Susceptibility of Red Mason Bee Larvae to Bacterial Threats Due to Microbiome Exchange with Imported Pollen Provisions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling and Provisioning Experiment
2.2. 16S rRNA Gene Metabarcoding
2.3. Analysis
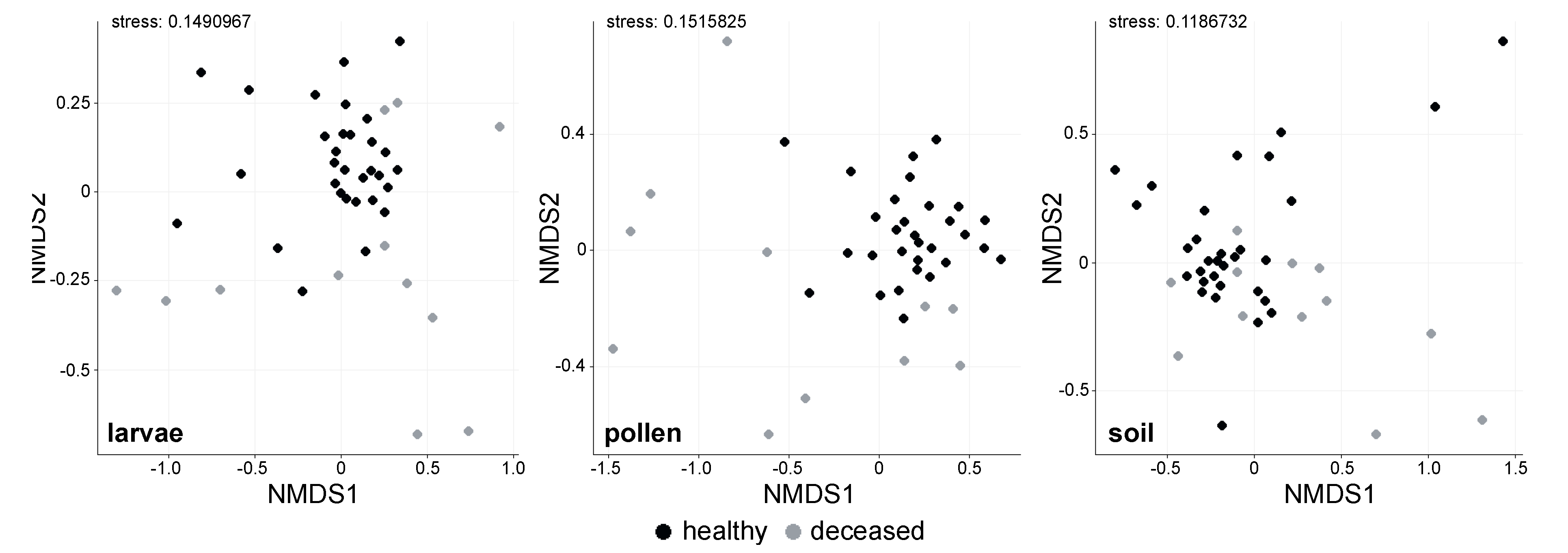
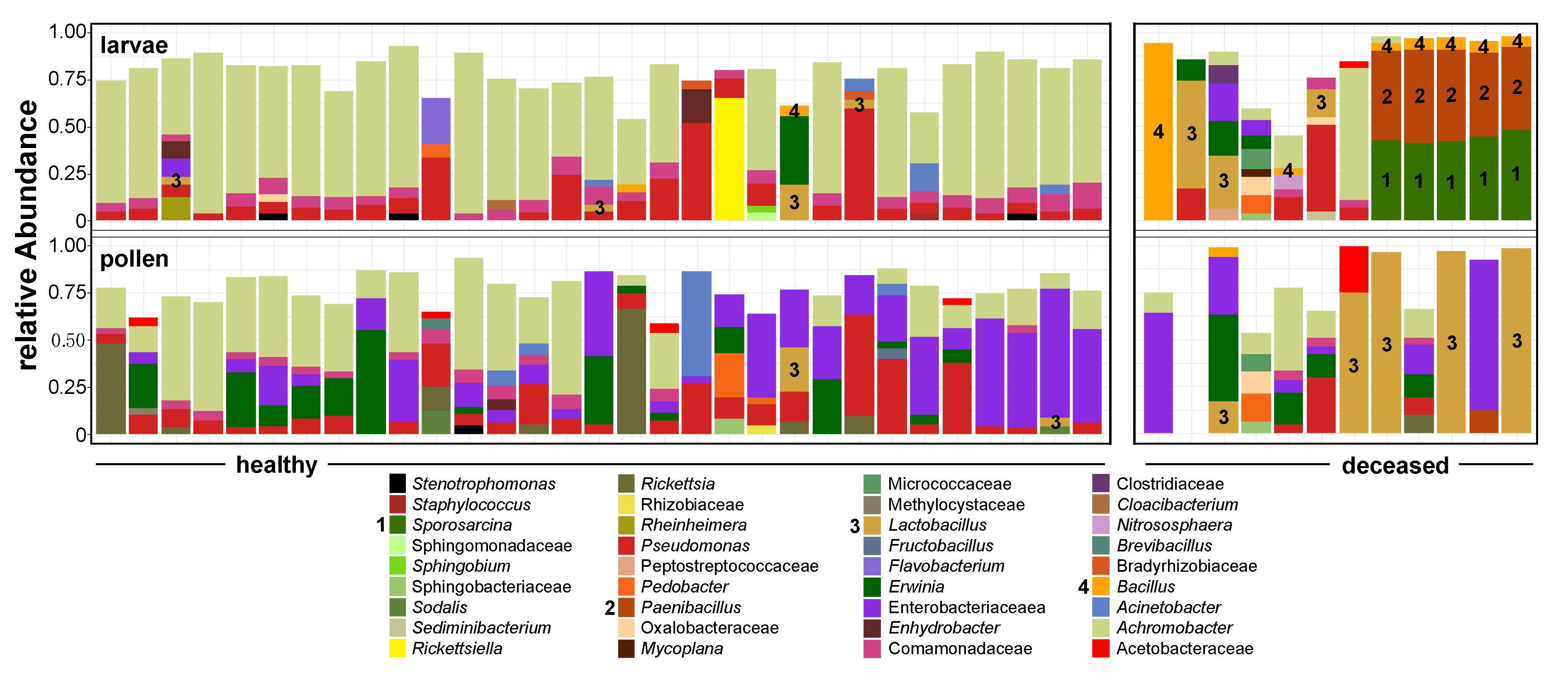
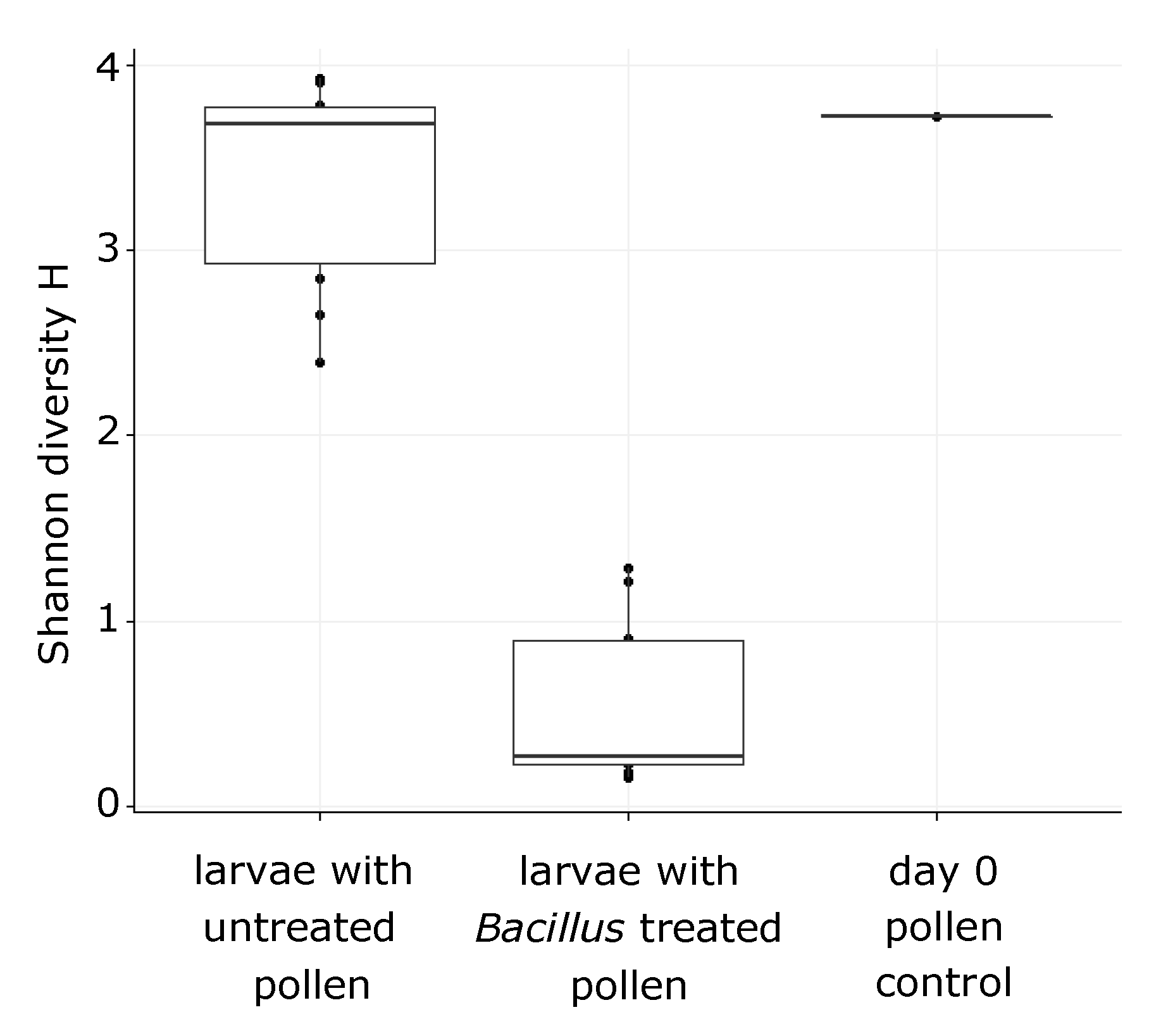
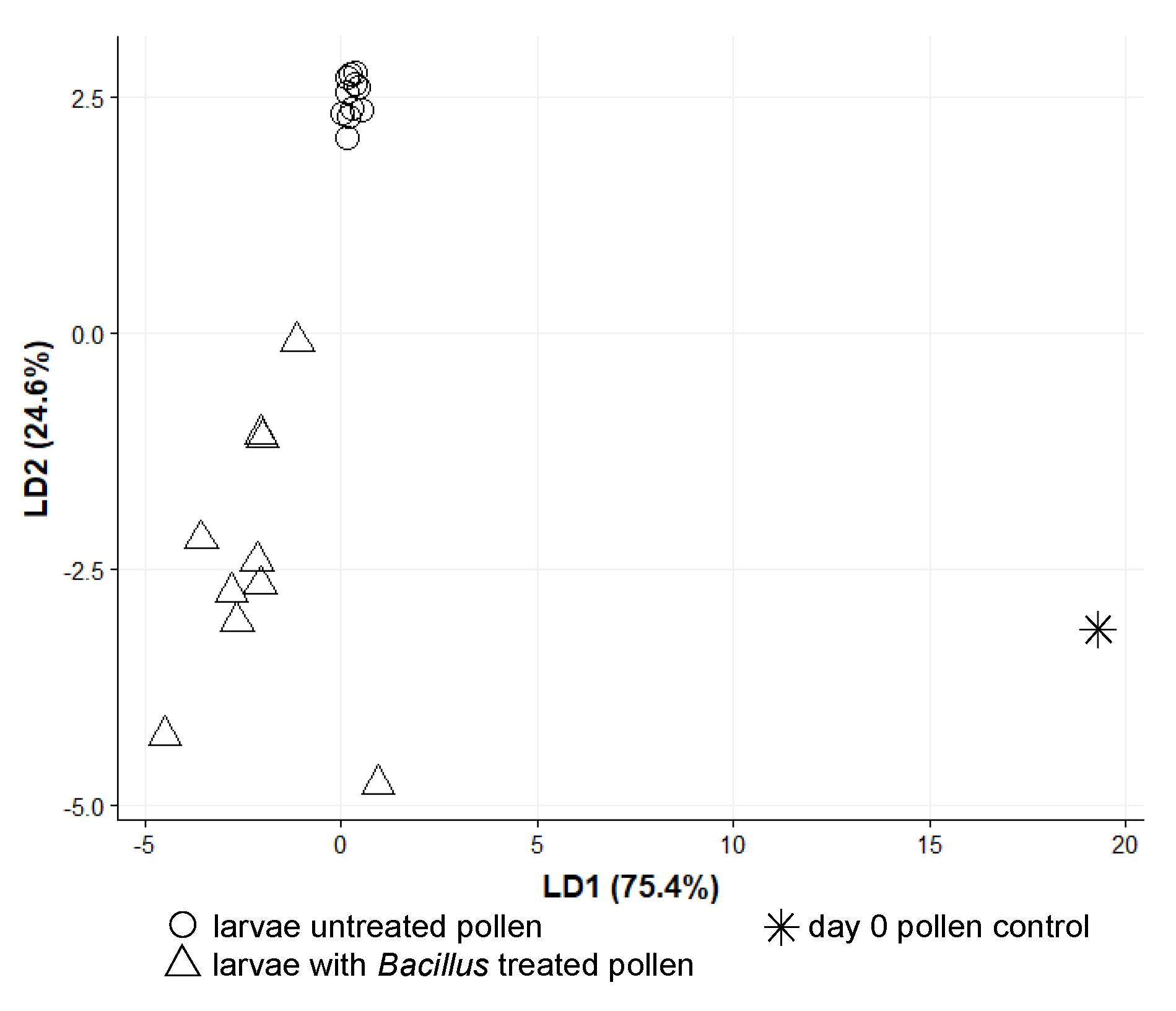
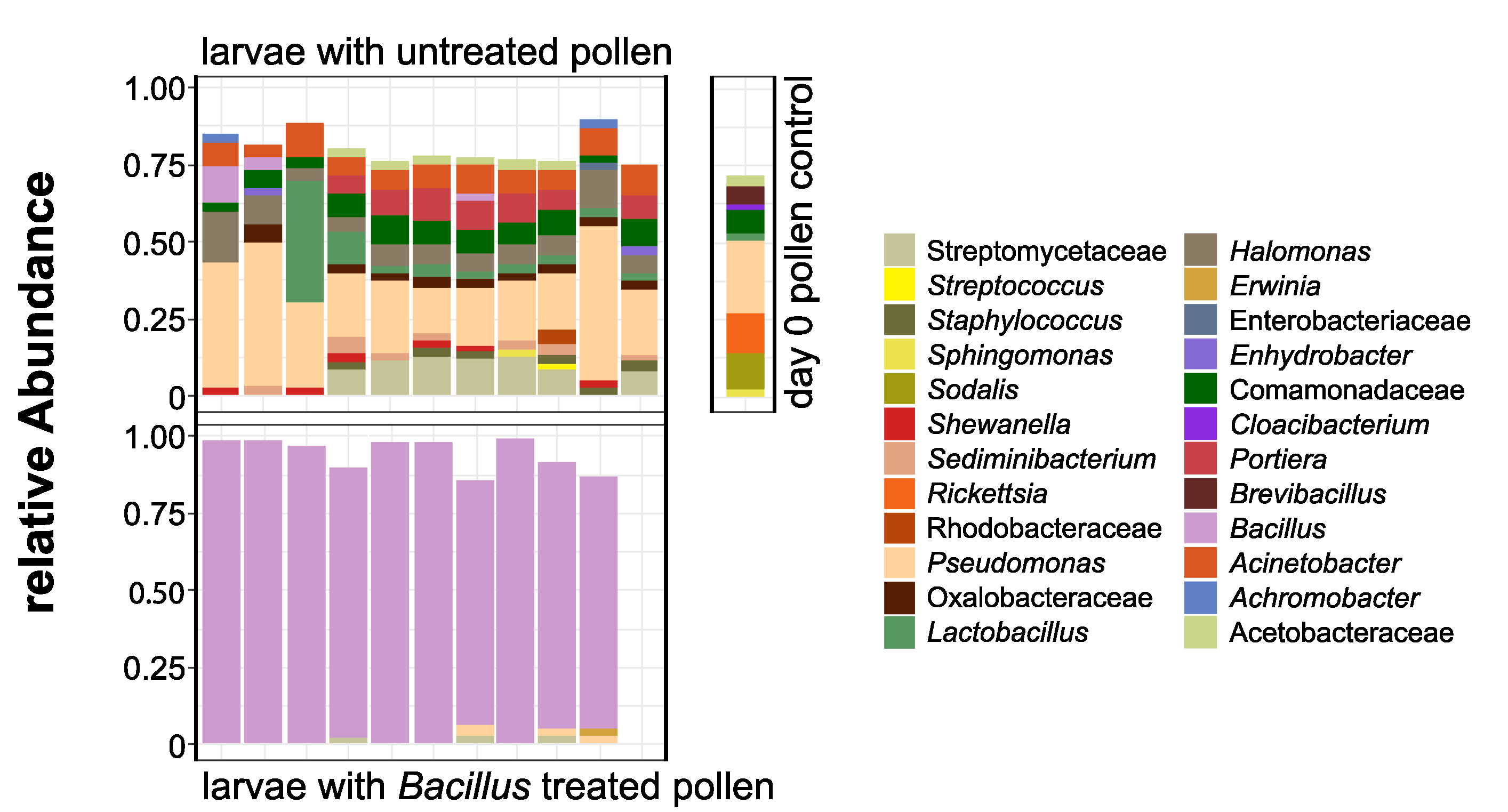
3. Results
3.1. Bacterial Alpha Diversity and Composition of O. bicornis Nest Chambers
3.2. O. bicornis Larvae Provisioning Experiment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Garibaldi, L.A.; Steffan-Dewenter, I.; Winfree, R.; Aizen, M.A.; Bommarco, R.; Cunningham, S.A.; Kremen, C.; Carvalheiro, L.G.; Harder, L.D.; Afik, O.; et al. Wild pollinators enhance fruit set of crops regardless of honey bee abundance. Science 2013, 340, 1608–1611. [Google Scholar] [CrossRef]
- Dainese, M.; Martin, E.A.; Aizen, M.A.; Albrecht, M.; Bartomeus, I.; Bommarco, R.; Carvalheiro, L.G.; Chaplin-Kramer, R.; Gagic, V.; Garibaldi, L.A.; et al. A global synthesis reveals biodiversity-mediated benefits for crop production. Sci. Adv. 2019, 5, eaax0121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aizen, M.A.; Garibaldi, L.A.; Cunningham, S.A.; Klein, A.M. Long-Term Global Trends in Crop Yield and Production Reveal No Current Pollination Shortage but Increasing Pollinator Dependency. Curr. Biol. 2008, 18, 1572–1575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potts, S.G.; Biesmeijer, J.C.; Kremen, C.; Neumann, P.; Schweiger, O.; Kunin, W.E. Global pollinator declines: Trends, impacts and drivers. Trends Ecol. Evol. 2010, 25, 345–353. [Google Scholar] [CrossRef] [PubMed]
- Steffan-Dewenter, I.; Potts, S.G.; Packer, L. Pollinator diversity and crop pollination services are at risk. Trends Ecol. Evol. 2005, 20, 651–652. [Google Scholar] [CrossRef]
- Rundlöf, M.; Andersson, G.K.S.; Bommarco, R.; Fries, I.; Hederström, V.; Herbertsson, L.; Jonsson, O.; Klatt, B.K.; Pedersen, T.R.; Yourstone, J.; et al. Seed coating with a neonicotinoid insecticide negatively affects wild bees. Nature 2015, 521, 77–80. [Google Scholar] [CrossRef]
- González-Varo, J.P.; Biesmeijer, J.C.; Bommarco, R.; Potts, S.G.; Schweiger, O.; Smith, H.G.; Steffan-Dewenter, I.; Szentgyörgyi, H.; Woyciechowski, M.; Vilà, M. Combined effects of global change pressures on animal-mediated pollination. Trends Ecol. Evol. 2013, 28, 524–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schenk, M.; Krauss, J.; Holzschuh, A. Desynchronizations in bee-plant interactions cause severe fitness losses in solitary bees. J. Anim. Ecol. 2018, 87, 139–149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strohm, E. How can cleptoparasitic drosophilid flies emerge from the closed brood cells of the red Mason bee? Physiol. Entomol. 2011, 36, 77–83. [Google Scholar] [CrossRef]
- Fünfhaus, A.; Ebeling, J.; Genersch, E. Bacterial pathogens of bees. Curr. Opin. Insect Sci. 2018, 26, 89–96. [Google Scholar] [CrossRef]
- Genersch, E. American Foulbrood in honeybees and its causative agent, Paenibacillus larvae. J. Invertebr. Pathol. 2010, 103, S10–S19. [Google Scholar] [CrossRef] [PubMed]
- McKee, B.A.; David Goodman, R.; Alan Hornitzky, M. The transmission of European foulbrood (Melissococcus plutonius) to artificially reared honey bee larvae (Apis mellifera). J. Apic. Res. 2004, 43, 93–100. [Google Scholar] [CrossRef]
- Mouches, C.; Bové, J.M.; Albisetti, J.; Clark, T.B.; Tully, J.G. A Spiroplasma of serogroup IV causes a May-disease-like disorder of honeybees in Southwestern France. Microb. Ecol. 1982, 8, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Mouches, C.; Bové, J.M.; Tully, J.G.; Rose, D.L.; McCoy, R.E.; Carle-Junca, P.; Garnier, M.; Saillard, C. Spiroplasma apis, a new species from the honey-bee Apis mellifera. Ann. De L’institut Pasteur. Microbiol. 1983, 134, 383–397. [Google Scholar] [CrossRef]
- Sinpoo, C.; Disayathanoowat, T.; Williams, P.H.; Chantawannakul, P. Prevalence of infection by the microsporidian Nosema spp. in native bumblebees (Bombus spp.) in northern Thailand. PLoS ONE 2019, 14, e0213171. [Google Scholar] [CrossRef] [Green Version]
- Plischuk, S.; Salvarrey, S.; Arbulo, N.; Santos, E.; Skevington, J.H.; Kelso, S.; Revainera, P.D.; Maggi, M.D.; Invernizzi, C.; Lange, C.E. Pathogens, parasites, and parasitoids associated with bumble bees (Bombus spp.) from Uruguay. Apidologie 2017, 48, 298–310. [Google Scholar] [CrossRef] [Green Version]
- Folly, A.J.; Koch, H.; Stevenson, P.C.; Brown, M.J. Larvae act as a transient transmission hub for the prevalent bumblebee parasite Crithidia bombi. J. Invertebr. Pathol. 2017, 148, 81–85. [Google Scholar] [CrossRef]
- Meeus, I.; Vercruysse, V.; Smagghe, G. Molecular detection of Spiroplasma apis and Spiroplasma melliferum in bees. J. Invertebr. Pathol. 2012, 109, 172–174. [Google Scholar] [CrossRef]
- Keller, A.; Grimmer, G.; Steffan-Dewenter, I. Diverse microbiota identified in whole intact nest chambers of the red mason bee Osmia bicornis (Linnaeus 1758). PLoS ONE 2013, 8, e78296. [Google Scholar] [CrossRef] [Green Version]
- Lozo, J.; Berić, T.; Terzić-Vidojević, A.; Stanković, S.; Fira, D.; Stanisavljević, L. Microbiota associated with pollen, bee bread, larvae and adults of solitary bee Osmia cornuta (Hymenoptera: Megachilidae). Bull. Entomol. Res. 2015, 105, 470–476. [Google Scholar] [CrossRef]
- McFrederick, Q.S.; Rehan, S.M. Characterization of pollen and bacterial community composition in brood provisions of a small carpenter bee. Mol. Ecol. 2016, 25, 2302–2311. [Google Scholar] [CrossRef] [PubMed]
- Voulgari-Kokota, A.; Ankenbrand, M.J.; Grimmer, G.; Steffan-Dewenter, I.; Keller, A. Linking pollen foraging of megachilid bees to their nest bacterial microbiota. Ecol. Evol. 2019, 9, 10788–10800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voulgari-Kokota, A.; Grimmer, G.; Steffan-Dewenter, I.; Keller, A. Bacterial community structure and succession in nests of two megachilid bee genera. Fems Microbiol. Ecol. 2019, 95, fiy218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McFrederick, Q.S.; Rehan, S.M. Wild bee pollen usage and microbial communities co-vary across landscapes. Microb. Ecol. 2019, 77, 513–522. [Google Scholar] [CrossRef]
- Ravoet, J.; De Smet, L.; Meeus, I.; Smagghe, G.; Wenseleers, T.; de Graaf, D.C. Widespread occurrence of honey bee pathogens in solitary bees. J. Invertebr. Pathol. 2014, 122, 55–58. [Google Scholar] [CrossRef]
- Bramke, K.; Müller, U.; McMahon, D.P.; Rolff, J. Exposure of Larvae of the Solitary Bee Osmia bicornis to the Honey Bee Pathogen Nosema ceranae Affects Life History. Insects 2019, 10, 380. [Google Scholar] [CrossRef] [Green Version]
- Strobl, V.; Yañez, O.; Straub, L.; Albrecht, M.; Neumann, P. Trypanosomatid parasites infecting managed honeybees and wild solitary bees. Int. J. Parasitol. 2019, 49, 605–613. [Google Scholar] [CrossRef]
- Saeed, A.; White, J.A. Surveys for maternally-inherited endosymbionts reveal novel and variable infections within solitary bee species. J. Invertebr. Pathol. 2015, 132, 111–114. [Google Scholar] [CrossRef]
- Sickel, W.; Ankenbrand, M.; Grimmer, G.; Holzschuh, A.; Härtel, S.; Lanzen, J.; Steffan-Dewenter, I.; Keller, A. Increased efficiency in identifying mixed pollen samples by meta-barcoding with a dual-indexing approach. BMC Ecol. 2015, 15, 20. [Google Scholar] [CrossRef] [Green Version]
- Voulgari-Kokota, A.; McFrederick, Q.S.; Steffan-Dewenter, I.; Keller, A. Drivers, Diversity, and Functions of the Solitary-Bee Microbiota. Trends Microbiol. 2019, 27, 1034–1044. [Google Scholar] [CrossRef]
- Dharampal, P.S.; Carlson, C.; Currie, C.R.; Steffan, S.A. Pollen-borne microbes shape bee fitness. Proc. R. Soc. B Biol. Sci. 2019, 286, 20182894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McFrederick, Q.S.; Wcislo, W.T.; Taylor, D.R.; Ishak, H.D.; Dowd, S.E.; Mueller, U.G. Environment or kin: Whence do bees obtain acidophilic bacteria? Mol. Ecol. 2012, 21, 1754–1768. [Google Scholar] [CrossRef] [PubMed]
- Bosch, J. The nesting behaviour of the mason bee Osmia cornuta (Latr) with special reference to its pollinating potential (Hymenoptera, Megachilidae). Apidologie 1994, 25, 84–93. [Google Scholar] [CrossRef] [Green Version]
- Becker, M.; Keller, A. Laboratory rearing of solitary bees and wasps. Insect Sci. 2016, 23, 918–923. [Google Scholar] [CrossRef]
- Junker, R.R.; Romeike, T.; Keller, A.; Langen, D. Density-dependent responses by bumblebees to flower dwelling bacteria. Apidologie 2014, 45, 467–477. [Google Scholar] [CrossRef] [Green Version]
- Good, A.P.; Gauthier, M.-P.L.; Vannette, R.L.; Fukami, T. Honey Bees Avoid Nectar Colonized by Three Bacterial Species, But Not by a Yeast Species, Isolated from the Bee Gut. PLoS ONE 2014, 9, e86494. [Google Scholar] [CrossRef]
- Burbach, K.; Seifert, J.; Pieper, D.H.; Camarinha-Silva, A. Evaluation of DNA extraction kits and phylogenetic diversity of the porcine gastrointestinal tract based on Illumina sequencing of two hypervariable regions. MicrobiologyOpen 2016, 5, 70–82. [Google Scholar] [CrossRef]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a Dual-Index Sequencing Strategy and Curation Pipeline for Analyzing Amplicon Sequence Data on the MiSeq Illumina Sequencing Platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [Green Version]
- Aronesty, E. Comparison of Sequencing Utility Programs. Open Bioinform. J. 2013, 7. [Google Scholar] [CrossRef]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. UCHIME2: Improved chimera prediction for amplicon sequencing. bioRxiv 2016, 074252. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Flyvbjerg, H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 2015, 31, 3476–3482. [Google Scholar] [CrossRef] [Green Version]
- Maidak, B.L.; Olsen, G.J.; Larsen, N.; Overbeek, R.; McCaughey, M.J.; Woese, C.R. The Ribosomal Database Project (RDP). Nucleic Acids Res. 1996, 24, 82–85. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: http://www.r-project.org (accessed on 1 October 2016).
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickham, H. ggplot2. Wires Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Lahti, L.; Shetty, S.; Blake, T. others Tools for microbiome analysis in R. Microbiome Package Version 0.99. 2017. Available online: http://microbiome.github.com/microbiome (accessed on 1 October 2016).
- Ripley, B.; Venables, B.; Bates, D.M.; Hornik, K.; Gebhardt, A.; Firth, D.; Ripley, M.B. Package ‘mass.’. 2013. Available online: http://www.cran.r-project.org/package=MASS (accessed on 1 October 2016).
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Stevens, M.H.H.; Oksanen, M.J.; Suggests, M. The vegan package. Community Ecol. Package 2007, 10, 631–637. [Google Scholar]
- Legendre, P.; Legendre, L.F. Numerical Ecology; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Friedman, J.; Alm, E.J. Inferring Correlation Networks from Genomic Survey Data. PLoS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller, A.; Brandel, A.; Becker, M.C.; Balles, R.; Abdelmohsen, U.R.; Ankenbrand, M.J.; Sickel, W. Wild bees and their nests host Paenibacillus bacteria with functional potential of avail. Microbiome 2018, 6, 229. [Google Scholar] [CrossRef] [Green Version]
- Junker, R.R.; Keller, A. Microhabitat heterogeneity across leaves and flower organs promotes bacterial diversity. FEMS Microbiol. Ecol. 2015, 91, fiv097. [Google Scholar] [CrossRef]
- Engel, P.; Kwong, W.K.; McFrederick, Q.; Anderson, K.E.; Barribeau, S.M.; Chandler, J.A.; Cornman, R.S.; Dainat, J.; de Miranda, J.R.; Doublet, V.; et al. The Bee Microbiome: Impact on Bee Health and Model for Evolution and Ecology of Host-Microbe Interactions. mBio 2016, 7, e02164-15. [Google Scholar] [CrossRef] [Green Version]
- Anderson, K.E.; Ricigliano, V.A. Honey bee gut dysbiosis: A novel context of disease ecology. Curr. Opin. Insect Sci. 2017, 22, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Maes, P.W.; Rodrigues, P.A.P.; Oliver, R.; Mott, B.M.; Anderson, K.E. Diet-related gut bacterial dysbiosis correlates with impaired development, increased mortality and Nosema disease in the honeybee (Apis mellifera). Mol. Ecol. 2016, 25, 5439–5450. [Google Scholar] [CrossRef] [PubMed]
- Grady, E.N.; MacDonald, J.; Liu, L.; Richman, A.; Yuan, Z.-C. Current knowledge and perspectives of Paenibacillus: A review. Microb Cell Fact 2016, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forsgren, E. European foulbrood in honey bees. J. Invertebr. Pathol. 2010, 103, S5–S9. [Google Scholar] [CrossRef]
- Menegatti, C.; Da Paixão Melo, W.G.; Carrão, D.B.; De Oliveira, A.R.M.; Do Nascimento, F.S.; Lopes, N.P.; Pupo, M.T. Paenibacillus polymyxa associated with the stingless bee Melipona scutellaris produces antimicrobial compounds against entomopathogens. J. Chem. Ecol. 2018, 44, 1158–1169. [Google Scholar] [CrossRef]
- Sáez-Nieto, J.A.; Medina-Pascual, M.J.; Carrasco, G.; Garrido, N.; Fernandez-Torres, M.A.; Villalón, P.; Valdezate, S. Paenibacillus spp. isolated from human and environmental samples in Spain: Detection of 11 new species. New Microbes New Infect. 2017, 19, 19–27. [Google Scholar] [CrossRef]
- Georgieva, Y.H.; Groudeva, V.I.; Shishiniova, M.D. Taxonomic Identification of Bacteria, Associated with Bulgarian Populations of Entomopathogenic Nematodes from Genus Steinernema (Rhabditida, Steinernematidae) II. Biotechnol. Biotechnol. Equip. 2005, 19, 68–73. [Google Scholar] [CrossRef] [Green Version]
- Waltmann, A.; Willcox, A.C.; Balasubramanian, S.; Borrini Mayori, K.; Mendoza Guerrero, S.; Salazar Sanchez, R.S.; Roach, J.; Condori Pino, C.; Gilman, R.H.; Bern, C.; et al. Hindgut microbiota in laboratory-reared and wild Triatoma infestans. PLoS Negl. Trop. Dis. 2019, 13, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Jurat-Fuentes, J.L.; Jackson, T.A. Chapter 8—Bacterial Entomopathogens. In Insect Pathology, 2nd ed.; Vega, F.E., Kaya, H.K., Eds.; Academic Press: San Diego, CA, USA, 2012; pp. 265–349. ISBN 978-0-12-384984-7. [Google Scholar]
- Poudel, R.; Jumpponen, A.; Schlatter, D.C.; Paulitz, T.C.; Gardener, B.B.M.; Kinkel, L.L.; Garrett, K.A. Microbiome Networks: A Systems Framework for Identifying Candidate Microbial Assemblages for Disease Management. Phytopathology 2016, 106, 1083–1096. [Google Scholar] [CrossRef]
- McFrederick, Q.S.; Thomas, J.M.; Neff, J.L.; Vuong, H.Q.; Russell, K.A.; Hale, A.R.; Mueller, U.G. Flowers and Wild Megachilid Bees Share Microbes. Microb. Ecol. 2017, 73, 188–200. [Google Scholar] [CrossRef]
- Vásquez, A.; Forsgren, E.; Fries, I.; Paxton, R.J.; Flaberg, E.; Szekely, L.; Olofsson, T.C. Symbionts as Major Modulators of Insect Health: Lactic Acid Bacteria and Honeybees. PLoS ONE 2012, 7, e33188. [Google Scholar] [CrossRef]
- Killer, J.; Dubná, S.; Sedláček, I.; Švec, P. Lactobacillus apis sp. nov., from the stomach of honeybees (Apis mellifera), having an in vitro inhibitory effect on the causative agents of American and European foulbrood. Int. J. Syst. Evol. Microbiol. 2014, 64, 152–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zemenick, A.T.; Vannette, R.L.; Rosenheim, J.A. Linked networks reveal dual roles of insect dispersal and species sorting for bacterial communities in flowers. bioRxiv 2019, 847376. [Google Scholar] [CrossRef]
- Gaube, P.; Junker, R.R.; Keller, A. Changes amid constancy: Flower and leaf microbiomes along land use gradients and between bioregions. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Miller, D.L.; Smith, E.A.; Newton, I.L.G. A bacterial symbiont protects honey bees from fungal disease. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Steffan, S.A.; Dharampal, P.S.; Danforth, B.N.; Gaines-Day, H.R.; Takizawa, Y.; Chikaraishi, Y. Omnivory in bees: Elevated trophic positions among all major bee families. Am. Nat. 2019. [Google Scholar] [CrossRef]
- Helm, B.R.; Payne, S.; Rinehart, J.P.; Yocum, G.D.; Bowsher, J.H.; Greenlee, K.J. Micro-computed tomography of pupal metamorphosis in the solitary bee Megachile rotundata. Arthropod Struct. Dev. 2018, 47, 521–528. [Google Scholar] [CrossRef]
- Rothman, J.A.; Andrikopoulos, C.; Cox-Foster, D.; McFrederick, Q.S. Floral and Foliar Source Affect the Bee Nest Microbial Community. Microb. Ecol. 2018. [Google Scholar] [CrossRef] [Green Version]

© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Voulgari-Kokota, A.; Steffan-Dewenter, I.; Keller, A. Susceptibility of Red Mason Bee Larvae to Bacterial Threats Due to Microbiome Exchange with Imported Pollen Provisions. Insects 2020, 11, 373. https://doi.org/10.3390/insects11060373
Voulgari-Kokota A, Steffan-Dewenter I, Keller A. Susceptibility of Red Mason Bee Larvae to Bacterial Threats Due to Microbiome Exchange with Imported Pollen Provisions. Insects. 2020; 11(6):373. https://doi.org/10.3390/insects11060373
Chicago/Turabian StyleVoulgari-Kokota, Anna, Ingolf Steffan-Dewenter, and Alexander Keller. 2020. "Susceptibility of Red Mason Bee Larvae to Bacterial Threats Due to Microbiome Exchange with Imported Pollen Provisions" Insects 11, no. 6: 373. https://doi.org/10.3390/insects11060373
APA StyleVoulgari-Kokota, A., Steffan-Dewenter, I., & Keller, A. (2020). Susceptibility of Red Mason Bee Larvae to Bacterial Threats Due to Microbiome Exchange with Imported Pollen Provisions. Insects, 11(6), 373. https://doi.org/10.3390/insects11060373





