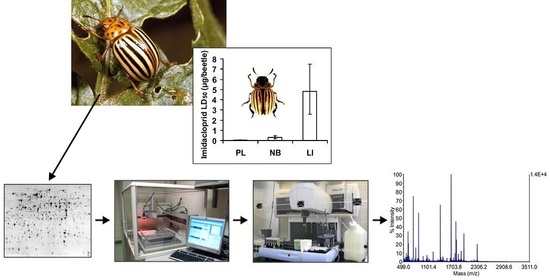
Proteomic Analyses Detect Higher Expression of C-Type Lectins in Imidacloprid-Resistant Colorado Potato Beetle Leptinotarsa decemlineata Say
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. CPB Strains
2.2. Insecticide Exposure
2.3. Cytochrome P450 and b5 Activity
2.4. Proteomic Analysis
2.5. Imaging and Mass Spectrometric Analysis
2.6. Protein Identification
3. Results
3.1. Imidacloprid Susceptibility
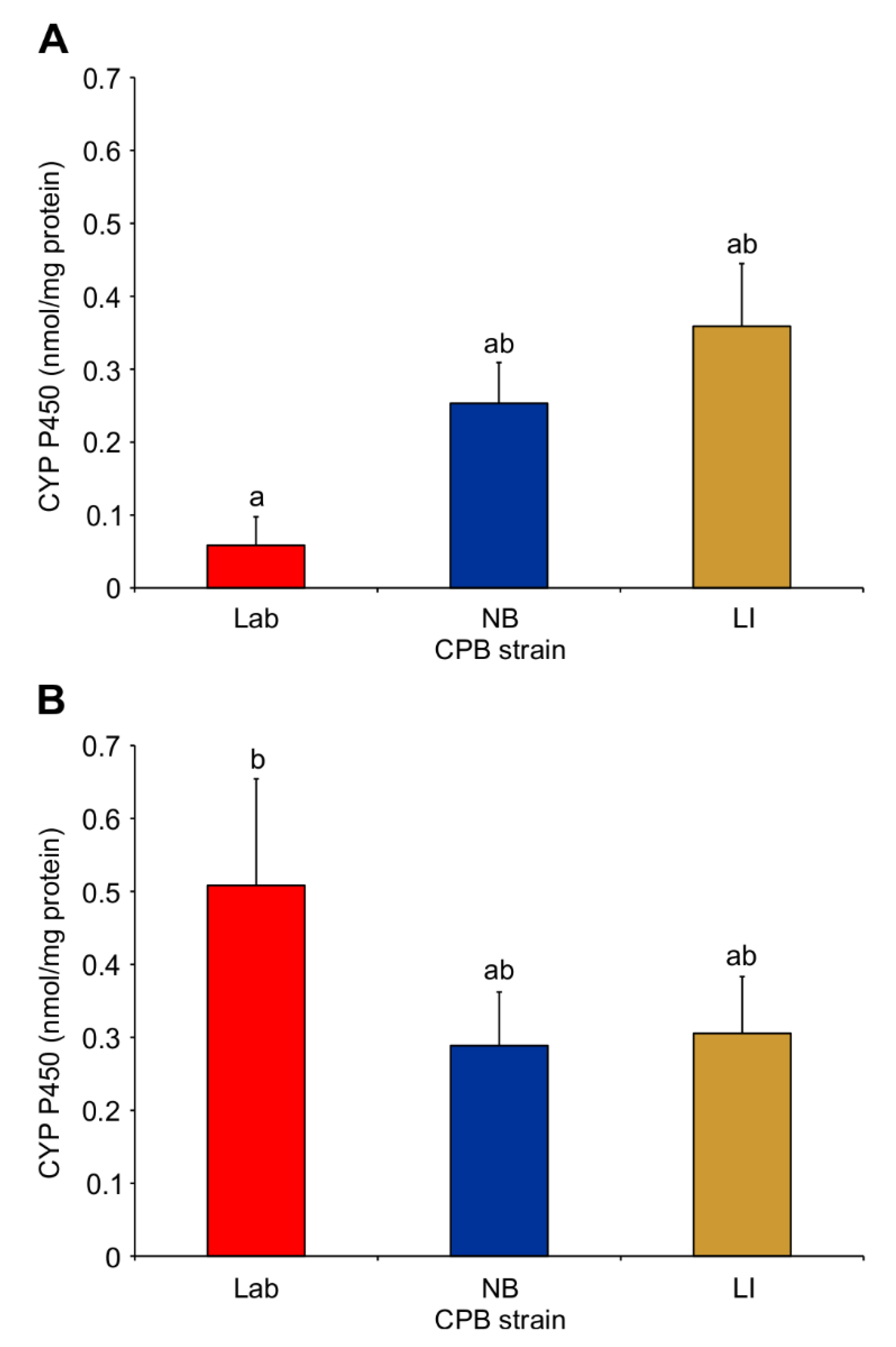
3.2. Cytochrome P450 and b5 Activity
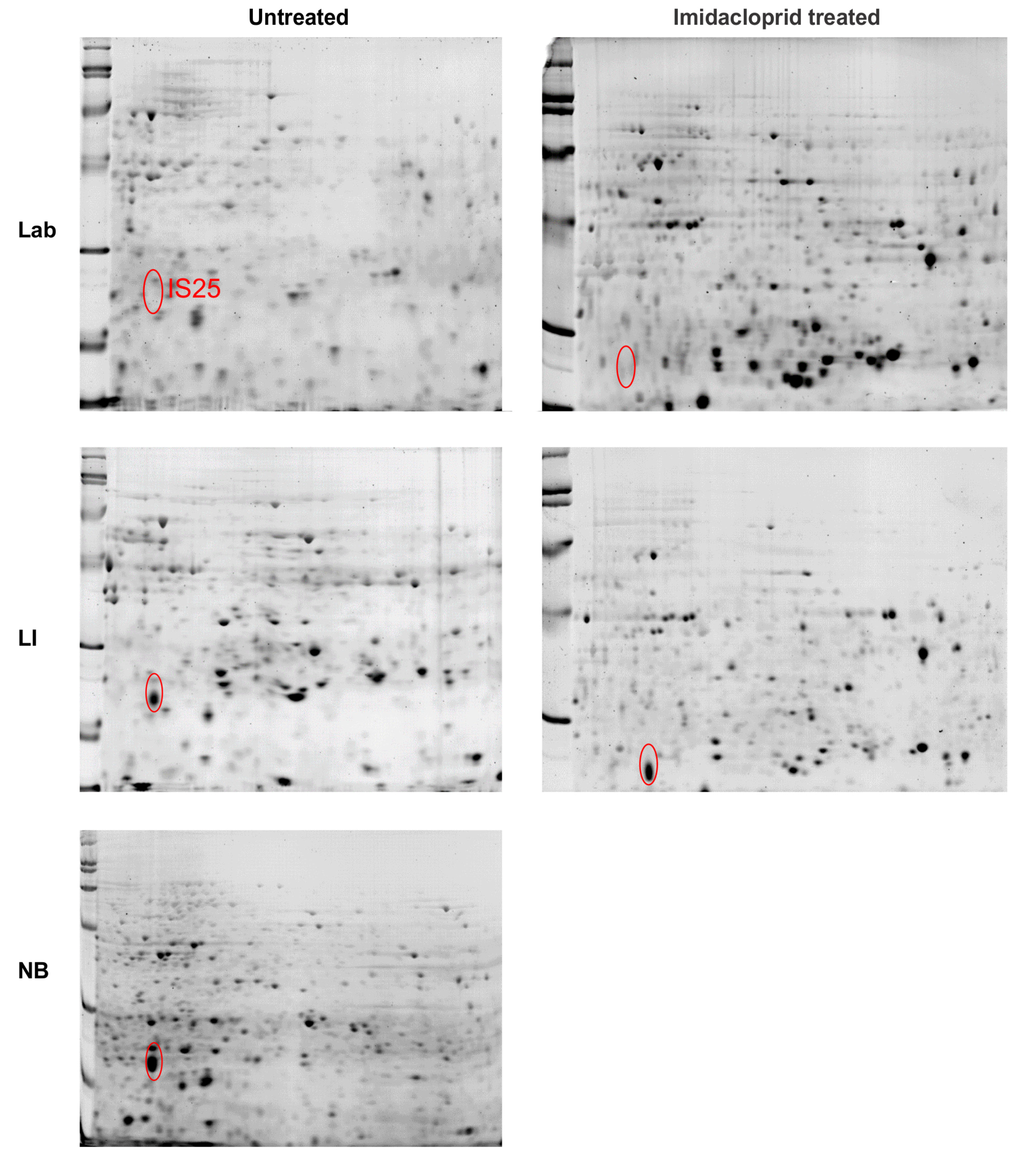
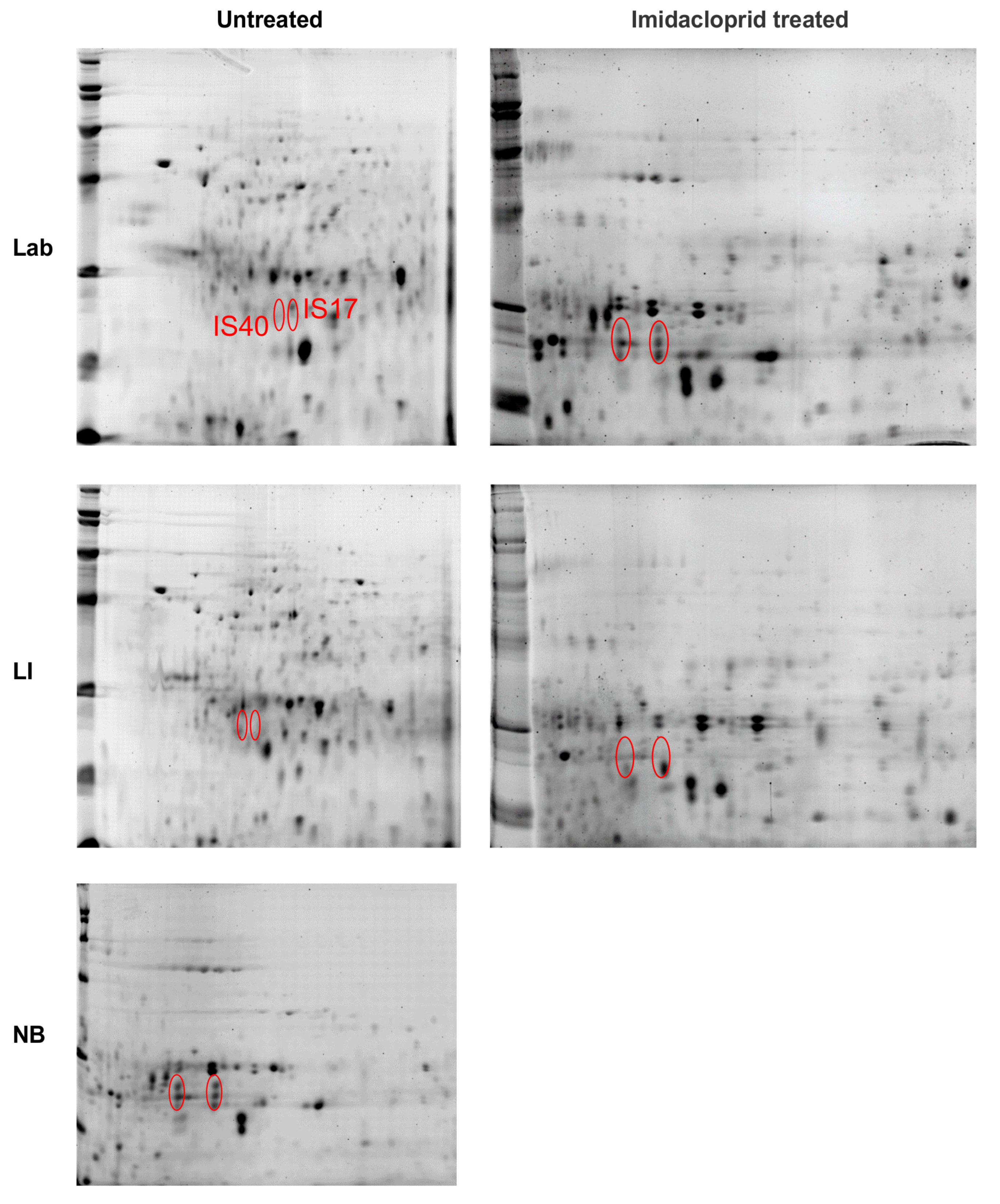
3.3. Proteomic Analyses
3.3.1. Abdominal Contents
3.3.2. Midgut
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alyokhin, A.; Chen, Y.H.; Udalov, M.; Benkovskaya, G.; Lindstrom, L. Evolutionary considerations in potato pest management. In Insect Pests of Potato: Global Perspectives on Biology and Management; Giordanengo, P., Vincent, C., Alyokhin, A., Eds.; Academic Press: Oxford, UK, 2013. [Google Scholar]
- Mota-Sanchez, D.; Hollingworth, R.M.; Grafius, E.J.; Moyer, D.D. Resistance and cross-resistance to neonicotinoid insecticides and spinosad in the Colorado potato beetle, Leptinotarsa decemlineata (Say) (Coleoptera: Chrysomelidae). Pest Manag. Sci. 2006, 62, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Alyokhin, A.; Dively, G.; Patterson, M.; Mahoney, M.; Rogers, D.; Wollam, J. Susceptibility of imidacloprid-resistant Colorado potato beetles to non-neonicotinoid insecticides in the laboratory and field trials. Am. J. Potato Res. 2006, 83, 485. [Google Scholar] [CrossRef]
- Scott, I.M.; Tolman, J.H.; MacArthur, D.C. Insecticide resistance and cross-resistance development in Colorado potato beetle Leptinotarsa decemlineata Say (Coleoptera: Chrysomelidae) populations in Canada 2008–2011. Pest Manag. Sci. 2015, 71, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Crossley, M.S.; Rondon, S.I.; Schoville, S.D. A comparison of resistance to imidacloprid in Colorado potato beetle (Leptinotarsa decemlineata say) populations collected in the northwest and midwest U.S. Am. J. Potato Res. 2018, 95, 495–503. [Google Scholar] [CrossRef]
- Kaplanoglu, E.; Chapman, P.; Scott, I.M.; Donly, C. Overexpression of a cytochrome P450 and a UDP-glycosyltransferase is associated with imidacloprid resistance in the Colorado potato beetle, Leptinotarsa decemlineata. Sci. Rep. 2017, 7, 1762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinkevich, F.D.; Su, C.; Lazo, T.A.; Hawthorne, D.J.; Tingey, W.M.; Naimov, S.; Scott, J.G. Multiple evolutionary origins of knockdown resistance (kdr) in pyrethroid-resistant Colorado potato beetle, Leptinotarsa decemlineata. Pestic. Biochem. Physiol. 2012, 104, 192–200. [Google Scholar] [CrossRef]
- Baker, M.B.; Alyokhin, A.; Porter, A.H.; Ferro, D.N.; Dastur, S.R.; Galal, N. Persistence and inheritance of costs of resistance to imidacloprid in Colorado potato beetle. J. Econ. Entomol. 2007, 100, 1871–1879. [Google Scholar] [CrossRef]
- Chen, J.; Alyokhin, A.; Mota-Sanchez, D.; Baker, M.; Whalon, M. Variation in fitness among geographically isolated Colorado potato beetle (Coleoptera: Chrysomelidae) populations. Ann. Entomol. Soc. Am. 2014, 107, 128–135. [Google Scholar] [CrossRef] [Green Version]
- Dawkar, V.V.; Chikate, Y.R.; More, T.H.; Gupta, V.S.; Giri, A.P. The expression of proteins involved in digestion and detoxification are regulated in Helicoverpa armigera to cope up with chlorpyrifos insecticide. Insect Sci. 2016, 23, 68–77. [Google Scholar] [CrossRef]
- Hou, M.-Z.; Shen, G.-M.; Wei, D.; Li, Y.-L.; Dou, W.; Wang, J.-J. Characterization of Bactrocera dorsalis serine proteases and evidence for their indirect role in insecticide tolerance. Int. J. Mol. Sci. 2014, 15, 3272–3286. [Google Scholar] [CrossRef] [Green Version]
- Silva, L.B.; Reis, A.P.; Pereira, E.J.G.; Oliveira, M.G.A.; Guedes, R.N.C. Altered cysteine proteinase activity in insecticide-resistant strains of the maize weevil: Purification and characterization. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2010, 157, 80–87. [Google Scholar] [CrossRef] [PubMed]
- Clements, J.; Olson, J.M.; Sanchez-Sedillo, B.; Bradford, B.; Groves, R.L. Changes in emergence phenology, fatty acid composition, and xenobiotic-metabolizing enzyme expression is associated with increased insecticide resistance in the Colorado potato beetle. Insect Biochem. Physiol. 2019, 103, e21630. [Google Scholar] [CrossRef] [PubMed]
- Huseth, A.S.; Groves, R.L. Effect of insecticide management history on emergence phenology and neonicotinoid resistance in Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). J. Econ. Entomol. 2013, 106, 2491–2505. [Google Scholar] [CrossRef] [PubMed]
- Schoville, S.D.; Chen, Y.H.; Andersson, M.N.; Benoit, J.B.; Bhandari, A.; Bowsher, J.H.; Brevik, K.; Cappelle, K.; Chen, M.-J.M.; Childers, A.K.; et al. A model species for agricultural pest genomics: The genome of the Colorado potato beetle, Leptinotarsa decemlineata (Coleoptera: Chrysomelidae). Sci. Rep. 2018, 8, 1–18. [Google Scholar] [CrossRef] [Green Version]
- Gruden, K.; Kuipers, A.G.J.; Gunčar, G.; Slapar, N.; Štrukelj, B.; Jongsma, M.A. Molecular basis of Colorado potato beetle adaptation to potato plant defence at the level of digestive cysteine proteinases. Insect Biochem. Mol. Biol. 2004, 34, 365–375. [Google Scholar] [CrossRef]
- Dastranj, M.; Gharechahi, J.; Salekdeh, G.H. Insect pest proteomics and its potential application in pest control management. In Agricultural Proteomics Volume 2: Environmental Stresses; Salekdeh, G.H., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 267–287. [Google Scholar]
- Veenstra, T.D.; Yates, J.R. Proteomics for Biological Discovery; Wiley-Liss: Hoboken, NJ, USA, 2006. [Google Scholar]
- Erban, T.; Stara, J. Methodology for glutathione S-transferase purification and localization in two-dimensional gel electrophoresis performed on the pollen beetle, Meligethes aeneus (Coleoptera: Nitidulidae). J. Asia-Pac. Entomol. 2014, 17, 369–373. [Google Scholar] [CrossRef]
- Baggerman, G.; Boonen, K.; Verleyen, P.; De Loof, A.; Schoofs, L. Peptidomic analysis of the larval Drosophila melanogaster central nervous system by two-dimensional capillary liquid chromatography quadrupole time-of-flight mass spectrometry. J. Mass Spectrom. 2005, 40, 250–260. [Google Scholar] [CrossRef]
- Guerrera, I.C.; Kleiner, O. Application of mass spectrometry in proteomics. Biosci. Rep. 2005, 25, 71–93. [Google Scholar] [CrossRef]
- Cañas, B.; Piñeiro, C.; Calvo, E.; López-Ferrer, D.; Gallardo, J.M. Trends in sample preparation for classical and second generation proteomics. J. Chromatogr. A 2007, 1153, 235–258. [Google Scholar] [CrossRef]
- Hillenkamp, F.; Peter-Katalinic, J. (Eds.) Maldi MS: A Practical Guide to Instrumentation, Methods and Applications; Wiley-VCH: Weinheim, Germany, 2007. [Google Scholar]
- Francis, F.; Gerkens, P.; Harmel, N.; Mazzucchelli, G.; De Pauw, E.; Haubruge, E. Proteomics in Myzus persicae: Effect of aphid host plant switch. Insect Biochem. Mol. Biol. 2006, 36, 219–227. [Google Scholar] [CrossRef] [Green Version]
- Sharma, R.; Komatsu, S.; Noda, H. Proteomic analysis of brown planthopper: Application to the study of carbamate toxicity. Insect Biochem. Mol. Biol. 2004, 34, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Shi, P.; Chen, G.; Du, J. Effects of azadirachtin on hemolymph protein expression in Ostrinia furnacalis (Lepidoptera: Crambidae). Ann. Entomol. Soc. Am. 2007, 100, 245–250. [Google Scholar] [CrossRef]
- Cox, B.; Kislinger, T.; Emili, A. Integrating gene and protein expression data: Pattern analysis and profile mining. Methods 2005, 35, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Scott, J.G.; Liu, N.; Wen, Z. Insect cytochromes p450: Diversity, insecticide resistance and tolerance to plant toxins. Comp. Biochem. Physiol. Part C Pharmacol. Toxicol. Endocrinol. 1998, 121, 147–155. [Google Scholar] [CrossRef]
- Lee, S.S.T.; Scott, J.G. An improved method for preparation, stabilization, and storage of house fly (Diptera: Muscidae) microsomes. J. Econ. Entomol. 1989, 82, 1559–1563. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Lu, Y.; Su, F.; Zhu, K.; Zhu, M.; Li, Q.; Hu, Q.; Zhang, J.; Zhang, R.; Yu, X.-Q. Comparative genomic analysis of C-type lectin-domain genes in seven holometabolous insect species. Insect Biochem. Mol. Biol. 2020, 126, 103451. [Google Scholar] [CrossRef]
- Xia, X.; You, M.-S.; Rao, X.-J.; Yu, X.-Q. Insect c-type lectins in innate immunity. Dev. Comp. Immunol. 2018, 83, 70–79. [Google Scholar] [CrossRef]
- Bi, J.; Ning, M.; Li, J.; Zhang, P.; Wang, L.; Xu, S.; Zhong, Y.; Wang, Z.; Song, Q.-S.; Li, B. A C-type lectin with dual-CRD from Tribolium castaneum is induced in response to bacterial challenge. Pest Manag. Sci. 2020, 76, 3965–3974. [Google Scholar] [CrossRef]
- Meng, Q.; Zhang, J.-H.; Zhang, H.; Zhou, G.-L.; Ni, R.-Y.; Zhao, Y.-N.; Qin, Q.-L.; Zou, Z. Comparative analysis of C-type lectin domain proteins in the ghost moth, Thitarodes xiaojinensis (Lepidoptera: Hepialidae). Insect Sci. 2019, 26, 453–465. [Google Scholar] [CrossRef] [Green Version]
- Pees, B.; Yang, W.; Zárate-Potes, A.; Schulenburg, H.; Dierking, K. High innate immune specificity through diversified C-type lectin-like domain proteins in invertebrates. J. Innate Immun. 2016, 8, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-J.; Wang, W.-W.; Liu, Y.; Chai, L.-Q.; Wang, G.-X.; Liu, X.-S.; Wang, Y.-F.; Wang, J.L. Steroid hormone 20-hydroxyecdysone promotes ctl1-mediated cellular immunity in Helicoverpa armigera. Insect Sci. 2020. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.-K.; Tian, M.-L.; Dong, Y.-P.; Ren, C.-B.; Du, Y.; Hu, J. The C-type lectin IML-10 promotes hemocytic encapsulation by enhancing aggregation of hemocytes in the Asian corn borer Ostrinia furnacalis. Insect Biochem. Mol. Biol. 2020, 118, 103314. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wang, G.; Zhuo, X.; Liu, Y.; Tang, L.; Liu, X.; Wang, J. C-type lectin-mediated microbial homeostasis is critical for Helicoverpa armigera larval growth and development. PLoS Pathog. 2020, 16, e1008901. [Google Scholar] [CrossRef] [PubMed]
- Vézilier, J.; Nicot, A.; De Lorgeril, J.; Gandon, S.; Rivero, A. The impact of insecticide resistance on Culex pipiens immunity. Evol. Appl. 2013, 6, 497–509. [Google Scholar] [CrossRef]
- Batool, K.; Alam, I.; Jin, L.; Xu, J.; Wu, C.; Wang, J.; Huang, E.; Guan, X.; Yu, X.-Q.; Zhang, L. Ctlga9 interacts with ALP1 and APN receptors to modulate Cry11Aa toxicity in Aedes aegypti. J. Agric. Food Chem. 2019, 67, 8896–8904. [Google Scholar] [CrossRef]
- Wang, W.; Lv, Y.; Fang, F.; Hong, S.; Guo, Q.; Hu, S.; Zou, F.; Shi, L.; Lei, Z.; Ma, K.; et al. Identification of proteins associated with pyrethroid resistance by iTRAQ-based quantitative proteomic analysis in Culex pipiens pallens. Parasites Vectors 2015, 8, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Yan, D.-K.; Hu, M.; Tang, Y.-X.; Fan, J.-Q. Proteomic analysis reveals resistance mechanism against chlorpyrifos in Frankliniella occidentalis (Thysanoptera: Thripidae). J. Econ. Entomol. 2015, 108, 2000–2008. [Google Scholar] [CrossRef]
- Kang, S.; Lee, H.J.; Kim, Y.H.; Kwon, D.H.; Oh, J.H.; Kim, B.J.; Lim, K.J.; Lee, S.; Hwang, S.Y.; Lee, S.H. Proteomics-based identification and characterization of biotype-specific carboxylesterase 2 putatively associated with insecticide resistance in Bemisia tabaci. J. Asia-Pac. Entomol. 2012, 15, 389–396. [Google Scholar] [CrossRef]
- Meng, J.; Zhang, C.; Chen, X.; Cao, Y.; Shang, S. Differential protein expression in the susceptible and resistant Myzus persicae (Sulzer) to imidacloprid. Pestic. Biochem. Physiol. 2014, 115, 1–8. [Google Scholar] [CrossRef]
- García-Robles, I.; De Loma, J.; Capilla, M.; Roger, I.; Boix-Montesinos, P.; Carrión, P.; Marcos Vicente, M.; López-Galiano, M.; Dolores Real, M.; Rausell, C. Proteomic insights into the immune response of the Colorado potato beetle larvae challenged with Bacillus thuringiensis. Dev. Comp. Immunol. 2020, 104, 103525. [Google Scholar] [CrossRef] [PubMed]
- Merzendorfer, H.; Kim, H.S.; Chaudhari, S.S.; Kumari, M.; Specht, C.A.; Butcher, S.; Brown, S.J.; Robert Manak, J.; Beeman, R.W.; Kramer, K.J.; et al. Genomic and proteomic studies on the effects of the insect growth regulator diflubenzuron in the model beetle species Tribolium castaneum. Insect Biochem. Mol. Biol. 2012, 42, 264–276. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Digestion Type | Hit # | Accession | Score −10lgP | Percent Coverage | Average Mass (Da) | 1 Protein Description |
|---|---|---|---|---|---|---|
| Trypsin | 1 | gi|1285030099 | 317.44 | 62 | 19,715 | ladderlectin-like |
| 2 | gi|1285031125 | 209.63 | 31 | 19,830 | E-selectin-like | |
| 3 | gi|1285019956 | 70.14 | 14 | 12,120 | glutamine synthetase-like | |
| AspN | 1 | gi|1285030099 | 300.21 | 56 | 19,715 | ladderlectin-like |
| 2 | gi|1285031125 | 110.03 | 26 | 19,830 | E-selectin-like | |
| 3 | gi|1285029928 | 54.49 | 9 | 18,910 | chromobox protein homolog 1-like | |
| Chymotrypsin | 1 | gi|1285030099 | 77.46 | 16 | 19,715 | ladderlectin-like |
| Digestion Type | Hit # | Accession | Score −10lgP | Percent Coverage | Average Mass (Da) | Protein Description | Species |
|---|---|---|---|---|---|---|---|
| Trypsin | 1 | gi|1285030099 | 208.61 | 40 | 19,715 | ladderlectin-like | Leptinotarsa decemlineata |
| 2 | gi|121531620 | 183.54 | 32 | 29,420 | digestive cysteine protease intestain, partial | Leptinotarsa decemlineata | |
| 3 | gi|1285031125 | 175.44 | 33 | 19,830 | E-selectin-like | Leptinotarsa decemlineata | |
| AspN | 1 | gi|1285030099 | 203.11 | 37 | 19,715 | ladderlectin-like | Leptinotarsa decemlineata |
| 2 | gi|121531620 | 102.9 | 9 | 29,420 | digestive cysteine protease intestain, partial | Leptinotarsa decemlineata | |
| 3 | gi|1285022454 | 41.51 | 3 | 53,879 | dihydrolipoyl dehydrogenase, mitochondrial | Leptinotarsa decemlineata | |
| Chymotrypsin | 1 | gi|1796093563 | 22.06 | 1 | 62,642 | dihydrolipoyl dehydrogenase | Stenotrophomonas maltophilia |
| Digestion Type | Hit # | Accession | Score −10lgP | Percent Coverage | Average Mass (Da) | 1 Protein Description |
|---|---|---|---|---|---|---|
| Trypsin | 1 | gi|121531620 | 86.58 | 8 | 29,420 | digestive cysteine protease intestain, partial |
| 2 | gi|815932195 | 71.24 | 4 | 33,107 | alpha-amylase, partial | |
| 3 | gi|1285027788 | 50.99 | 2 | 47,891 | protein disulfide-isomerase-like, partial |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Scott, I.M.; Hatten, G.; Tuncer, Y.; Clarke, V.C.; Jurcic, K.; Yeung, K.K.-C. Proteomic Analyses Detect Higher Expression of C-Type Lectins in Imidacloprid-Resistant Colorado Potato Beetle Leptinotarsa decemlineata Say. Insects 2021, 12, 3. https://doi.org/10.3390/insects12010003
Scott IM, Hatten G, Tuncer Y, Clarke VC, Jurcic K, Yeung KK-C. Proteomic Analyses Detect Higher Expression of C-Type Lectins in Imidacloprid-Resistant Colorado Potato Beetle Leptinotarsa decemlineata Say. Insects. 2021; 12(1):3. https://doi.org/10.3390/insects12010003
Chicago/Turabian StyleScott, Ian M., Gabrielle Hatten, Yazel Tuncer, Victoria C. Clarke, Kristina Jurcic, and Ken K.-C. Yeung. 2021. "Proteomic Analyses Detect Higher Expression of C-Type Lectins in Imidacloprid-Resistant Colorado Potato Beetle Leptinotarsa decemlineata Say" Insects 12, no. 1: 3. https://doi.org/10.3390/insects12010003