764 (lipase3), PF00657 (GDSL), and hormone-sensitive lipase (PF06350) [1,2]. All predicted lipase genes were manually validated by BLASTp (blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins), and their signal peptides were predicted by the online program SignalP (www.cbs.dtu.dk/services/SignalP) [31].
2.2. Expression Patterns Analysis
Our laboratory previously generated a number of RNA-seq libraries from P. puparum (eggs, larvae, female pupae, male pupae, female adults, male adults, female venom glands, female carcass without venom glands, ovaries, larval salivary glands) (access to the released transcriptome data via GECT01000000). Trinity and Cufflinks were used to assemble the transcriptomic raw data and calculate the expression levels of genes [32], which were represented as fragments per kilobase of exon model per million mapped fragments (FPKM). It is proportional to the number of cDNA fragments originating from it. Using a tool of the website BMK-Cloud (www.biocloud.net), two heatmap figures displaying expression profiles of lipases across P. puparum different developmental stages and tissues were constructed, respectively. The Venn diagram was drawn using an online program (bioinformatics.psb.ugent.be/webtools/Venn/).
2.3. RNA Extraction and cDNA Synthesis
To validate the expression patterns of lipases in P. puparum, we collected a total of 15 groups of P. puparum samples: six sample groups of different developmental stages or genders (embryos, larvae, female pupae, male pupae, female adults, and male adults), three larval tissues (midguts, salivary glands, and carcass), and six female tissues (heads, thoraxes, midguts, ovaries, venom glands, and carcass), as described previously [33]. They were pooled into a centrifuge tube with TRIzol reagent (Invitrogen, Carlsbad, CA, US) separately for the total RNA extractions. The PrimeScript™ One-Step RT-PCR Kit (Takara, Kusatsu, Japan) was subsequently used for reverse transcriptions according to the manufacturer’s instructions, and the synthesized cDNA was stored at −80 °C for further experiments.
2.4. RT-qPCR
SYBR Green Supermix Kit (Takara, Kusatsu, Japan) was used to perform a real-time quantitative PCR (RT-qPCR) on the BIO-RAD CFX96™ Real-Time System. RT-qPCR reaction was set up as follows: 95 °C for 30 s, 40 cycles at 95 °C for 5 s to denature DNA, and 60 °C for 34 s to anneal. The relative expression levels were conducted based on the 2–ΔΔCt method [34]. A stably expressed reference gene 18s rRNA was selected as an internal control. All primers used were designed on Primer3web (primer3.ut.ee) (Supplementary Materials Table S2).
2.5. Identification of the Catalytic Triads in P. puparum Lipases
To search the catalytic residues of neutral and acid lipase protein sequences in P. puparum, sequences of recognized canine pancreatic (NP_001003319) and gastric (NP_001003209) lipases were downloaded from NCBI and used to perform multiple sequence alignments, respectively [35]. Alignments were constructed by the Clustal Omega program (www.ebi.ac.uk/Tools/msa/clustalo), and then displayed and edited by the software GeneDoc.
2.6. Search for the β9 loop and Lid Motifs in P. puparum Lipases
For the analysis of β9 loop and lid motifs, eight neutral lipase sequences from holometabolous insects, namely Anopheles gambiae (AgamN14a and AgamN14b), Apis mellifera (AmelN9 and AmelN10), D. melanogaster (CG5966 and CG6847), Bombyx mori (BmorN12), and Tribolium castaneum (TcasN6) were used for a sequence alignment with all predicted neutral lipases in P. puparum. Sequence alignments were constructed as described above. The β9 loop and lid motifs of neutral lipases in P. puparum were identified according to the alignment results. Taking CG5966 as an example, its β9 loop was contained between residues His254–His273 and the lid domain was defined by a disulfide bridge between Cys287 and Cys310.
2.7. Phylogenetic Analysis
A number of neutral and acid lipase protein sequences of holometabolous insects [36] were used to analyze the predicted lipases in P. puparum. Muscle v3.8.31 was used for alignments, and trimAl v1.2 was used for the automated trimming of multiple sequence alignments [37,38]. Phylogenetic trees were constructed by RAxML with 1000 bootstrap replications [39]. According to the lowest BIC and AIC score conculcated by ProtTest v3.4 [40,41], their best models of amino acid replacement were selected as LG. The online tool iTOL v3 was used for tree visualization [42].
3. Results
3.1. Genome-Scale Identification of Encoding Lipases in Parasitoid Wasps
A total of 64 genes were identified as potential lipase encoding genes in P. puparum on the genome-scale (Supplementary Materials Table S3). Among them, 32 encoded for neutral lipases, 26 for acid lipases, 3 for lipase3, 2 for GDSL, and 1 for hormone-sensitive lipase. Considering the existence of isoforms of the same gene that could be repeatedly counted, here we lined out all genes encoding lipases tandemly arranged on the same scaffold and aligned with the same lipase gene in N. vitripennis, a closely related species of P. puparum in the same family of Pteromalidae. PPU13509 and PPU13510 on scaffold 13 were both identical to XP_008213053.2, PPU11430 and PPU11431 on scaffold 26 corresponded to XP_016840181.1, PPU09230 and PPU09231 on scaffold 150 to XP_016838373.1, and PPU04615 and PPU04616 on scaffold 0 to XP_016836798.1.
We also annotated the lipase encoding genes of eight parasitoid wasps and one ant species using their genomes to compare the number of each lipase gene family among insect species (Table 1). The total number of lipase genes varied from 26 in A. mellifera to 68 in P. puparum. N. vitripennis, the sister species of P. puparum, had a similar number of lipid genes in each family as P. puparum and a relatively large total number of predicted lipases (62). T. pretiosum had 59 lipases in total. For the other parasitoid wasps, the total number of lipase genes in D. alloeum, C. floridanum, and F. arisanus were all less than 50 each (48, 46, and 38, respectively). Interestingly, the total number of lipase genes in insect species seems to be related to the diversity of their food. There were 31 lipase genes found in the fig wasp C. solmsi, which lays eggs in figs as its hosts. The silkworm B. more, which mainly eats mulberry leaves, had only 29 lipase genes in total. The western honeybee A. mellifera, which lives in a highly standardized caste environment and develops by consuming a relatively specialized diet, had the lowest number of lipase genes (26). On the contrary, the omnivorous insects generally had a relatively large number of lipase genes (over 50): 62 in the fire ant S. invicta, 56 in the fruit fly D. melanogaster, and 54 in the red flour beetle T. castaneum.

Table 1.
Numbers of predicted lipase genes from each lipase family of selected insect species.
Additionally, it seems that the lipase numbers in the lipase3 family among parasitoid wasps are generally larger than in the other species, which suggests that parasitoid wasps process a relatively different lipid metabolism, utilization, and absorption due to their unique life cycle. All insect species in our comparison had only one hormone-sensitive gene but lacked the lipase2 family gene.
3.2. Expression Patterns of Encoding Lipases in P. puparum
Using a total of ten RNA-seq libraries data of P. puparum (embryos, 2-day larvae, female pupae, male pupae, female adults, male adults, venom glands, female carcasses, ovaries, and larval salivary glands), we calculated FPKM of each putative encoding lipase in each sample (Supplementary Materials Table S4) to represent its relative expression level. FPKM values of all predicted lipase genes ranged from 0 to 2022.12. Two heatmaps were constructed based on these values to display the lipase expression patterns across different developmental stages and tissues, respectively.
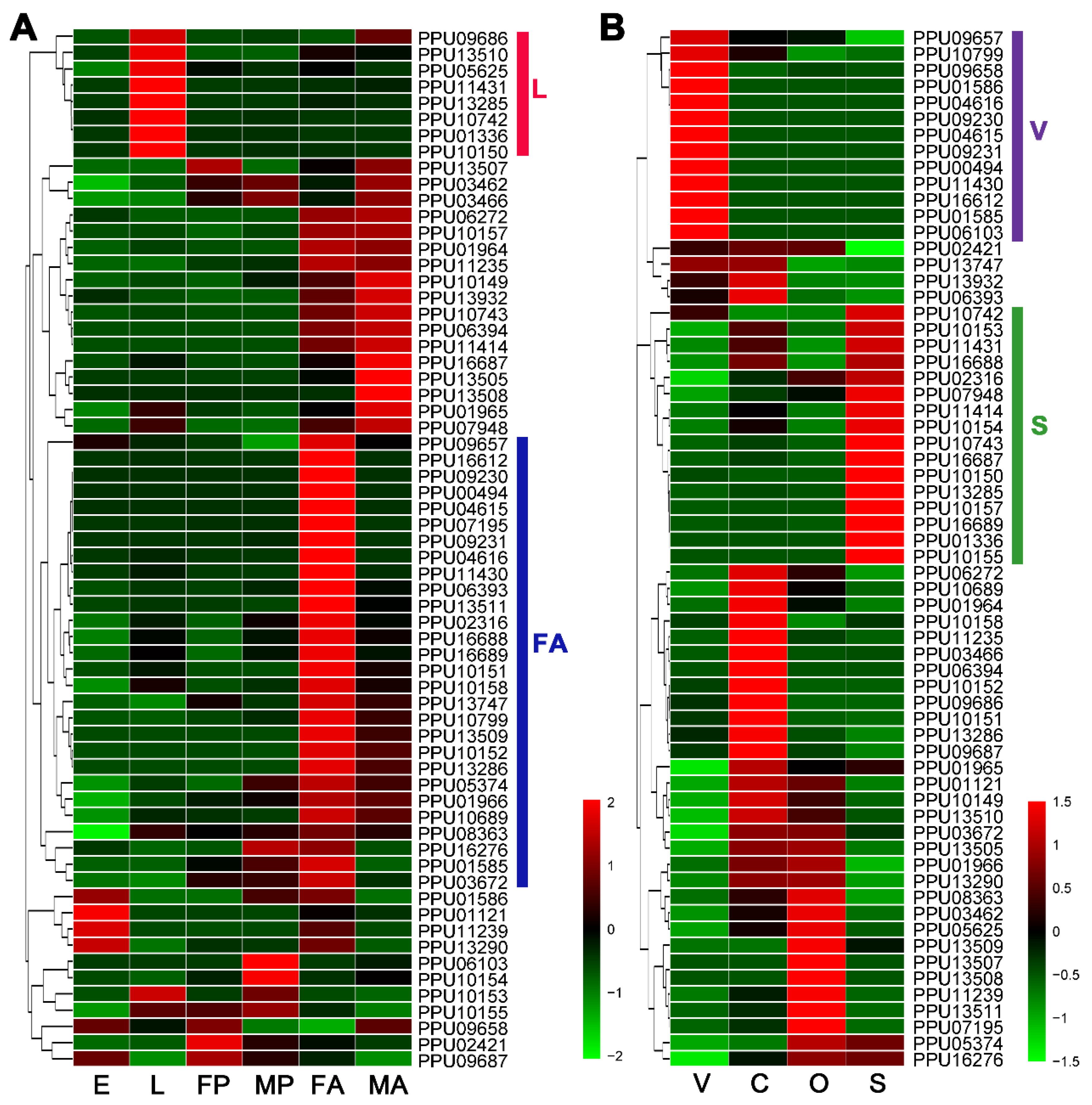
According to the heatmap based on the expression levels of developmental stages (Figure 1A), there were eight lipase genes with especially high expression at the larval stage (L group) and 28 lipase genes with particularly high expression in female adults (FA group). In addition, 13 lipase genes highly expressed particularly in venom glands were sorted into the V group, and 16 lipases with especially high-expression in larval salivary belonged to the S group (Figure 1B).
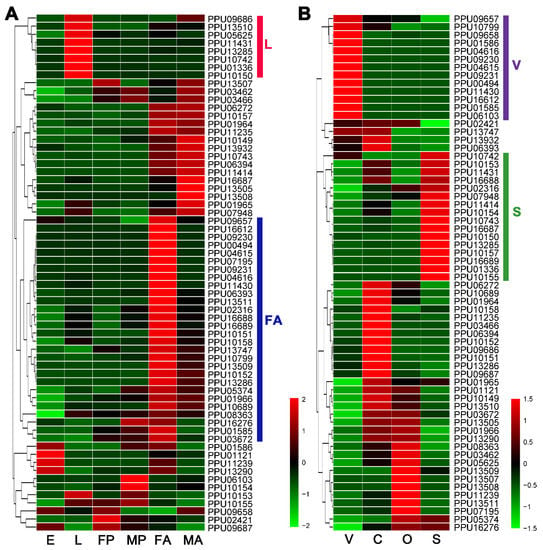
Figure 1.
Heatmap analysis of P. puparum lipase genes across different developmental stages (A) and tissues (B). Log2FPKM values are indicted by a green–red bar where the red represents higher expression levels, the green represents the lower. Lipases are sorted into different groups according to their expression profiles. Colored bars at right represent the group names. E = eggs, L = larvae, FP = female pupae, MP = male pupae, FA = female adults, MA = male adults, V = female venom glands, C = carcass (female adults without venom glands), O = ovaries, S = larval salivary glands.
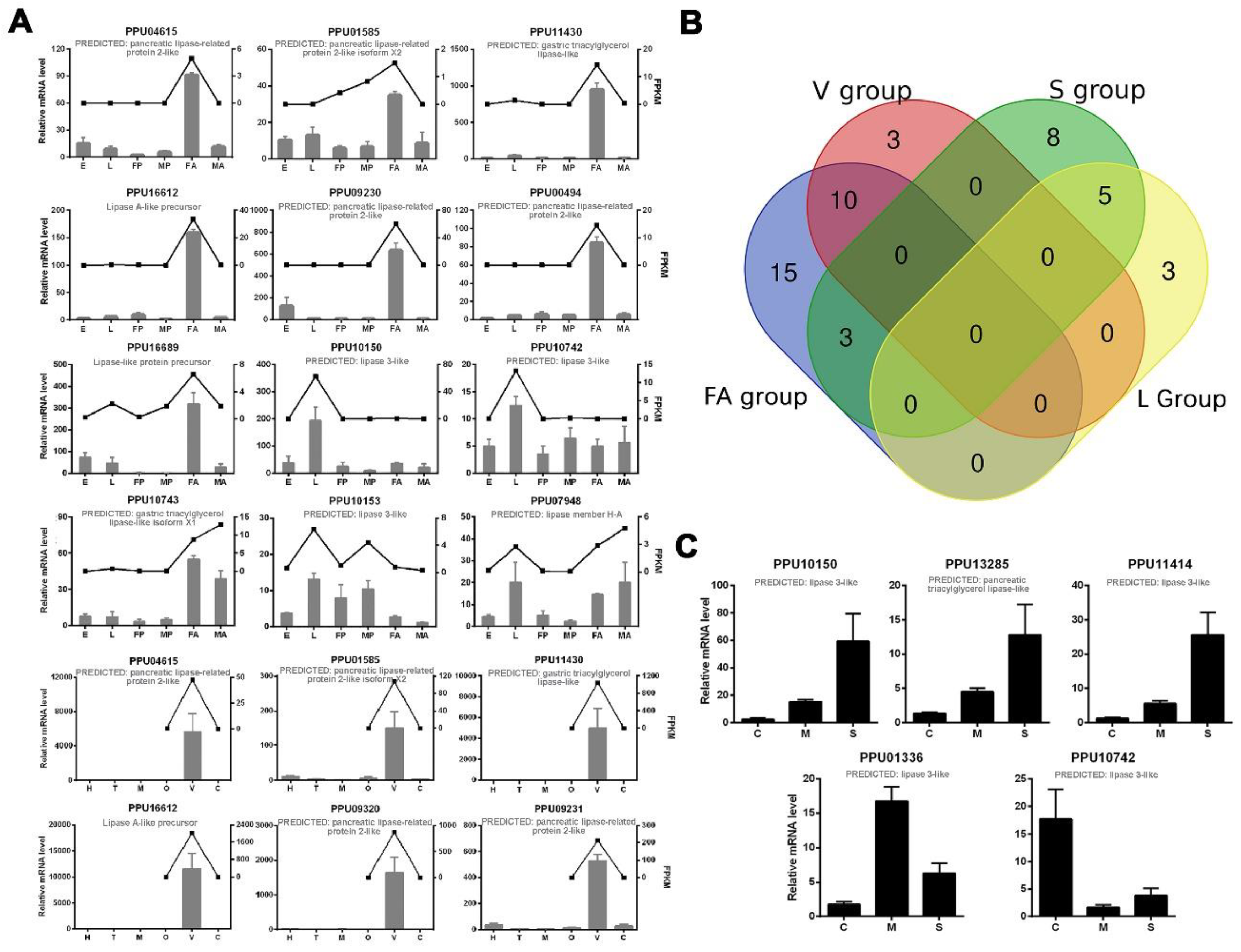
A number of predicted lipase genes of P. puparum were selected for qPCR analysis, and the results were mostly consistent with our transcriptomics analysis (Figure 2A). For further strict screening of venom-specific and salivary-specific lipases in P. puparum, a Venn diagram was drawn (Figure 2B). Ten lipases showed high-expression both at the female adult stage and in the venom gland tissue. Among them, two (PPU10799 and PPU09657) with a Log2 (venom-gland FPKM/carcass FPKM) value less than one were no longer considered to be putative venom-specific lipases in P. puparum. In the same way, five lipases had high-expression at the larvae stage and in the larval salivary gland tissue. We finally determined four of them as putative salivary-specific lipases with one exception, namely PPU10742. Although it was highly expressed in particular in the larval salivary gland, its FPKM was low at 0.5173. All the rest of the lipase genes in the S group had low expression levels (FPKM less than 10) at the larvae stage but high expression levels at the female adult stage. They were thus not considered as salivary-specific lipases in P. puparum.
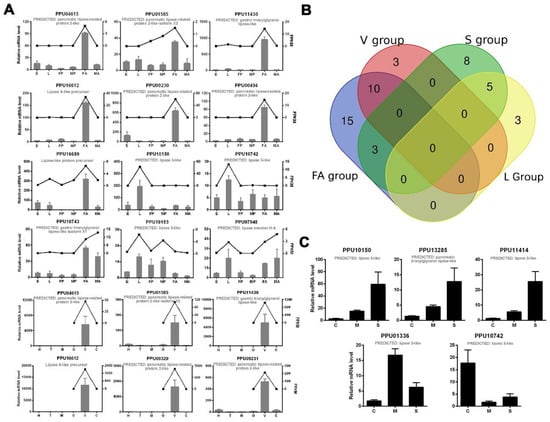
Figure 2.
qPCR results of selected lipase genes in P. puparum. (A) Total RNA extracted from P. puparum across different developmental stages and female tissues were used to perform qPCR. Lines display the FPKM values in transcriptome analysis, corresponding to the Y-axis at right. qPCR results of the selected lipases were mostly in accord with the transcriptome data. E = eggs, L = larvae, FP = female pupae, MP = male pupae, FA = female adults, MA = male adults, H = heads, T = thoraxes, M = midguts, O = ovaries, V = female venom glands, C = carcass (female adults without venom glands). (B) Venn diagram of potential venom lipases and salivary lipases. The blue circle indicates the lipases with especially high expression in female adults (FA group), the red circle indicates lipases with especially high expression in venom glans (V group), the green circle indicates lipases with especially high expression in larval salivary glands (S group), and the yellow circle indicates lipases with especially high expression at the larvae stage (L group). (C). Total RNA was extracted from the larval tissues of P. puparum. C = carcass, M = midguts, S = salivary glands; 18s rRNA of P. puparum was used as a housekeeping gene. Error bars represent the standard errors of the mean from three biological replicates.
To investigate the transcription levels of lipases in other larval tissues of P. puparum, we also performed qPCR for those putative salivary lipases using larval carcass, midgut, and salivary glands. Our results showed that three lipase genes were particularly highly expressed in larval salivary glands, while PPU01336 has a higher expression level in the midgut (Figure 2C). However, this lipase was not removed from putative salivary lipases. In P. puparum larvae, the salivary gland tightly adheres to midguts and thus the midgut samples usually mixed with the unremoved salivary gland. The higher expression level in midgut suggested that PPU01336 has high expression in midgut as well, but the comparison of expression between the salivary gland and midguts was still unclear. For further analysis, the positions of the signal peptides of those certain lipases were predicted, and their peptide-spectrum matches (PSMs) acquired from P. puparum venom proteome analysis [11] were displayed in Table 2. Nearly all putative venom lipases had at least two peptide-spectrum matches against the venom proteome except for PPU04615, with the lowest FPKM in the venom gland compared to others.

Table 2.
Information of the lipases with especially high expression in venom glands and larval salivary glands of P. puparum.
While 75% (6/8) of putative venom lipases were predicted as pancreatic lipase-relative protein 2-like (PLRP2-like), no predicted PLRP2-like was found among putative salivary lipases. Half of the putative salivary lipases were identified as lipase3-like. For mammals, PLRP2 usually presents at the suckling–weaning transition to digest breast milk fats and carry through adulthood, participating in the cytotoxic activity of T-cells [43]. For endoparasitoids P. puparum, we suggest that some PLRP2-like proteins in venom probably contribute to the regulation of lipid metabolism in hosts and benefit the early development of endoparasitoid young. Subsequently, the larval salivary lipases may take over the duty of regulating the host lipid environment. The venom and salivary lipases may play different roles at different developmental stages of wasps.
3.3. Incomplete Catalytic Triads in Predicted Lipases of P. puparum
Enzymes in numerous families, including the α/β hydrolase fold superfamily, serine proteases, and brain acetylhydrolase, require a catalytic triad to drive the catalytic mechanisms [2]. For the neutral and acid lipases in mammals, a catalytic triad usually consists of Ser–Asp–His residues and produces a charge relay system to achieve the reaction mechanisms [44,45]. While many substitutions at these activity sites could lead to a non-functional enzyme, a minority could maintain the enzyme functions [35,46]. The frequent replacements occurred as Cys or Thr instead of Ser, Glu or His instead of Asp, and Lys instead of His [35].
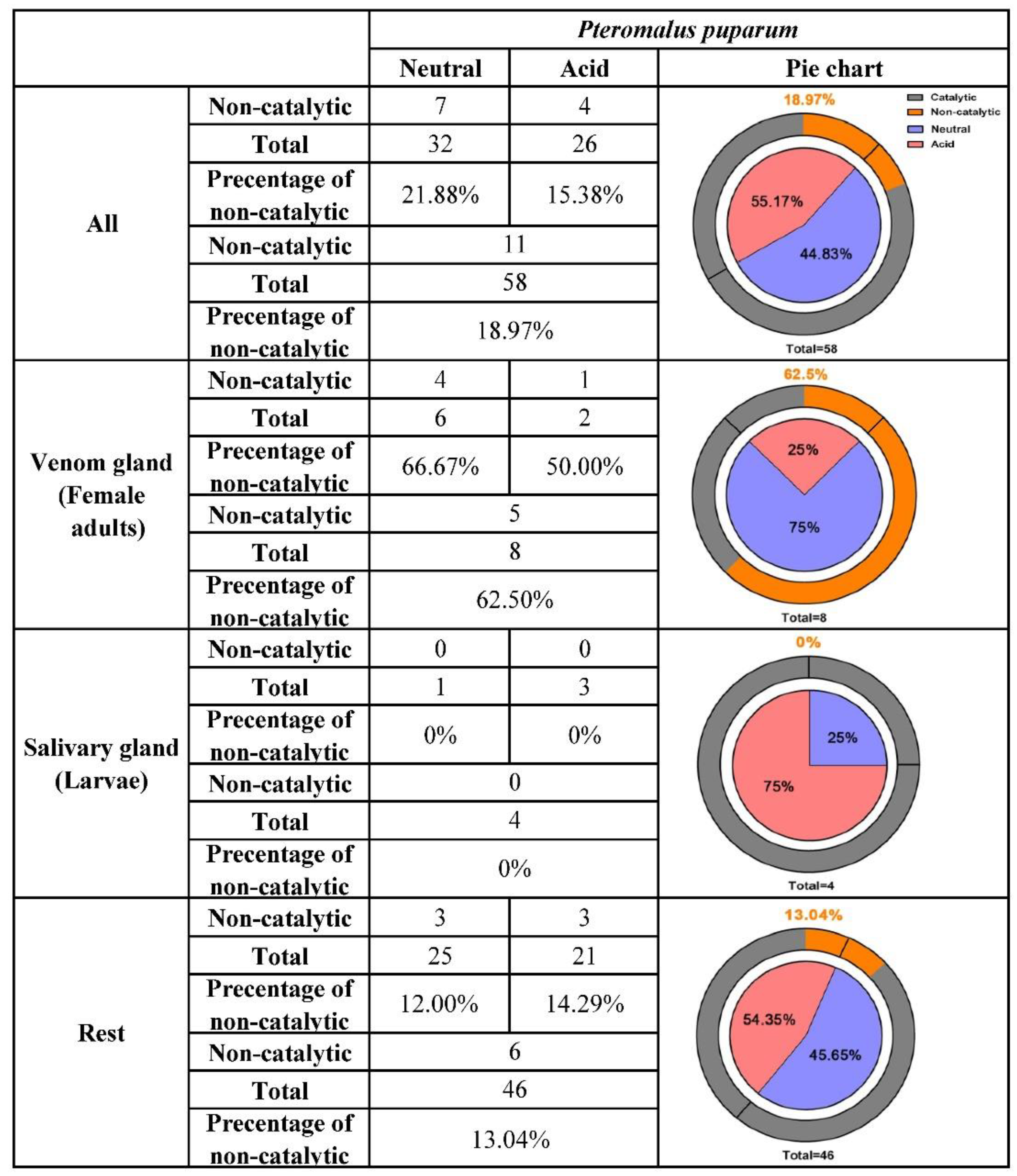
In insects, an average of 10% neutral and acid lipases have generally lost the motif critical for catalytic function [36]. However, in P. puparum, this percentage was high at 18.97% (Figure 3). Eleven out of 58 putative neutral and acid lipases were found without a consensus catalytic triad. Interestingly, 72.73% of them (8 out of 11) with an inconsistent triad, belonging to the V group mentioned above, had especially high expression in the venom glands. The remaining three were highly expressed in the ovaries. It seems that all lipases without a typical catalytic triad were likely responsible for the development of wasp eggs. Focusing on the putative venom-specific lipases from the strict selection, we observed that 62.5% of them had incomplete catalytic triads. Most of them lost or altered a residue at His positions (including PPU00494, PPU01585, and PPU04615), while PPU04616 lost a residue at the Ser position. The only putative venom lipase gene with a conservative substitution (Glu) in place of Asp was PPU16612, which had the highest expression in venom glands among all lipases. This conservative replacement was not uncommon and had been found in two closely related lipases from mosquito salivary as well [47]. Among the putative salivary lipases, none was with replaced residues at the activity sites. The percentage of lipases with incomplete catalytic triads decreased to 13.04% when putative venom and salivary lipases were not considered.
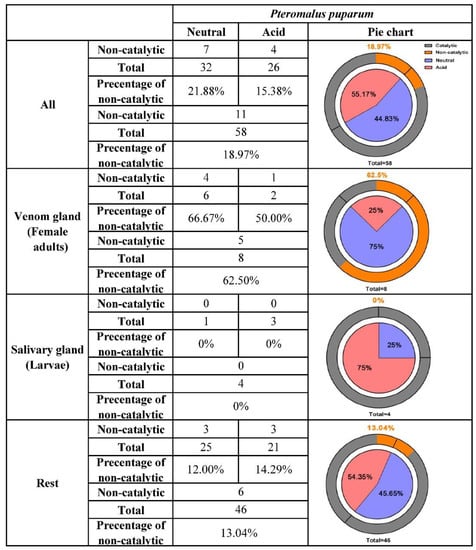
Figure 3.
Summary of the neutral and acid lipase characteristics and their distribution in P. puparum. For the pie charts in the right columns, the blue scale in the inner circle represented the percentage of neutral lipases while the pink scale represented the percentage of acid lipases. Lipases without catalytic activity were displayed in the orange scale of the outer circle and its percentage was shown at the top of each pie chart.
Additionally, the percentages of neutral and acid lipases were likely polarized in the venom glands and salivary glands of P. puparum. A total of 32 neutral and 26 acid lipases were annotated in the P. puparum genome, occupying 55.17% and 44.83%, respectively. Among the putative venom lipases, 75% were neutral and 25% were acid. Neutral (25%) and acid lipases (75%) were found among the putative salivary lipases in P. puparum. While the putative venom and salivary lipases were not considered, the percentages of neutral and acid lipases were similar to the original levels (54.35% against 45.65%). In brief, many putative venom lipases are neutral while many putative salivary lipases are acid.
3.4. Identification of β9 Loop and Lids of Lipases in P. puparum
A β9 loop and a lid are two substantial features in mammalian neutral lipases, usually covering the active site (the catalytic triad) of lipases and involved in substrate recognition [48,49]. Although the minimum lengths of loops and lids required to generate functions are still unclear [49], many pieces of evidence have indicated the lipases with smaller numbers of residues in β9 loops or lids had weak TAG hydrolytic activity [50]. For a phosphatidylserine phospholipase A1 with both a short β9 loop (13 amino acids) and a small lid (12 amino acids), no TAG hydrolytic activity was detected in [51].
We constructed an alignment using serval recognized neutral lipase genes of holometabolous insects and all putative neutral lipase genes in P. puparum (Supplementary Materials Figure S1). As is known, the β9 loop with > 15 residues in length is essential for the TAG hydrolytic activity of lipases in insects [36]. The length of the β9 loop in neutral lipases of P. puparum ranged from 15 to 23 residues, except a quite short sequence PPU01585 with neither β9 loop nor lid (Table 3). Nearly all putative venom lipases had relative long β9 loops (19 to 21 residues), and the only putative neutral salivary lipase had a similar size β9 loop with 20-residue length. For the lids, at least 18 residues are usually required to present the normal TAG hydrolytic activity [36]. Over half of the predicted neutral lipase in P. puparum had lids smaller than 18 residues, as well as most neutral lipases with incomplete catalytic triads, which were highly expressed in venom glands (V group). It suggests that they may have weak TAG hydrolytic activity. Coincidently, the long β9 loops and shortened lids have been also found in the venom lipases from some parasitoid wasps. The lipase DelePLA from the endoparasitoid wasp Diversinervus elegans has 18 residues in β9 loop and 13 residues in the lid domain [17]. c6971 from Ooencyrtus telenomicida has 18 and 16 residues in its β9 loop and lid domain, respectively [12]. For some social wasps and ants, their venom-associated phospholipase A1s (PLA1s) have been observed with both shorten β9 loops and small lid domains, indicating that these lipases lack TAG hydrolytic activity either [52,53,54].

Table 3.
Predicted activity site residues, β9 loop and lid domains of P. puparum neutral lipases.
Among 125 neutral lipases from 19 insect species analyzed by Horne [36], only 8 have both β9 loops greater than 15 and lids greater than 18 residues in length, meeting the consensus requirement for TAG activity in the mammalian enzymes. However, many more neutral lipases with predicted TAG hydrolytic activity were found in P. puparum. There were 14 neutral lipases meeting the β9 loop and lid criteria in P. puparum, including venom lipases PPU09230 and PPU09231. These two lipases had only 205 and 288 amino acids in length, but the longest lids among all neutral lipases, with 45 and 49 residues, respectively. They were both identified with a conserved catalytic triad.
3.5. Phylogenetic Analysis of Lipases in P. puparum
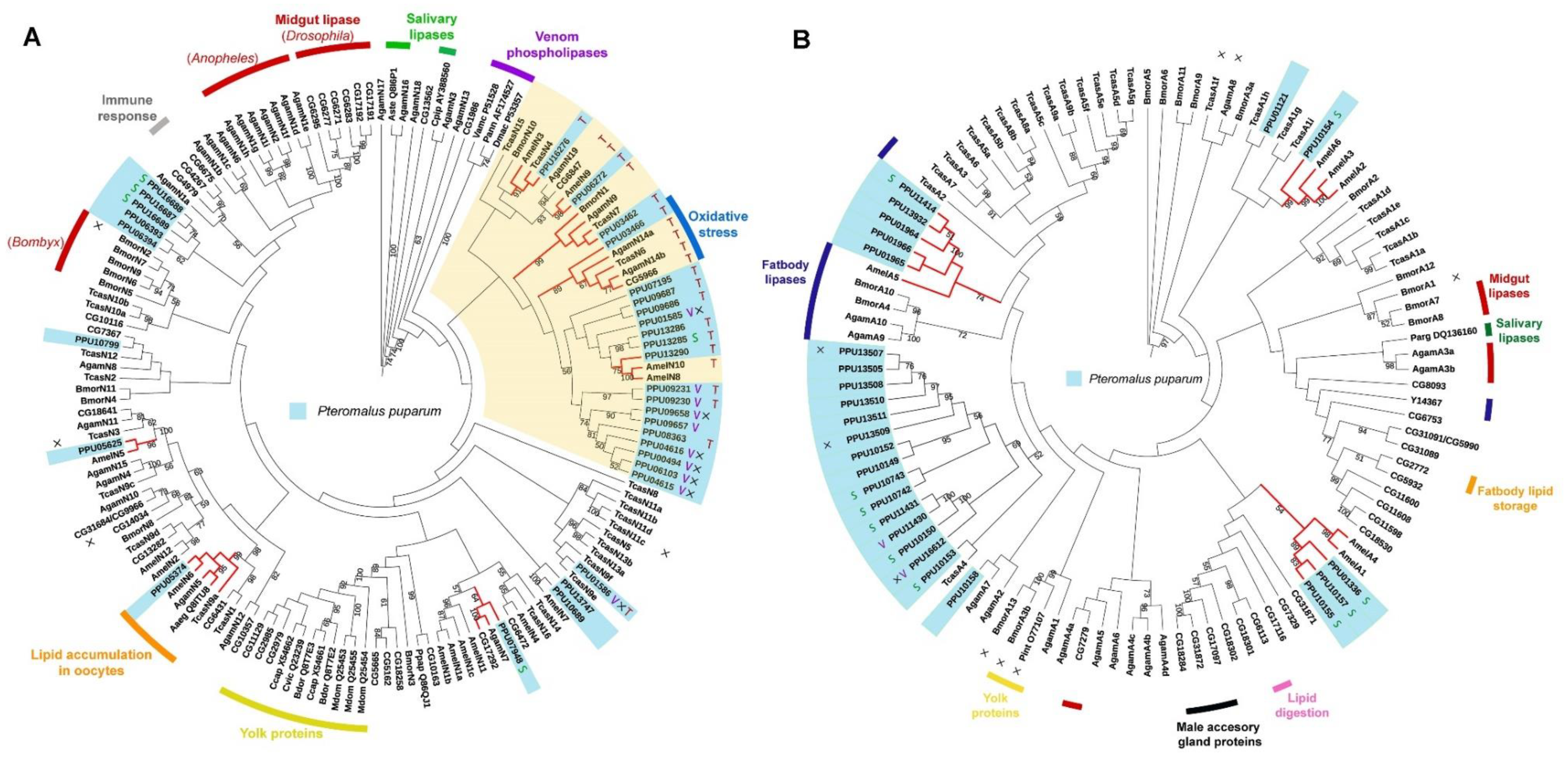
Two phylogenetic trees were constructed using the predicted neutral and acid lipases of P. puparum. According to the neutral lipase phylogeny (Figure 4A), PPU05374 is closed to a neutral lipase from the yellow fever mosquito Aedes aegypti (Aaeg_ Q8ITU8AF303984) in a single phylogenetic clade. Aaeg_Q8ITU8 AF303984 has been deposited in the NCBI database with an annotated function as lipid accumulation in oocytes. In this clade, orthologues were also found in numerous insect genomes including A. mellifera (AmelN6), A. gambiae (AgamN5), and T. castaneum (TcasN9a). Moreover, a neutral lipase of P. puparum PPU03466, clustered with D. melanogaster CG5966, a T. castaneum lipase (TcasN6), and two A. gambiae lipases (AgamN14a and AgamN14b) were found. While the expression of CG5966 increased under oxidative stress [55], the upregulation of TcasN6 [56] and AgamN14b [57] was observed during an immunological challenge. They were thus supposed to be responsible for membrane damage and new membrane construction. It indicates that PPU03466 in this cluster probably has a similar response. In the plasma membrane of M. sexta ovarian follicles, a 100 kDa membrane protein was found with DAG hydrolyze activity in hemolymph lipophorin [58]. PPU06272 and CG6847 were the only lipases with molecular weight >100 kDa in P. puparum and D. melanogaster genomes, respectively. CG6847 has been predicted as a midgut intracellular TAG lipase or a lipoprotein lipase involved in TAG mobilization from hemolymphatic lipophorin [36]. They were closely located in a conserved phylogenetic clade with AmelN9 and AgamN19, suggesting that they likely have a conserved function related to DAG hydrolysis. Besides, several orthologues were also found in our phylogenetic analysis. PPU05625 is the ortholog of AmelN5, PPU07948 is closed to AgamN7 and CG17292, and PPU03462 has orthologues such as BmorN1, AgamN9, and TcasN7. As orthologues, they probably have a similar function, although most of their functions are still unclarified.
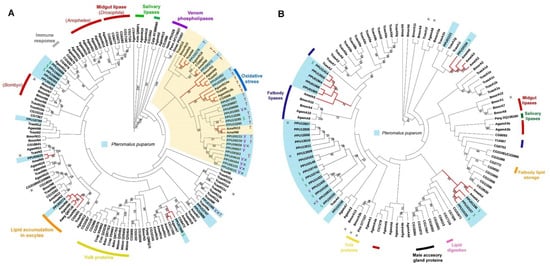
Figure 4.
Phylogenetic trees of lipase proteins from P. puparum and other recognized insect lipases. (A) Neighbor-joining tree of neutral lipases. The yellow clade includes a cluster of lipases with predicted TAG hydrolytic activity. (B) Neighbor-joining tree of acid lipases. Lipases with especially high expression in venom glands and salivary glands are represented as letters V and S, respectively. Indications were used to display whether the proteins are catalytically inactive (×) or have putative TAG hydrolytic activity (T) predicted by the lengths of their respective β9 loops and lids. Branches in red represent the orthologues of P. puparum lipases. All predicated lipase proteins were displayed in a light blue box. Only bootstrap values (from 1000 replicates) over 50 were displayed as text. The functions or predicted functions of the lipases were presented at the outer circle in different colors.
Interestingly, predicted lipase genes with especially high expression in venom glands (V group) clustered except for PPU01585 and PPU01586. Furthermore, nearly all those lipases with incomplete catalytic triads were located in a conserved clade. PPU08363 is the only lipase with a complete catalytic triad in this clade but has low expression levels across all different developmental stages and tissues. However, this non-catalytic clade was located far from the yolk protein clade whose lipases are generally with incomplete catalytic triads. Additionally, most neutral lipases with predicted TAG activity were observed in the yellow clade. A set of paralogs with predicted TAG hydrolytic activity likely evolved novel functions by gene duplications within the P. puparum genome.
In the acid lipase phylogenetic tree (Figure 4B), five P. puparum lipases formed an orthologous group with a fatbody lipase in A. mellifera (AmelA5). Three acid lipases with high expression levels in P. puparum larval salivary glands (PPU01336, PPU10157, and PPU10155) were identified as orthologs of acid lipases from A. mellifera (AmelA1 and AmelA4). PPU10154 was located in a conserved clade with A. mellifera lipase AmelA6 whose relevant ESTs (BI505714 and BI509544) were isolated from A. mellifera brain tissue [59]. Intriguingly, two putative acid venom lipases of P. puparum PPU11430 and PPU16612 were located in a closed relationship with PPU11431 and PPU10150, which were the putative salivary lipases of P. puparum. This suggests that those four acid lipases probably have a similar function in regulating the host lipid metabolism post-parasitization.
4. Discussion
A total of 64 predicted lipase genes were identified in the P. puparum genome. They belong to five lipase families, including neutral, acid, lipase3, GDSL, and hormone-sensitive. To compare the numbers of lipases among different parasitoid wasp species, we also examined eight parasitoid wasp genomes to identify their lipases. Our results showed that P. puparum had the greatest number of lipases among all investigated insects. Based on data from ten RNA-seq libraries of P. puparum, the expression profiles of all predicted lipases were further analyzed. While 13 lipases had especially high expression in venom glands, 16 lipases had especially high expression in salivary glands. They were sorted into the V group and S group, respectively. Combining with their expression profiles at different developmental stages, we finally determined eight putative venom lipase genes and four putative salivary lipase genes. All 12 lipases were considered as key components participating in regulating the host lipid metabolism post-parasitized.
Lipases always have an α/β hydrolase fold and the helical portion known as a lid to cover the active site [60]. Once a lipase contacts lipid under suitable conditions, the active site opens and hydrolysis begins. For most lipases, the amino acid residues at this active site are Ser, Asp, and His. They consist of a classic catalytic triad and represent the catalytic activity of lipases [61,62]. Interestingly, the percentage of non-catalytic lipases in P. puparum was higher than the average (10%) in other insect species. Many lipases in venom glands of P. puparum were predicted as non-catalytic lipases. Some non-catalytic lipases were found with high expression in ovaries. It seems that those non-catalytic lipases were related to the development of wasp eggs. During the embryonic development, yolk proteins could function as some non-catalytic roles in lipid binding, lipid transport, and energy storage [63]. Numerous non-catalytic neutral lipases have been used as yolk proteins in some higher Diptera and acid lipases in Lepidoptera [64]. In some insect species, vitellogenins (the precursors of yolk proteins) have been replaced with non-catalytic lipase-like proteins. Since functional vitellogenin has been found in Hymenoptera [65], few lipase-like proteins performing a yolk protein function have been investigated within this order [22]. In P. puparum, non-catalytic lipases were mostly neutral and highly expressed in venom glands in particular. We assumed that while P. puparum injects venom along with eggs into hosts, these non-catalytic venom lipases probably act as yolk proteins, binding host lipids to deliver energy for egg development in parasitized hosts. Nevertheless, more experiments have to be performed to validate our hypothesis.
Nevertheless, venom lipases with an incomplete catalytic triad were not widely found in parasitoid wasps. Among all recognized venom lipases as reported, an acid lipase NV21220 from N. vitripennis (Pteromalidae) was the only one with an incomplete catalytic triad (Ser137–Glu401–Asp434). Two well-characterized venom lipases in O. telenomicida (c6971) and D. elegans (DelePLA) have been reported with a high consensus catalytic triad previously [12,17]. In other words, the venom proteins with incomplete catalytic triads may not widely exist in parasitoid wasps. They frequently present in P. puparum in particular, which has the largest number of predicted lipase genes among all eight parasitoid wasps in our analysis. It suggests that different parasitoid wasps probably use a variety of strategies to regulate and utilize host lipids.
Two important features responsible for the significant TAG hydrolytic activity of lipases are a β9 loop and the lid, which cover the active site. According to our prediction based on the lengths of β9 loops and the lids, nearly all of non-catalytic neutral lipases of P. puparum have short lids indicating weak TAG hydrolytic activity. It seems that the incomplete catalytic triads are interconnected with the small lids. However, some reported venom-associated lipases in other wasps generally have both classic catalytic triads and small lids. Moreover, P. puparum has a greater number of lipases with predicted TAG hydrolytic activity, compared to those reported lipases from other insect species. This number in P. puparum is even greater than the total number in all Holometabolous insects analyzed by Horne et al. [36]. Based on our analysis of the phylogenetic dendrogram, most of these lipases in P. puparum are in-paralogues and likely have evolved novel functions. They clustered into a sister clade of the venom lipases clade in P. puparum, suggesting that they are probably responsible for the unique life cycle of P. puparum as well.
5. Conclusions
It is generally believed that many parasitoid wasps have lost the capability to synthesize lipids themselves and have to obtain lipids from hosts during development. Our genome-scale identification and expression analysis of P. puparum lipases partially contribute to the understanding of lipases in parasitoid wasps, which is essential to figure out how parasitoid wasps utilize host lipids for development. The frequent observation of non-catalytic lipases in P. puparum venom suggests that an uncommon function of lipases has been evolved in parasitoid wasps. We speculate that future studies may need to focus on various molecular mechanisms of lipases inside parasitoid wasp venom. Although the TAG hydrolytic activity of lipases was not experimentally validated, it was predicted by analyzing sequences’ characteristics, and a general feature in venom lipases was discussed. We look forward to forthcoming insights in the insect lipase database and predict that an increasing number of insect lipases could be probably developed and explored in different application fields.
Supplementary Materials
The following are available online at https://www.mdpi.com/2075-4450/11/4/227/s1. Supplementary file 1. Seven well-characterized lipase protein sequences from six lipase families in D. melanogaster genome. They were used as probes to identify lipase genes in P. puparum. Figure S1: Alignments of eight recognized insect lipase sequences and all putative neutral lipases annotated from P. puparum. Red boxes indicate the predicated β9 loop domains and lid domains. The numbers at the right show the position of the sequence. Table S1: The lipases identified in the venom of various parasitoid wasp species. Table S2: Primers of the selected lipase sequences for qPCR. Table S3: The information of all identified lipases of P. puparum. Table S4: FPKM values of lipases from different libraries of P. puparum.
Author Contributions
Conceptualization and methodology, G.Y. and Q.F.; software and formal analysis, J.W.; investigation, J.S., J.W., H.Y., and F.W.; writing—original draft preparation, J.W.; writing—review and editing, G.Y. and Q.S.; All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Key Program of the National Natural Science Foundation of China (grant number 31830074), the National Key R&D Program of China (No. 2017YFD0200400), the Major International (Regional) Joint Research Project of the National Natural Science Foundation of China (grant number 31620103915), the Program for Chinese Innovation Team in Key Areas of Science and Technology of the Ministry of Science and Technology of the People’s Republic of China (grant number 2016RA4008), the Program for Chinese Outstanding Talents in Agricultural Scientific Research of the Ministry of Agriculture and Rural Affairs of the People’s Republic of China, and the Natural Science Foundation of Zhejiang Province of China (grant number LY18C140001).
Acknowledgments
Jiale Wang acknowledges Todd A. Schlenke at the University of Arizona for the inspiring discussion and the China Scholarship Council for its support during Jiale’s study period in the US.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Derewenda, Z.S. Structure and function of lipases. Adv. Protein Chem. 1994, 45, 1–52. [Google Scholar]
- Holmquist, M. Alpha/beta-hydrolase fold enzymes: Structures, functions and mechanisms. Curr. Protein Pept. Sci. 2000, 1, 209–235. [Google Scholar] [CrossRef]
- Yuan, C.H.; Tsai, M.D. Pancreatic phospholipase A(2): New views on old issues. BBA-Mol. Cell Biol. Lipids 1999, 1441, 215–222. [Google Scholar] [CrossRef]
- Carriere, F.; Withers-Martinez, C.; van Tilberugh, H.; Roussel, A.; Cambillau, C.; Verger, R. Structural basis for the substrate selectivity of pancreatic lipases and some related proteins. BBA-Rev. Biomembr. 1998, 1376, 417–432. [Google Scholar] [CrossRef]
- Gargouri, Y.; Moreau, H.; Verger, R. Gastric lipases—Biochemical and physiological-studies. Biochim. Biophys. Acta 1989, 1006, 255–271. [Google Scholar] [CrossRef]
- Akoh, C.C.; Lee, G.C.; Liaw, Y.C.; Huang, T.H.; Shaw, J.F. GDSL family of serine esterases/lipases. Prog. Lipid Res. 2004, 43, 534–552. [Google Scholar] [CrossRef]
- Holm, C.; Kirchgessner, T.G.; Svenson, K.L.; Fredrikson, G.; Nilsson, S.; Miller, C.G.; Shively, J.E.; Heinzmann, C.; Sparkes, R.S.; Mohandas, T.; et al. Hormone-sensitive lipase—Sequence, expression, and chromosomal localization to 19 cent-q13.3. Science 1988, 241, 1503–1506. [Google Scholar] [CrossRef]
- Belfrage, P.; Fredrikson, G.; Stalfors, P.; Tornqvist, H. Lipases; Borgström, B., Brockman, H.I., Eds.; Elsevier: Amsterdam, The Netherlands, 1984; pp. 365–416. [Google Scholar]
- Hu, C. Life-history and occurrence of Pteromalus puparum L. in Hangzhou. Acta Entomol. Sin. 1984, 27, 302–307. [Google Scholar]
- Yan, Z.C.; Fang, Q.; Wang, L.; Liu, J.D.; Zhu, Y.; Wang, F.; Li, F.; Werren, J.H.; Ye, G.Y. Insights into the venom composition and evolution of an endoparasitoid wasp by combining proteomic and transcriptomic analyses. Sci. Rep. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Fang, Q.; Wang, L.; Hu, C.; Ye, G.Y. Proteomic analysis of the venom from the endoparasitoid wasp Pteromalus puparum (Hymenoptera: Pteromalidae). Arch. Insect Biochem. Physiol. 2010, 75, 28–44. [Google Scholar] [CrossRef]
- Cusumano, A.; Duvic, B.; Jouan, V.; Ravallec, M.; Legeai, F.; Peri, E.; Colazza, S.; Volkoff, A.N. First extensive characterization of the venom gland from an egg parasitoid: Structure, transcriptome and functional role. J. Insect Physiol. 2018, 107, 68–80. [Google Scholar] [CrossRef]
- Laurino, S.; Grossi, G.; Pucci, P.; Flagiello, A.; Bufo, S.A.; Bianco, G.; Salvia, R.; Vinson, S.B.; Vogel, H.; Falabella, P. Identification of major Toxoneuron nigriceps venom proteins using an integrated transcriptomic/proteomic approach. Insect Biochem. Mol. Biol. 2016, 76, 49–61. [Google Scholar] [CrossRef]
- Mathe-Hubert, H.; Colinet, D.; Deleury, E.; Belghazi, M.; Ravallec, M.; Poulain, J.; Dossat, C.; Poirie, M.; Gatti, J.L. Comparative venomics of Psyttalia lounsburyi and P. concolor, two olive fruit fly parasitoids: A hypothetical role for a GH1 beta-glucosidase. Sci. Rep. 2016, 6, 35873. [Google Scholar] [CrossRef]
- De Graaf, D.C.; Aerts, M.; Brunain, M.; Desjardins, C.A.; Jacobs, F.J.; Werren, J.H.; Devreese, B. Insights into the venom composition of the ectoparasitoid wasp Nasonia vitripennis from bioinformatic and proteomic studies. Insect Mol. Biol. 2010, 19, 11–26. [Google Scholar] [CrossRef]
- Sim, A.D.; Wheeler, D. The venom gland transcriptome of the parasitoid wasp Nasonia vitripennis highlights the importance of novel genes in venom function. BMC Genom. 2016, 17, 571. [Google Scholar] [CrossRef]
- Liu, N.Y.; Wang, J.Q.; Zhang, Z.B.; Huang, J.M.; Zhu, J.Y. Unraveling the venom components of an encyrtid endoparasitoid wasp Diversinervus elegans. Toxicon 2017, 136, 15–26. [Google Scholar] [CrossRef]
- Xin, B.; Liu, P.X.; Xu, X.R.; Zhang, S.; Zheng, Y.N. Identification of venom proteins of the indigenous endoparasitoid Chouioia cunea (Hymenoptera: Eulophidae). J. Econ. Entomol. 2017, 110, 2022–2030. [Google Scholar] [CrossRef]
- Vincent, B.; Kaeslin, M.; Roth, T.; Heller, M.; Poulain, J.; Cousserans, F.; Schaller, J.; Poirie, M.; Lanzrein, B.; Drezen, J.M.; et al. The venom composition of the parasitic wasp Chelonus inanitus resolved by combined expressed sequence tags analysis and proteomic approach. BMC Genom. 2010, 11, 693. [Google Scholar] [CrossRef]
- Heavner, M.E.; Gueguen, G.; Rajwani, R.; Pagan, P.E.; Small, C.; Govind, S. Partial venom gland transcriptome of a Drosophila parasitoid wasp, Leptopilina heterotoma, reveals novel and shared bioactive profiles with stinging Hymenoptera. Gene 2013, 526, 195–204. [Google Scholar] [CrossRef]
- Colinet, D.; Deleury, E.; Anselme, C.; Cazes, D.; Poulain, J.; Azema-Dossat, C.; Belghazi, M.; Gatti, J.L.; Poirie, M. Extensive inter- and intraspecific venom variation in closely related parasites targeting the same host: The case of Leptopilina parasitoids of Drosophila. Insect Biochem. Mol. Biol. 2013, 43, 601–611. [Google Scholar] [CrossRef]
- Burke, G.R.; Strand, M.R. Systematic analysis of a wasp parasitism arsenal. Mol. Ecol. 2014, 23, 890–901. [Google Scholar] [CrossRef]
- Akanbi, T.O.; Adcock, J.L.; Barrow, C.J. Selective concentration of EPA and DHA using Thermomyces lanuginosus lipase is due to fatty acid selectivity and not regioselectivity. Food Chem. 2013, 138, 615–620. [Google Scholar] [CrossRef]
- Haba, E.; Bresco, O.; Ferrer, C.; Marques, A.; Busquets, M.; Manresa, A. Isolation of lipase-secreting bacteria by deploying used frying oil as selective substrate. Enzym. Microb. Technol. 2000, 26, 40–44. [Google Scholar] [CrossRef]
- Hiol, A.; Jonzo, M.D.; Rugani, N.; Druet, D.; Sarda, L.; Comeau, L.C. Purification and characterization of an extracellular lipase from a thermophilic Rhizopus oryzae strain isolated from palm fruit. Enzym. Microb. Technol. 2000, 26, 421–430. [Google Scholar] [CrossRef]
- Olusesan, A.T.; Azura, L.K.; Forghani, B.; Abu Bakar, F.; Mohamed, A.K.S.; Radu, S.; Manap, M.Y.A.; Saari, N. Purification, characterization and thermal inactivation kinetics of a non-regioselective thermostable lipase from a genotypically identified extremophilic Bacillus subtilis NS 8. New Biotechnol. 2011, 28, 738–745. [Google Scholar] [CrossRef]
- Wang, J.; Schlenke, T.; Ye, G. Lipidomics reveals how the endoparasitoid wasp Pteromalus puparum manipulates host energy stores for its young. Entomology 2019, in press. [Google Scholar]
- Gao, X.K.; Zhang, S.; Luo, J.Y.; Lu, L.M.; Zhang, L.J.; Cui, J.J. Lipidomics and RNA-seq study of lipid regulation in Aphis gossypii parasitized by Lysiphlebia japonica. Sci. Rep. 2017, 7, 1–13. [Google Scholar]
- Kim, Y. Genome level analysis of Pteromalus puparum transcriptome: Preface. Arch. Insect Biochem. 2020, 103, e21641. [Google Scholar] [CrossRef]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, 279–285. [Google Scholar] [CrossRef]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- Yang, L.; Lin, Z.; Fang, Q.; Wang, J.L.; Yan, Z.C.; Zou, Z.; Song, Q.S.; Ye, G.Y. The genomic and transcriptomic analyses of serine proteases and their homologs in an endoparasitoid, Pteromalus puparum. Dev. Comp. Immunol. 2017, 77, 56–68. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT > method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Dodson, G.; Wlodawer, A. Catalytic triads and their relatives. Trends Biochem. Sci. 1998, 23, 347–352. [Google Scholar] [CrossRef]
- Horne, I.; Haritos, V.S.; Oakeshott, J.G. Comparative and functional genomics of lipases in holometabolous insects. Insect Biochem. Mol. Biol. 2009, 39, 547–567. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. ProtTest 3: Fast selection of best-fit models of protein evolution. Bioinformatics 2011, 27, 1164–1165. [Google Scholar] [CrossRef]
- Abascal, F.; Zardoya, R.; Posada, D. ProtTest: Selection of best-fit models of protein evolution. Bioinformatics 2005, 21, 2104–2105. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, 242–245. [Google Scholar] [CrossRef]
- Lowe, M.E. Properties and function of pancreatic lipase related protein 2. Biochimie 2000, 82, 997–1004. [Google Scholar] [CrossRef]
- Brenner, S. The molecular evolution of genes and proteins—A tale of 2 serines. Nature 1988, 334, 528–530. [Google Scholar] [CrossRef]
- Hide, W.A.; Chan, L.; Li, W.H. Structure and evolution of the lipase superfamily. J. Lipid Res. 1992, 33, 167–178. [Google Scholar]
- Paetzel, M.; Dalbey, R.E. Catalytic hydroxyl/amine dyads within serine proteases. Trends Biochem. Sci. 1997, 22, 28–31. [Google Scholar] [CrossRef]
- Valenzuela, J.G.; Francischetti, I.M.B.; Pham, V.M.; Garfield, M.K.; Ribeiro, J.M.C. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem. Mol. 2003, 33, 717–732. [Google Scholar] [CrossRef]
- Winkler, F.K.; Darcy, A.; Hunziker, W. Structure of human pancreatic lipase. Nature 1990, 343, 771–774. [Google Scholar] [CrossRef]
- Aoki, J.; Inoue, A.; Makide, K.; Saiki, N.; Arai, H. Structure and function of extracellular phospholipase A(1) belonging to the pancreatic lipase gene family. Biochimie 2007, 89, 197–204. [Google Scholar] [CrossRef]
- Hirata, K.; Dichek, H.L.; Cioffi, J.A.; Choi, S.Y.; Leeper, N.J.; Quintana, L.; Kronmal, G.S.; Cooper, A.D.; Quertermous, T. Cloning of a unique lipase from endothelial cells extends the lipase gene family. J. Biol. Chem. 1999, 274, 14170–14175. [Google Scholar] [CrossRef]
- Aoki, J.; Nagai, Y.; Hosono, H.; Inoue, K.; Arai, H. Structure and function of phosphatidylserine-specific phospholipase A1. Biochim. Biophys. Acta 2002, 1582, 26–32. [Google Scholar] [CrossRef]
- Hoffman, D.R.; Sakell, R.H.; Schmidt, M. Sol i 1, the phospholipase allergen of imported fire ant venom. J. Allergy Clin. Immunol. 2005, 115, 611–616. [Google Scholar] [CrossRef]
- Sukprasert, S.; Rungsa, P.; Uawonggul, N.; Incamnoi, P.; Thammasirirak, S.; Daduang, J.; Daduang, S. Purification and structural characterisation of phospholipase A(1) (Vespapase, Ves a 1) from Thai banded tiger wasp (Vespa affinis) venom. Toxicon 2013, 61, 151–164. [Google Scholar] [CrossRef]
- Perez-Riverol, A.; Pereira, F.D.C.; Lasa, A.M.; Fernandes, L.G.R.; dos Santos-Pinto, J.R.A.; Justo-Jacomini, D.L.; de Azevedo, G.O.; Bazon, M.L.; Palma, M.S.; Zollner, R.D.; et al. Molecular cloning, expression and IgE-immunoreactivity of phospholipase Al, a major allergen from Polybia paulista (Hymenoptera: Vespidae) venom. Toxicon 2016, 124, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Zou, S.; Meadows, S.; Sharp, L.; Jan, L.Y.; Jan, Y.N. Genome-wide study of aging and oxidative stress response in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2000, 97, 13726–13731. [Google Scholar] [CrossRef]
- Park, Y.; Aikins, J.; Wang, L.J.; Beeman, R.W.; Oppert, B.; Lord, J.C.; Brown, S.J.; Lorenzen, M.D.; Richards, S.; Weinstock, G.M.; et al. Analysis of transcriptome data in the red flour beetle, Tribolium castaneum. Insect Biochem. Mol. 2008, 38, 380–386. [Google Scholar] [CrossRef]
- Dana, A.N.; Hillenmeyer, M.E.; Lobo, N.F.; Kern, M.K.; Romans, P.A.; Collins, F.H. Differential gene expression in abdomens of the malaria vector mosquito, Anopheles gambiae, after sugar feeding, blood feeding and Plasmodium berghei infection. BMC Genom. 2006, 7, 119. [Google Scholar] [CrossRef]
- Van Antwerpen, R.; Salvador, K.; Tolman, K.; Gentry, C. Uptake of lipids by developing oocytes of the hawkmoth Manduca sexta. The possible role of lipoprotein lipase. Insect Biochem. Mol. 1998, 28, 399–408. [Google Scholar] [CrossRef]
- Whitfield, C.W.; Band, M.R.; Bonaldo, M.F.; Kumar, C.G.; Liu, L.; Pardinas, J.R.; Robertson, H.M.; Soares, M.B.; Robinson, G.E. Annotated expressed sequence tags and cDNA microarrays for studies of brain and behavior in the honey bee. Genome Res. 2002, 12, 555–566. [Google Scholar] [CrossRef]
- Nardini, M.; Dijkstra, B.W. Alpha/beta hydrolase fold enzymes: The family keeps growing. Curr. Opin. Struct. Biol. 1999, 9, 732–737. [Google Scholar] [CrossRef]
- Ollis, D.L.; Cheah, E.; Cygler, M.; Dijkstra, B.; Frolow, F.; Franken, S.M.; Harel, M.; Remington, S.J.; Silman, I.; Schrag, J.; et al. The alpha/beta-hydrolase fold. Protein Eng. 1992, 5, 197–211. [Google Scholar] [CrossRef]
- Van Pouderoyen, G.; Eggert, T.; Jaeger, K.-E.; Dijkstra, B.W. The crystal structure of Bacillus subtili lipase: A minimal α/β hydrolase fold enzyme. J. Mol. Biol. 2001, 309, 215–226. [Google Scholar] [CrossRef] [PubMed]
- Bownes, M. Why is there sequence similarity between insect yolk proteins and vertebrate lipases. J. Lipid Res. 1992, 33, 777–790. [Google Scholar] [PubMed]
- Hagedorn, H.H.; Maddison, D.R.; Tu, Z.J. The evolution of vitellogenins, cyclorrhaphan yolk proteins and related molecules. Adv. Insect Physiol. 1998, 27, 335–384. [Google Scholar]
- Amdam, G.V.; Norberg, K.; Hagen, A.; Omholt, S.W. Social exploitation of vitellogenin. Proc. Natl. Acad. Sci. USA 2003, 100, 1799–1802. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).