Sublethal Exposure Effects of the Neonicotinoid Clothianidin Strongly Modify the Brain Transcriptome and Proteome in the Male Moth Agrotis ipsilon
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Chemicals
2.3. Clothianidin Intoxication
2.4. Transcriptomic Analyses
2.5. Abundance Estimation and Differential Expression Analysis
2.6. Proteomics Analysis
3. Results
3.1. Brain Transcriptome Assembly and Annotation
3.2. DMSO Exposure Affects Both A. ipsilon Brain Transcriptome and Proteome Profiles
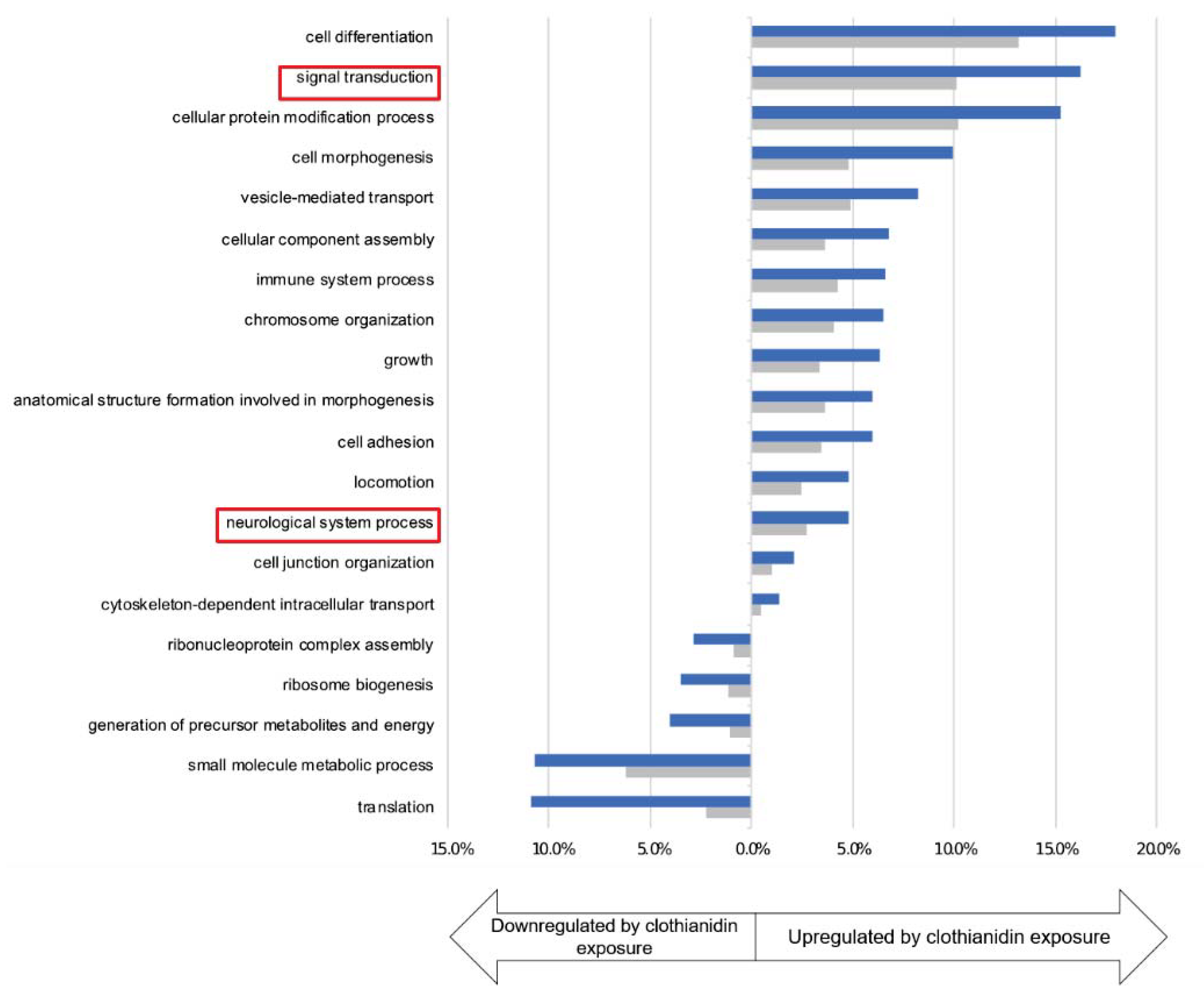
3.3. Clothianidin Exposure Affects Protein Biosynthesis and Metabolic Processes
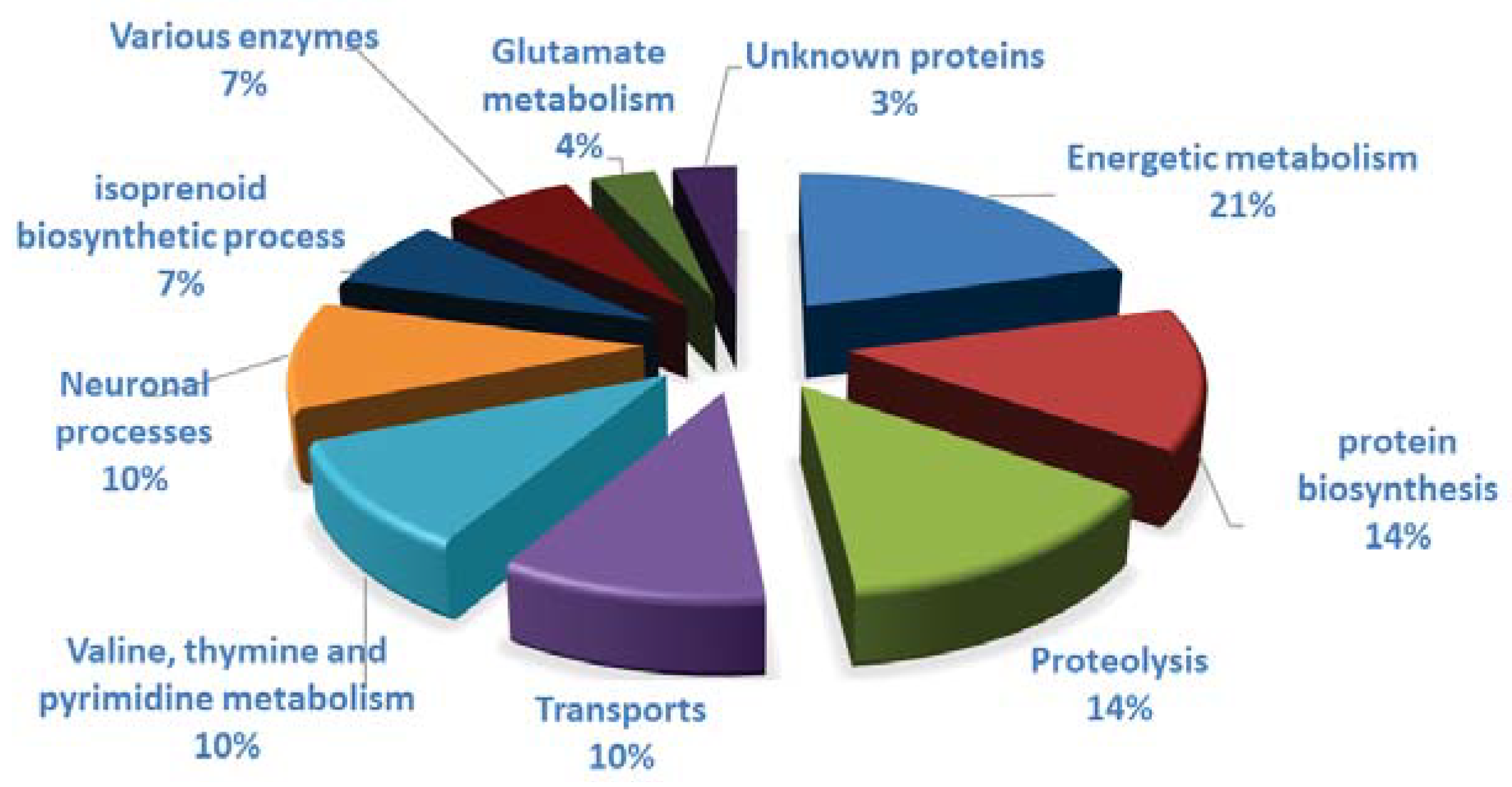
3.4. Clothianidin Exposure Affects Detoxification Enzymes
3.5. Clothianidin Exposure Affects Neuronal Processes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Veres, A.; Wyckhuys, K.A.G.; Kiss, J.; Tóth, F.; Burgio, G.; Pons, X.; Avilla, C.; Vidal, S.; Razinger, J.; Bazok, R.; et al. An update of the Worldwide Integrated Assessment (WIA) on systemic pesticides. Part 4: Alternatives in major cropping systems. Environ. Sci. Pollut. Res. Int. 2020, 27, 29867–29899. [Google Scholar] [CrossRef]
- Jeschke, P.; Nauen, R.; Schindler, M.; Elbert, A. Overview of the status and global strategy for neonicotinoids. J. Agric. Food Chem. 2011, 59, 2897–2908. [Google Scholar] [CrossRef]
- Casida, J.E.; Durkin, K.A. Neuroactiv e Insecticides: Targets, Selectivity, Resistance, and Secondary Effects. Annu. Rev. Entomol. 2013, 58, 99–117. [Google Scholar] [CrossRef]
- Matsuda, K.; Buckingham, S.D.; Kleier, D.; Rauh, J.J.; Grauso, M.; Sattelle, D.B. Neonicotinoids: Insecticides acting on insect nicotinic acetylcholine receptors. Trends Pharmacol. Sci. 2001, 22, 573–580. [Google Scholar] [CrossRef]
- Goulson, D. REVIEW: An overview of the environmental risks posed by neonicotinoid insecticides. J. Appl. Ecol. 2013, 50, 977–987. [Google Scholar] [CrossRef]
- Blacquière, T.; Smagghe, G.; Van Gestel, C.A.M.; Mommaerts, V. Neonicotinoids in bees: A review on concentrations, side-effects and risk assessment. Ecotoxicology 2012, 21, 973–992. [Google Scholar] [CrossRef] [PubMed]
- Desneux, N.; Decourtye, A.; Delpuech, J.-M. The Sublethal Effects of Pesticides on Beneficial Arthropods. Annu. Rev. Entomol. 2007, 52, 81–106. [Google Scholar] [CrossRef]
- Lürling, M.; Scheffer, M. Info-disruption: Pollution and the transfer of chemical information between organisms. Trends Ecol. Evol. 2007, 22, 374–379. [Google Scholar] [CrossRef]
- Calabrese, E.J.; Baldwin, L.A. Defining hormesis. Hum. Exp. Toxicol. 2002, 21, 91–97. [Google Scholar] [CrossRef]
- Calabrese, E.J. The Frequency of U-Shaped Dose Responses in the Toxicological Literature. Toxicol. Sci. 2001, 62, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, E.J. The road to linearity: Why linearity at low doses became the basis for carcinogen risk assessment. Arch. Toxicol. 2009. [Google Scholar] [CrossRef]
- Cutler, G.C. Insects, Insecticides and Hormesis: Evidence and Considerations for Study. Dose-Response 2012, 11, 154–177. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Zhao, Y.; Zhang, Z.; Xu, C.; Mu, W. Sublethal and Hormesis Effects of Clothianidin on the Black Cutworm (Lepidoptera: Noctuidae). J. Econ. Entomol. 2018, 111, 2809–2816. [Google Scholar] [CrossRef] [PubMed]
- Guedes, R.N.C.; Cutler, G.C. Insecticide-induced hormesis and arthropod pest management. Pest Manag. Sci. 2014, 70, 690–697. [Google Scholar] [CrossRef]
- Dewer, Y.; Pottier, M.A.; Lalouette, L.; Maria, A.; Dacher, M.; Belzunces, L.P.; Kairo, G.; Renault, D.; Maibeche, M.; Siaussat, D. Behavioral and metabolic effects of sublethal doses of two insecticides, chlorpyrifos and methomyl, in the Egyptian cotton leafworm, Spodoptera littoralis (Boisduval) (Lepidoptera: Noctuidae). Environ. Sci. Pollut. Res. 2016, 23, 3086–3096. [Google Scholar] [CrossRef]
- Lalouette, L.; Pottier, M.A.; Wycke, M.A.; Boitard, C.; Bozzolan, F.; Maria, A.; Demondion, E.; Chertemps, T.; Lucas, P.; Renault, D.; et al. Unexpected effects of sublethal doses of insecticide on the peripheral olfactory response and sexual behavior in a pest insect. Environ. Sci. Pollut. Res. 2016, 23, 3073–3085. [Google Scholar] [CrossRef] [PubMed]
- Rabhi, K.K.; Esancy, K.; Voisin, A.; Crespin, L.; Le Corre, J.; Tricoire-Leignel, H.; Anton, S.; Gadenne, C. Unexpected effects of low doses of a neonicotinoid insecticide on behavioral responses to sex pheromone in a pest insect. PLoS ONE. 2014, 9. [Google Scholar] [CrossRef]
- Tricoire-Leignel, H.; Thany, S.H.; Gadenne, C.; Anton, S. Pest insect olfaction in an insecticide-contaminated environment: Info-disruption or hormesis effect. Front. Physiol. 2012, 3, 58. [Google Scholar] [CrossRef]
- Enders, L.S.; Rault, L.C.; Heng-moss, T.M.; Siegfried, B.D.; Miller, J. Transcriptional responses of soybean aphids to sublethal insecticide exposure. Insect Biochem. Mol. Biol. 2019, 103285. [Google Scholar] [CrossRef]
- Dondero, F.; Negri, A.; Boatti, L.; Marsano, F.; Mignone, F.; Viarengo, A. Transcriptomic and proteomic effects of a neonicotinoid insecticide mixture in the marine mussel (Mytilus galloprovincialis, Lam.). Sci. Total Environ. 2010, 408, 3775–3786. [Google Scholar] [CrossRef] [PubMed]
- Diesner, M.; Gallot, A.; Binz, H.; Gaertner, C.; Vitecek, S.; Kahnt, J.; Schachtner, J.; Jacquin-Joly, E.; Gadenne, C. Mating-Induced Differential Peptidomics of Neuropeptides and Protein Hormones in Agrotis ipsilon Moths. J. Proteome Res. 2018, 17, 1397–1414. [Google Scholar] [CrossRef] [PubMed]
- Gadenne, C.; Barrozo, R.B.; Anton, S. Plasticity in Insect Olfaction: To Smell or Not to Smell? Annu. Rev. Entomol. 2016, 61, 317–333. [Google Scholar] [CrossRef]
- Rabhi, K.K.; Deisig, N.; Demondion, E.; Le Corre, J.; Robert, G.; Tricoire-Leignel, H.; Lucas, P.; Gadenne, C.; Anton, S. Low doses of a neonicotinoid insecticide modify pheromone response thresholds of central but not peripheral olfactory neurons in a pest insect. Proc. R. Soc. B Biol. Sci. 2016, 283. [Google Scholar] [CrossRef]
- Arthidoro de Castro, M.B.; Martinez, L.C.; Cossolin, J.F.S.; Serra, R.S.; Serrão, J.E. Cytotoxic effects on the midgut, hypopharyngeal, glands and brain of Apis mellifera honey bee workers exposed to chronic concentrations of lambda-cyhalothrin. Chemosphere 2020, 248, 126075. [Google Scholar] [CrossRef]
- Moreira, D.R.; Sinópolis Gigliolli, A.A.; Falco, J.R.P.; Julio, A.H.F.; Volnistem, E.A.; Chagas, F.D.; Toledo, V.A.A.; Ruvolo-Takasusuki, M.C.C. Toxicity and effects of the neonicotinoid thiamethoxam on Scaptotrigona bipunctata Lepeletier, 1836 (Hymenoptera: Apidae). Environ. Toxicol. 2018, 33, 463–475. [Google Scholar] [CrossRef]
- Surendra Nath, B.; Surendra Kumar, R.P. Toxic impact of organophosphorus insecticides on acetylcholinesterase activity in the silkworm, Bombyx mori L. Ecotoxicol. Environ. Saf. 1999, 42, 157–162. [Google Scholar] [CrossRef]
- Gadenne, C.; Renou, M.; Sreng, L. Hormonal control of pheromone responsiveness in the male black cutworm Agrotis ipsilon. Experientia 1993, 49, 721–724. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Chevreux, B.; Suhai, S. Genome Sequence Assembly Using Trace Signals and Additional Sequence Information. Comput. Sci. Biol. Proc. Ger. Conf. Bioinf. 1999, 99, 45–56. [Google Scholar]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Eddy, S.R. A Probabilistic Model of Local Sequence Alignment That Simplifies Statistical Significance Estimation. PLoS Comput. Biol. 2008, 4, e1000069. [Google Scholar] [CrossRef] [PubMed]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; Fau-von Heijne, G.; von Heijne, G.; Fau-Sonnhammer, E.L.; Sonnhammer, E.L.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Bigot, L.; Shaik, H.A.; Bozzolan, F.; Party, V.; Lucas, P.; Debernard, S.; Siaussat, D. Peripheral regulation by ecdysteroids of olfactory responsiveness in male Egyptian cotton leaf worms, Spodoptera littoralis. Insect Biochem. Mol. Biol. 2012, 42, 22–31. [Google Scholar] [CrossRef]
- Dépagne, J.; Chevalier, F. Technical updates to basic proteins focalization using IPG strips. Proteome Sci. 2012, 10, 54. [Google Scholar] [CrossRef]
- Ruiz-Delgado, G.J.; Mancías-Guerra, C.; Tamez-Gómez, E.L.; Rodríguez-Romo, L.N.; López-Otero, A.; Hernández-Arizpe, A.; Gómez-Almaguer, D.; Ruiz-Argüelles, G.J. Dimethyl Sulfoxide-Induced Toxicity in Cord Blood Stem Cell Transplantation: Report of Three Cases and Review of the Literature. Acta Haematol. 2009, 122, 1–5. [Google Scholar] [CrossRef]
- Harbo, J.R. Sterility in honey bees caused by dimethyl sulfoxide. J. Hered. 1986, 77, 129–130. [Google Scholar] [CrossRef]
- Milchreit, K.; Ruhnke, H.; Wegener, J.; Bienefeld, K. Effects of an insect growth regulator and a solvent on honeybee (Apis mellifera L.) brood development and queen viability. Ecotoxicology 2016, 25, 530–537. [Google Scholar] [CrossRef]
- Jacob, S.W.; de la Torre, J.C. Pharmacology of dimethyl sulfoxide in cardiac and CNS damage. Pharmacol. Rep. 2009. [Google Scholar] [CrossRef]
- Drummond, J.; Williamson, S.M.; Fitchett, A.E.; Wright, G.A.; Judge, S.J. Spontaneous honeybee behaviour is altered by persistent organic pollutants. Ecotoxicology 2017, 26, 141–150. [Google Scholar] [CrossRef]
- Haider, S.; Pal, R. Integrated Analysis of Transcriptomic and Proteomic Data. Curr. Genom. 2013, 14, 91–110. [Google Scholar] [CrossRef]
- Kumar, D.; Bansal, G.; Narang, A.; Basak, T.; Abbas, T.; Dash, D. Integrating transcriptome and proteome profiling: Strategies and applications. Proteomics 2016, 16, 2533–2544. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Yang, D.; Ding, J.; Zhang, J.; Pan, L.; Wang, X.; Yin, Q.; Gao, Y.; Chen, Z.; Chen, D. Resistance of mosquitoes to insecticide. Chinese. J. Hyg. Insectic. Equip. 2007, 13, 256–258. [Google Scholar]
- Casida, J.E. Neonicotinoid metabolism: Compounds, substituents, pathways, enzymes, organisms, and relevance. J. Agric. Food Chem. 2011, 59, 2923–2931. [Google Scholar] [CrossRef]
- Iwasa, T.; Motoyama, N.; Ambrose, J.T.; Roe, R.M. Mechanism for the differential toxicity of neonicotinoid insecticides in the honey bee, Apis mellifera. Crop Prot. 2004, 23, 371–378. [Google Scholar] [CrossRef]
- Karunker, I.; Benting, J.; Lueke, B.; Ponge, T.; Nauen, R.; Roditakis, E.; Vontas, J.; Gorman, K.; Denholm, I.; Morin, S. Over-expression of cytochrome P450 CYP6CM1 is associated with high resistance to imidacloprid in the B and Q biotypes of Bemisia tabaci (Hemiptera: Aleyrodidae). Insect Biochem. Mol. Biol. 2008, 38, 634–644. [Google Scholar] [CrossRef]
- Daborn, P.; Boundy, S.; Yen, J.; Pittendrigh, B. DDT resistance in Drosophila correlates with Cyp6g1 over-expression and confers cross-resistance to the neonicotinoid imidacloprid. Mol. Genet. Genom. 2001, 266, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Magesh, V.; Zhu, Z.; Tang, T.; Chen, S.; Li, L.; Wang, L.; Varma, K.K.; Wu, Y. Toxicity of Neonicotinoids to honey bees and detoxification mechanism in honey bees. IOSR J. Environ. Sci. Toxicol. Food Technol. 2017, 11, 102–110. [Google Scholar] [CrossRef]
- Yang, N.; Xie, W.; Yang, X.; Wang, S.; Wu, Q.; Li, R.; Pan, H.; Liu, B.; Shi, X.; Fang, Y. Transcriptomic and proteomic responses of sweetpotato whitefly, Bemisia tabaci, to thiamethoxam. PLoS ONE 2013, 8, e61820. [Google Scholar] [CrossRef]
- Chen, C.; Shi, X.; Desneux, N.; Han, P.; Gao, X. Detection of insecticide resistance in Bradysia odoriphaga Yang et Zhang (Diptera: Sciaridae) in China. Ecotoxicology 2017, 26, 868–875. [Google Scholar] [CrossRef] [PubMed]
- Alyokhin, A.; Baker, M.; Mota-Sanchez, D.; Dively, G.; Grafius, E. Colorado potato beetle resistance to insecticides. Am. J. Potato Res. 2008, 85, 395–413. [Google Scholar] [CrossRef]
- Yang, Y.X.; Yu, N.; Zhang, J.H.; Zhang, Y.X.; Liu, Z.W. Induction of P450 genes in Nilaparvata lugens and Sogatella furcifera by two neonicotinoid insecticides. Insect Sci. 2017, 25, 401–408. [Google Scholar] [CrossRef]
- Penning, T.M. The aldo-keto reductases (AKRs): Overview. Chem. Biol. Interact. 2015, 234, 236–246. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Webb, T.J.; Powls, R.; Rees, H.H. Purification and Characterisation of Haemolymph 3-Dehydroecdysone 3β-Reductase in Relation to Ecdysteroid Biosynthesis in the Cotton Leafworm Spodoptera littoralis. Eur. J. Biochem. 1996, 242, 394–401. [Google Scholar] [CrossRef]
- Yamamoto, K.; Wilson, D.K. Identification, characterization, and crystal structure of an aldo–keto reductase (AKR2E4) from the silkworm Bombyx mori. Arch. Biochem. Biophys. 2013, 538, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Yamada, N. Identification of a diazinon-metabolizing glutathione S-transferase in the silkworm, Bombyx mori. Sci. Rep. 2016, 6, 30073. [Google Scholar] [CrossRef] [PubMed]
- Marchitti, S.A.; Brocker, C.; Stagos, D.; Vasiliou, V. Non-P450 aldehyde oxidizing enzymes: The aldehyde dehydrogenase superfamily. Expert Opin. Drug Metab. Toxicol. 2008, 4, 697–720. [Google Scholar] [CrossRef] [PubMed]
- Fry, J.D.; Saweikis, M. Aldehyde dehydrogenase is essential for both adult and larval ethanol resistance in Drosophila melanogaster. Genet. Res. (Camb.) 2006, 87, 87–92. [Google Scholar] [CrossRef]
- Lumjuan, N.; Wicheer, J.; Leelapat, P.; Choochote, W.; Somboon, P. Identification and characterisation of Aedes aegypti aldehyde dehydrogenases involved in pyrethroid metabolism. PLoS ONE 2014, 9, e102746. [Google Scholar]
- Doorn, J.A.; Florang, V.R.; Schamp, J.H.; Vanle, B.C. Aldehyde dehydrogenase inhibition generates a reactive dopamine metabolite autotoxic to dopamine neurons. Parkinsonism Relat. Disord. 2014, 20, S73–S75. [Google Scholar] [CrossRef]
- Fitzmaurice, A.G.; Rhodes, S.L.; Cockburn, M.; Ritz, B.; Bronstein, J.M. Aldehyde dehydrogenase variation enhances effect of pesticides associated with Parkinson disease. Neurology 2014, 82, 419–426. [Google Scholar] [CrossRef]
- Volders, K.; Scholz, S.; Slabbaert, J.R.; Nagel, A.C.; Verstreken, P.; Creemers, J.W.M.; Callaerts, P.; Schwärzel, M. Drosophila rugose is a functional homolog of mammalian Neurobeachin and affects synaptic architecture, brain morphology, and associative learning. J. Neurosci. 2012, 32, 15193–15204. [Google Scholar] [CrossRef]
- Akbergenova, Y.; Cunningham, K.L.; Zhang, Y.V.; Weiss, S.; Littleton, J.T. Characterization of developmental and molecular factors underlying release heterogeneity at Drosophila synapses. Elife 2018, 7, e38268. [Google Scholar] [CrossRef]
- Faurobert, E.; Chen, C.-K.; Hurley, J.B.; Teng, D.H.-F. Drosophila neurocalcin, a fatty acylated, Ca-binding protein that associates with membranes and inhibits in vitro phosphorylation of bovine rhodopsin. J. Biol. Chem. 1996, 271, 10256–10262. [Google Scholar] [CrossRef]
- Gokhale, A.; Mullin, A.P.; Zlatic, S.A.; Easley, C.A.; Merritt, M.E.; Raj, N.; Larimore, J.; Gordon, D.E.; Peden, A.A.; Sanyal, S.; et al. The N-ethylmaleimide-sensitive factor and dysbindin interact to modulate synaptic plasticity. J. Neurosci. 2015, 35, 7643–7653. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Tian, Y.; Li, Q.; Chen, H.; Lv, H.; Xie, W.; Han, J. The neurexin/N-ethylmaleimide-sensitive factor (NSF) interaction regulates short term synaptic depression. J. Biol. Chem. 2015, 290, 17656–17667. [Google Scholar] [CrossRef]
- Nunes, P.; Haines, N.; Kuppuswamy, V.; Fleet, D.J.; Stewart, B.A. Synaptic vesicle mobility and presynaptic F-actin are disrupted in a N-ethylmaleimide-sensitive factor allele of Drosophila. Mol. Biol. Cell 2006, 17, 4709–4719. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Caballero, J.P.; Murillo, L.; List, O.; Bastiat, G.; Flochlay-Sigognault, A.; Guerino, F.; Lefrançois, C.; Lautram, N.; Lapied, B.; Apaire-Marchais, V. Nanoencapsulated deltamethrin as synergistic agent potentiates insecticide effect of indoxacarb through an unusual neuronal calcium-dependent mechanism. Pestic. Biochem. Physiol. 2019, 157, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Roedding, A.S.; Tong, S.Y.; Au-Yeung, W.; Li, P.P.; Warsh, J.J. Chronic oxidative stress modulates TRPC3 and TRPM2 channel expression and function in rat primary cortical neurons: Relevance to the pathophysiology of bipolar disorder. Brain Res. 2013, 1517, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Ffrench-Constant, R.H.; Rocheleau, T.A. Drosophila γ-aminobutyric acid receptor gene Rdl shows extensive alternative splicing. J. Neurochem. 1993, 60, 2323–2326. [Google Scholar] [CrossRef]
- Jonsson, N.N.; Klafke, G.; Corley, S.W.; Tidwell, J.; Berry, C.M.; Koh-Tan, H.H. Molecular biology of amitraz resistance in cattle ticks of the genus Rhipicephalus. Front. Biosci. 2018, 23, 796–810. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Puinean, A.M.; Williamson, M.S.; Smelt, C.L.C.; Millar, N.S.; Wu, Y. Mutations on M3 helix of Plutella xylostella glutamate-gated chloride channel confer unequal resistance to abamectin by two different mechanisms. Insect Biochem. Mol. Biol. 2017, 86, 50–57. [Google Scholar] [CrossRef]
- Karlstrom, R.O.; Wilder, L.P.; Bastiani, M.J. Lachesin: An immunoglobulin superfamily protein whose expression correlates with neurogenesis in grasshopper embryos. Development 1993, 118, 509–522. [Google Scholar]
- Abe, T.; Yamazaki, D.; Murakami, S.; Hiroi, M.; Nitta, Y.; Maeyama, Y.; Tabata, T. The NAV2 homolog Sickie regulates F-actin-mediated axonal growth in Drosophila mushroom body neurons via the non-canonical Rac-Cofilin pathway. Development 2014, 141, 4716–4728. [Google Scholar] [CrossRef]
- Jones, M.A.; Amr, S.; Ferebee, A.; Huynh, P.; Rosenfeld, J.A.; Miles, M.F.; Davies, A.G.; Korey, C.A.; Warrick, J.M.; Shiang, R.; et al. Genetic studies in Drosophila and humans support a model for the concerted function of CISD2, PPT1 and CLN3 in disease. Biol. Open 2014, 3, 342–352. [Google Scholar] [CrossRef] [PubMed]
- Koster, K.P.; Yoshii, A. Depalmitoylation by palmitoyl-protein thioesterase 1 in neuronal health and degeneration. Front. Synaptic Neurosci. 2019. [Google Scholar] [CrossRef]
- Bannan, B.A.; Van Etten, J.; Kohler, J.A.; Tsoi, Y.; Hansen, N.M.; Sigmon, S.; Fowler, E.; Buff, H.; Williams, T.S.; Ault, J.G.; et al. The drosophila protein palmitoylome characterizing palmitoyl-thioesterases and DHHC palmitoyl-transferases. Fly (Austin) 2008, 2, 198–214. [Google Scholar] [CrossRef] [PubMed]
- Aby, E.; Gumps, K.; Roth, A.; Sigmon, S.; Jenkins, S.E.; Kim, J.J.; Kramer, N.J.; Parfitt, K.D.; Korey, C.A. Mutations in palmitoyl-protein thioesterase 1 alter exocytosis and endocytosis at synapses in Drosophila larvae. Fly (Austin) 2013, 7, 267–279. [Google Scholar] [CrossRef]
- Chu-LaGraff, Q.; Blanchette, C.; O’Hern, P.; Denefrio, C. The Batten disease Palmitoyl Protein Thioesterase 1 gene regulates neural specification and axon connectivity during Drosophila embryonic development. PLoS ONE 2010, 5, e14402. [Google Scholar] [CrossRef]
- Hickey, A.J.; Chotkowski, H.L.; Singh, N.; Ault, J.G.; Korey, C.A.; MacDonald, M.E.; Glaser, R.L. Palmitoyl-protein thioesterase 1 deficiency in Drosophila melanogaster causes accumulation of abnormal storage material and reduced life span. Genetics 2006, 172, 2379–2390. [Google Scholar] [CrossRef]
- Ding, B.J.; Löfstedt, C. Analysis of the Agrotis segetum pheromone gland transcriptome in the light of Sex pheromone biosynthesis. BMC Genom. 2015, 16. [Google Scholar] [CrossRef]
- Abrieux, A.; Mhamdi, A.; Rabhi, K.K.; Egon, J.; Debernard, S.; Duportets, L.; Tricoire-Leignel, H.; Anton, S.; Gadenne, C. An Insecticide Further Enhances Experience-Dependent Increased Behavioural Responses to Sex Pheromone in a Pest Insect. PLoS ONE 2016, 11, e0167469. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Tian, F.; Wei, X.; Wu, Y.; Gao, X.; Xi, J.; Shang, Q. Thiamethoxam resistance in Aphis gossypii glover relies on multiple UDP-glucuronosyltransferases. Front. Physiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Yu, L.; Liu, D.; Hu, D. Synthesis and Insecticidal Activity of Mesoionic Pyrido [1, 2-α] pyrimidinone Derivatives Containing a Neonicotinoid Moiety. Molecules 2018, 23, 1217. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Zhang, C.; Chen, X.; Cao, Y.; Shang, S. Differential protein expression in the susceptible and resistant Myzus persicae (Sulzer) to imidacloprid. Pestic. Biochem. Physiol. 2014, 115, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Du Rand, E.E.; Smit, S.; Beukes, M.; Apostolides, Z.; Pirk, C.W.W.; Nicolson, S.W. Detoxification mechanisms of honey bees (Apis mellifera) resulting in tolerance of dietary nicotine. Sci. Rep. 2015, 5, 11779. [Google Scholar] [CrossRef] [PubMed]



| Diesner et al. | This Study | |
|---|---|---|
| Number of contigs | 50,750 | 17,986 |
| Median contig length (nt) | 618 bp | 1233 bp |
| Complete BUSCOs | 1062 (64.0%) | 1462 (88.2%) |
| Complete and single-copy BUSCOs | 891 (53.7%) | 1381 (83.3%) |
| Complete and duplicated BUSCOs | 171 (10.3%) | 81 (4.9%) |
| Fragmented BUSCOs | 309 (18.6%) | 99 (6.0%) |
| Missing BUSCOs | 287 (17.4%) | 97 (5.8%) |
| Functional Categories | Protein Name | Uniprot Reference | Species | Peptide Number | p Value * | Protein Quantity in Comparison to Control (%) |
|---|---|---|---|---|---|---|
| Glutamate metabolism | Glutamate dehydrogenase | A0A0G3VFZ9_CHISP | Chilo suppressalis | 4 | 0.009 | −17.1509 ↘ |
| Xenobiotic enzymes | Dimeric dihydrodiol dehydrogenase | S4P4A7_9NEOP | Pararge aegeria | 8 | 0.026 | −62.4242 ↘ |
| Aldehyde dehydrogenase X. mitochondrial-like | AL1B1_HUMAN | Amyelois transitella | 6 | 0.019 | −18.8077 ↘ | |
| Isoprenoid biosynthetic process | Geranylgeranyl diphosphate synthase | U3RD44_CHOFU | Choristoneura fumiferana | 15 | 0.010 | −21.7245 ↘ |
| Farnesyl diphosphate synthase 2 | A5A7A5_BOMMO | Bombyx mori | 7 | 0.026 | −62.4242 ↘ | |
| Neuronal processes | Palmitoyl-protein Thioesterase 2 | KGM_213554 | Danaus plexippus | 4 | 0.019 | −18.8077 ↘ |
| Fatty alcohol acetyltransferase | A0A088M9K3_AGRSE | Agrotis segetum | 13 | 0.006 | −43.1842 ↘ | |
| N-ethylmaleimide sensitive fusion protein | Q5G1P2_HELAM | Helicoverpa armigera | 11 | 0.035 | −15,9046 ↘ | |
| Valine. thymine and pyrimidine metabolism | Methylmalonate-semialdehyde dehydrogenase [acylating]. mitochondrial isoform X2 | MMSA_DROME | Papilio polytes | 2 | 0.019 | −18.8077 ↘ |
| 3-hydroxyisobutyryl-CoA hydrolase. mitochondrial | A0A068FL83_AGRSE | Agrotis segetum | 7 | 0.026 | −62.4242 ↘ | |
| Dihydropyrimidinase isoform X2 | A0A170WSE7_TRIIF | Bombyx mori | 3 | 0.028 | 16.72897 ↗ | |
| Transport | Transferrin | A7IT76_SPOLT | Spodoptera litura | 2 | 0.004 | −18.3537 ↘ |
| Apolipophorins | APLP_DROME | Papilio polytes | 3 | 0.028 | 16.72897 ↗ | |
| Rabenosyn-5 | A0A194QJD8_PAPXU | Papilio xuthus | 6 | 0.019 | −18.8077 ↘ | |
| Proteolysis | Protease m1 zinc metalloprotease | G6D6V6_DANPL | Danaus plexippus | 23 | 0.017 | 28.09224 ↗ |
| Protease m1 zinc metalloprotease | G6D6V6_DANPL | Danaus plexippus | 20 | 0.030 | 19.18782 ↗ | |
| Mitochondrial processing peptidase beta subunit | I4DK27_PAPXU | Papilio xuthus | 9 | 0.006 | −43.1842 ↘ | |
| Protein biosynthesis | Elongation factor 1-a | L7WID6_SPOLT | Spodoptera litura | 4 | 0.029 | −23.5498 ↘ |
| Elongation factor 1 alpha | E0D8P8_LOCMI | Locusta migratoria | 4 | 0.029 | −23.5498 ↘ | |
| UDP-N-acetylglucosamine-peptide N-acetylglucosaminyltransferase 110 kDa subunit | OGT1_HUMAN | Bombyx mori | 2 | 0.013 | −16.5289 ↘ | |
| Translation initiation factor 2 gamma subunit | Q684K4_9NEOP | Scoliopteryx libatrix | 3 | 0.029 | −23.5498 ↘ | |
| Energy metabolism | ATP synthase subunit b. mitochondrial | A0A194PTU2_PAPXU | Papilio xuthus | 2 | 0.034 | −33.3046 ↘ |
| ATP synthase | Q2F5T4_BOMMO | Bombyx mori | 5 | 0.020 | −18.8077 ↘ | |
| Aconitate hydratase. mitochondrial | Q86GF8_ANTYA | Antheraea yamamai | 5 | 0.004 | −18.3537 ↘ | |
| Pyruvate kinase | H9IZ23_BOMMO | Bombyx mori | 5 | 0.009 | −17.1509 ↘ | |
| NADH-ubiquinone oxidoreductase 75 kDa subunit. mitochondrial | NDUS1_DROME | Bombyx mori | 28 | 0.028 | 16.72897 ↗ | |
| UDP-glucose pyrophosphorylase | A0A0S1MMM3_ANTPE | Antheraea pernyi | 5 | 0.009 | −17.1509 ↘ | |
| Unknown | unknown | 17 | 0.003 | −28.2986 ↘ |
| * | Transcript ID | logFC Clothianidin vs. DMSO | logFC Clothianidin vs. Control | Annotation by Blast Research |
|---|---|---|---|---|
| Up | DN9632_c0_g1_i1 | 1.35 | 1.13 | Cytochrome P450 4C1 |
| DN9385_c0_g1_i1 | 0.78 | UDP-glucuronosyltransferase 2B7 | ||
| Down | DN27963_c0_g1_i1 | −1.49 | Probable cytochrome P450 6a13 | |
| DN20937_c0_g1_i1 | −1.42 | Cytochrome P450 4c3 | ||
| DN37703_c0_g1_i1 | −1.16 | Cytochrome P450 6B7 | ||
| DN34383_c0_g1_i1 | −0.96 | Cytochrome P450 4C1 | ||
| DN20657_c0_g1_i1 | −0.69 | Aldo-keto reductase AKR2E4 | ||
| DN5740_c0_g1_i1 | −0.63 | Glutathione S-transferase 1 | ||
| DN4625_c0_g1_i1 | −0.35 | Cytochrome P450 CYP12A2 | ||
| DN4767_c0_g1_i1 | −0.50 | Venom carboxylesterase-6 |
| GO Function | * | Transcript ID | logFC Clothianidin vs. DMSO | logFC Clothianidin vs. Control | Annotation by Blast Research |
|---|---|---|---|---|---|
| Synaptic function | Up | DN6306_c0_g1_i1 | 0.89 | Neurobeachin | |
| DN2580_c0_g1_i1 | 0.42 | Synaptotagmin 1 | |||
| DN4891_c0_g1_i1 | 0.33 | Neurocalcin homolog | |||
| Down | DN10953_c0_g1_i1 | −0.53 | Synaptic vesicle glycoprotein 2B | ||
| Exchanges across the neuronal membrane | Up | DN21572_c0_g1_i1 | 1.14 | Tyrosine-protein phosphatase 69D | |
| DN19704_c0_g1_i1 | 0.46 | Transient receptor potential cation channel trpm | |||
| DN6537_c0_g1_i1 | 0.56 | Sodium channel protein para | |||
| DN12028_c0_g1_i1 | 0.53 | Chloride channel protein 2 | |||
| DN27405_c0_g1_i1 | 1.21 | Voltage-dependent calcium channel type D subunit alpha-1 | |||
| DN1850_c0_g1_i1 | 1.31 | 1.59 | Sodium/calcium exchanger 3 | ||
| Neuronal activity | Up | DN6443_c0_g1_i1 | 0.49 | Sodium- and chloride-dependent GABA transporter 1 | |
| Down | DN392_c0_g1_i1 | −0.31 | Sodium- and chloride-dependent GABA transporter ine | ||
| Neuronal processes | Up | DN20357_c0_g1_i1 | 0.64 | Mushroom body large-type Kenyon cell-specific protein 1 | |
| DN7325_c0_g1_i1 | 0.96 | Krueppel homolog 1 | |||
| DN25883_c0_g1_i1 | 1.36 | Octopamine receptor beta-3R | |||
| DN49002_c0_g1_i1 | 0.84 | Glutamate-gated chloride channel | |||
| DN18143_c0_g1_i1 | 0.48 | Neuropeptide CCHamide-2 receptor | |||
| Neurogenesis/axonal growth | Up | DN33688_c0_g1_i1 | 0.88 | Lachesin | |
| DN10822_c0_g1_i1 | 0.49 | Protein sickie |
| Proteins | Transcripts | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Number of Detected Spots or Proteins | Proteins Showing Significant Variation in Quantity | Up | Down | Number of Analyzed Unigenes | Up | Down | Molecules or Condition | Species | References |
| 1760 | 49 | 6 | 23 | 17,986 | 2292 | 1646 | Clothianidin | Agrotis ipsilon | Present study |
| 700 | 12 | nc | nc | nc | 41 | 56 | Imidacloprid | Mytilus galloprovincialis | [20] |
| nc | 43 | 37 | Thiacloprid | ||||||
| nc | 26 | 23 | Mix | ||||||
| 29,146 to 31,467 | 646 to 658 | 284 to 533 | Clothianidin | Bradysia odoriphaga | [66] | ||||
| 35,222 | 349 | 271 | Thiamethoxam | Aphis gossypii | [89] | ||||
| 11,150 to 11,426 | 225 | 384 | Thiamethoxam | Apis mellifera | [64] | ||||
| 1005 | 52 | 38 | 14 | nc | 664 | 674 | Thiamethoxam | Bemisia tabaci | [65] |
| 821 | 143 | 35 | 108 | Mesoionic pyrido[1,2-]pyrimidinone | Aphis craccivora | [90] | |||
| >1300 | 28 | 14 | 14 | resistant strain to imidacloprid | Myzus persicae | [91] | |||
| 1470 | 155 | 96 | 59 | Nicotine | Apis mellifera | [92] | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meslin, C.; Bozzolan, F.; Braman, V.; Chardonnet, S.; Pionneau, C.; François, M.-C.; Severac, D.; Gadenne, C.; Anton, S.; Maibèche, M.; et al. Sublethal Exposure Effects of the Neonicotinoid Clothianidin Strongly Modify the Brain Transcriptome and Proteome in the Male Moth Agrotis ipsilon. Insects 2021, 12, 152. https://doi.org/10.3390/insects12020152
Meslin C, Bozzolan F, Braman V, Chardonnet S, Pionneau C, François M-C, Severac D, Gadenne C, Anton S, Maibèche M, et al. Sublethal Exposure Effects of the Neonicotinoid Clothianidin Strongly Modify the Brain Transcriptome and Proteome in the Male Moth Agrotis ipsilon. Insects. 2021; 12(2):152. https://doi.org/10.3390/insects12020152
Chicago/Turabian StyleMeslin, Camille, Françoise Bozzolan, Virginie Braman, Solenne Chardonnet, Cédric Pionneau, Marie-Christine François, Dany Severac, Christophe Gadenne, Sylvia Anton, Martine Maibèche, and et al. 2021. "Sublethal Exposure Effects of the Neonicotinoid Clothianidin Strongly Modify the Brain Transcriptome and Proteome in the Male Moth Agrotis ipsilon" Insects 12, no. 2: 152. https://doi.org/10.3390/insects12020152
APA StyleMeslin, C., Bozzolan, F., Braman, V., Chardonnet, S., Pionneau, C., François, M.-C., Severac, D., Gadenne, C., Anton, S., Maibèche, M., Jacquin-Joly, E., & Siaussat, D. (2021). Sublethal Exposure Effects of the Neonicotinoid Clothianidin Strongly Modify the Brain Transcriptome and Proteome in the Male Moth Agrotis ipsilon. Insects, 12(2), 152. https://doi.org/10.3390/insects12020152











