Integrative Taxonomy of the Spinous Assassin Bug Genus Sclomina (Heteroptera: Reduviidae: Harpactorinae) Reveals Three Cryptic Species Based on DNA Barcoding and Morphological Evidence
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimens and Acronyms
- BMNH, Department of Entomology, Natural History Museum, London, U.K.;
- CAU, Entomological Museum of China Agricultural University, Beijing, China;
- NKU, College of Life Sciences, Nankai University, Tianjin, China;
- NNU, Nanning Normal University, Nanning, Guangxi, China;
- KU, Department of Plant Protection, Kaili University, Kaili, Guizhou, China.
2.2. Taxonomy
2.3. Integrative Molecular Analysis Based on DNA Barcodes
2.3.1. DNA Extraction and Sequencing
2.3.2. Phylogenetic Analyses
2.3.3. Species Delimitation
2.3.4. Estimation of Divergence Time
2.4. Biological Study
3. Results
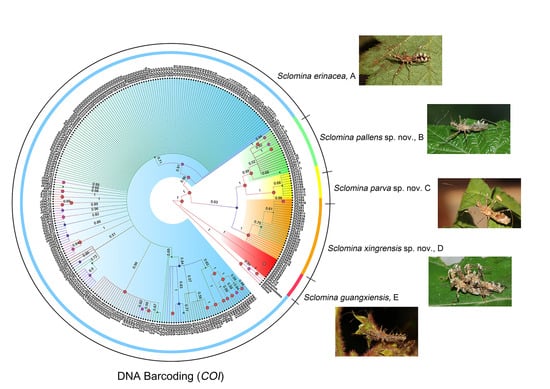
3.1. Phylogenetic Relationships
3.2. Species Delimitation
3.3. Estimation of Divergence Time
3.4. Biology
3.4.1. Life History
3.4.2. Egg and Nymph
3.4.3. Symbiotic Plants
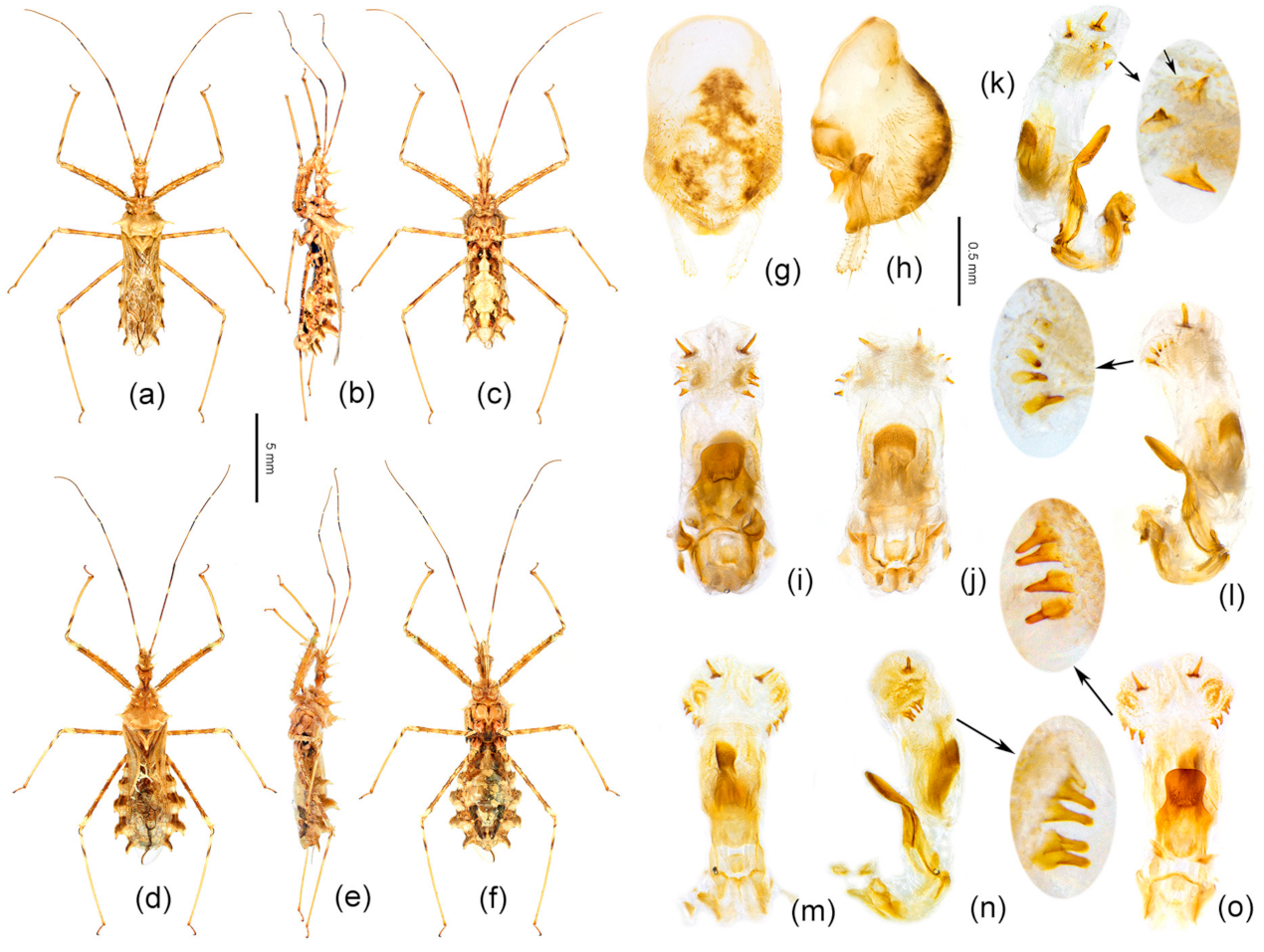
3.5. Taxonomy
- 1.
- Body length more than 14 mm; post-lateral angle of abdominal connexivum spine-shaped laterally produced (Figure 3a–p)—2
- -
- Body length less than 14 mm; post-lateral angle of abdominal connexivum Y-shaped laterally produced (Figure 3q–t)—3
- 2.
- Body yellowish-brown; spines on surface of head and pronotum not as strong; apical part of post-lateral angle of abdominal connexivum short spine-shaped (Figure 3a–o)—Sclomina erinacea Stål, 1861.
- -
- Body dark reddish-brown; spines on surface of head and pronotum long, slender, and strong; post-lateral angle of abdominal connexivum long spine-shaped (Figure 3p)—Sclomina guangxiensis Ren, 2001.
- 3.
- Body length less than 13 mm; apical part of endosoma without spines (Figure 3t)—Sclomina parva sp. nov.
- -
- Body length more than 13 mm but less than 14 mm; apical part of endosoma lateral, with a pair of large spines and three to five pairs of small spines (Figure 3q–s)—4
- 4.
- Two processes of post-lateral angle of connexivum somewhat acute; endosoma apically, with a pair of larger spines, and subapically with five smaller spines on one side and four smaller spines on the other side (Figure 3q,r)—Sclomina pallens sp. nov.
- -
- Two processes of post-lateral angle of connexivum round; endosoma apical with a pair of larger spines and subapically lateral with four smaller spines on one side and three or four smaller spines on the other side (Figure 3s)—Sclomina xingrensis sp. nov.
3.5.1. Sclomina erinacea Stål, 1861
3.5.2. Sclomina guangxiensis Ren, 2001
3.5.3. Sclomina pallens P. Zhao and Cai sp. nov.
3.5.4. Sclomina parva P. Zhao and Cai sp. nov.
3.5.5. Sclomina xingrensis P. Zhao and Cai sp. nov.
4. Discussion
4.1. Phylogeny and Species Delimitation
4.2. Evolutionary History
4.3. Symbiotic Relationship between Sclomina and Rubus: Mimicry and Protective Color
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Species | Sample Code | Locality, Elevation | Sampling Time | Geographical Coordinates | Symbiotic Species | Sample Size | Accession Number |
|---|---|---|---|---|---|---|---|
| Ingroup | |||||||
| Sclomina erinacea | AHFY | Fengyang Mountain, Anhui, China, 80 m | 2008-VI-5 | 32°42′58′′ N 117°39′44′′ E | / | 5 | MW387904–06 MW387643–44 |
| FJQZ | Dehua, Quanzhou, Fujian, China, 490 m | 2014-VI-26, 2013-IX-07, 2014-VII-27 | 25°29′39′′ N 118°14′10′′ E | / | 1 | MW387907 | |
| FJWY | Wuyishan, Fujian, China | 2009-VI-5 | 27°45′35′′ N 118°148′54′′ E | / | 7 | MW387908–09 MW387645–49 | |
| GDMZ | Longtan, Jiaoling, Meizhou, Guangdong, China, 355 m | 2019-IX-03 | 24°38′32′′ N 116°15′6′′ E | Rubus lambertianus Rubus tsangii | 7 | MW387910–16 | |
| GDQY | Yangshan, Qingyuan, Guangdong, China, 200 m | 2019-VIII-05 | 24°49′ N 112°51′58′′ E | Rubus lambertianus | 8 | MW387917–24 | |
| GDSG | Shaoguan, Guangdong, China, 54 m | 24°48′13′′ N 113°35′5′′ E | / | 3 | MW387650–52 | ||
| GXJX | Jinxiu, Guangxi, China, 800 m | 2019-V | 24°7′59′′ N 110°11′8′′ E | / | 9 | MW387653–55 MW387925–30 | |
| GXYL | Darongshan, Yulin, Guangxi, China, 260 m | 2018-VIII-19 | 22°50′42′′ N 110°17′22′′ E | Solanum aculeatissimum | 9 | MW387678–86 | |
| GZJP | Yandong, Jinping, Guizhou, China, 834 m | 2018-VIII-28 | 26°37′31′′ N 108°55′2′′ E | Rubus setchuenensis Rubus rosifolius Polygonum senticosum | 20 | MW387693–712 | |
| GZLP | Liping, Guizhou, China, 1150 m | 2012-VIII-12 | 26°14′19′′ N 109°18′38′′ E | / | 11 | MW387713–23 | |
| GZLS | Pingxiang, Fangxiang, Leishan, Guizhou, China, 1050 m | 2017-X-11 2018-VII-26 2018-VII-26 | 26°19′19′′ N 108°11′34′′ E | Rubus rosifolius (mainly) Rubus lambertianus Rhododendron rivulare (nymph mainly) | 34 | MW387724–57 | |
| GZML | Maolan, Guizhou, China, 600 m | 2013-X-12 | 25°14′14′′ N 108°1′50′′ E | / | 1 | MW387758 | |
| GZSB | Yuntaishan Mountain, Shibing, Guizhou, China, 980 m | 2018-VI-28 2019-IV-6 | 27°6′17′′ N 108°6′45′′ E | Rubus chiliadenus Rubus lambertianus (mainly) | 18 | MW387931–38 MW387759–68 | |
| HNBW | Bawangling, Hainan, China, 660 m | 2008-V-9 2007-X-21 | 19°8′10′′ N 109°15′28′′ E | / | 2 | MW387939–40 | |
| HNCD | Changde, Hunan, China | / | 29°2′35′′ N 111°41′8′′ E | / | 2 | MW387793–94 | |
| JXJG | Pingci, Jinggangshan, Jiangxi, China, 840 m | 2018-VIII-16 | 26°34′44′′ N 114°9′47′′ E | Rubus alceifolius (nymph mainly) Rubus lambertianus (few) Rubus rosifolius (few) Sambucus chinensis (adult) Boehmeria nivea (adult) Woodwardia prolifera (few, nymph and adult) Hibiscus mutabilis (few, adult) Artemisia anomala (few, adult) | 36 | MW387795–830 | |
| JXNC | Meiling, Nanchang, Jiangxi, China, 150 m | 2018-VIII-14 | 28°47′43′′ N 115°44′45′′ E | Rubus rosifolius (mainly) Macrothelypteris sp. Pilea sinofasciata Polygonum senticosum | 26 | MW387831–56 | |
| SCYA | Yaan, Sichuan, China, 650 m | 2018-IX-15 | 29°53′58′′ N 103°2′7′′ E | / | 1 | MW387857 | |
| TWNT | Yuchi, Nantou, Taiwan, China, 120 m | 2010-XI-11 2012-VI-27 2016-XI-5 | 24°56′23′′ N 121°15′35′′ E | / | 5 | MW387868–72 | |
| TWPD | Lianhuachi Research Center, Pindong, Taiwan, China, 33 m | 2012-VI-24 | 22°56′43′′ N 121°22′52′′ E | / | 3 | MW387873–75 | |
| TW | Lijialindao, Taidong, Taiwan, China, 18 m | 2012-VI-03 | 22°45′28′′ N 121°8′47′′ E | / | 6 | MW387862–67 | |
| VNM | Xuan Son National Park, Phu Tho Province, Vietnam, 400 m | 2004-X-14 | 21°10′13′′ N 104°58′21′′ E | / | 1 | MW387948 | |
| YNPB | Pingbian, Dweishan, Honghe, Yunnan, China, 1500 m | 2006-V-15 | 22°58′0′′ N 103°41′59′′ E | / | 1 | MW387949 | |
| ZJLS | Lishui, Zhejiang, China, 100 m | 28°28′15′′ N 119°55′7′′ E | / | 1 | MW387876 | ||
| ZJZC | Zhou Country, Ershibadu, Zhejiang, China, 400 m | 2018-VIII-12 | 28°20′24′′ N 118°36′15′′ E | Rubus tsangorum (mainly) Rubus tephrodes (mainly) Rubus lambertianus (nymph) Rubus tsangii (few) Rubus buergeri (few) | 27 | MW387877–903 | |
| Sclomina xingrensis sp. nov. | GZXR | Lishuping, Xingren, Guizhou, China, 1380 m | 2018-IX-08 2018-VII-26 | 25°24′30′′ N 105°10′60′′ E | Rubus lambertianus | 24 | MW387769–92 |
| Sclomina pallens sp. nov. | GZAL | Xianheping, Anlong, Guizhou, China, 1330 m | 2017-X-1 2018-VII-25 2018-IX-10 | 24°59′49′′ N 105°36′53′′ E | Rubus lambertianu Rubus rosifolius Rubus tephrodes | 6 | MW387687–92 |
| GXTL | Langping, Tianlin, Guangxi, China, 1110 m | 2017-VIII-22 | 24°28′34′′ N 106°21′34′′ E | Rubus lambertianus | 13 | MW387665–77 | |
| Sclomina parva sp. nov. | SXYX | Hongjunlin, Yangxian, Shannxi, China, 1090 m | 2018-VIII-6 2012-VIII-5 | 33º36′47′′ N 107°30′42′′ E | Rubus coreanus | 11 | MW387941–47 MW387858–61 |
| Sclomina guangxiensis | GXLZ | Nonggang, Longzhou, Guangxi, China, 260 m | 2017-VIII-22 2017-XI-11 | 22°28′24′′ N 106°57′11′′ E | Rubus lambertianus (mainly) Rubus amphidasys | 9 | MW387656–64 |
| Outgroup | / | / | / | / | / | / | |
| Endochus albomaculatus | Endochus | / | / | / | / | / | KC834737 |
| Sphedanolestes annulipes | Sphedanolestes | / | / | / | / | / | KJ629990 |
| Velinus annulatus | Velinus | / | / | / | / | / | KJ629888 |
| Scipinia horrida | Scipinia | / | / | / | / | / | NC037744 |
References
- Schuh, R.T.; Slater, J.A. Chapter 7. Mimicry and Protective coloration and Shape; Chapter 48. Reduviidae. In True Bugs of the World (Hemiptera: Heteroptera); Schuh, R.T., Slater, J.A., Eds.; Cornell University Press: Ithaca, New York, NY, USA, 1995; pp. 27–31, 150–161. [Google Scholar]
- Stål, C. Miscellanea hemipterologica. Stettiner Entomol. Zeitung 1861, 22, 129–153. [Google Scholar]
- Maldonado-Capriles, J. Systematic Catalogue of the Reduviidae of the World (Insecta: Heteroptera). Special Edition, i–x. Caribb. J. Sci. 1990, 1–694. [Google Scholar]
- Hsiao, T.Y.; Ren, S.Z. Reduviidae. In A Handbook for the Determination of the Chinese Hemiptera-Heteroptera (II) (In Chinese with English Summary); Hsiao, T.Y., Ren, S.Z., Zheng, L.Y., Jing, X.L., Zou, H.G., Liu, S.L., Eds.; Science Press: Beijing, China, 1981; pp. 390–538. [Google Scholar]
- Putshkov, P.V.; Putshkov, V.G. Family Reduviidae Latreille, 1807, assassin-bugs. In Catalogue of the Heteroptera of the Palaearctic Region, volume 2 Cimicomorpha I; Putshkov, P.V., Putshkov, V.G., Eds.; The Netherlands Entomological Society: Amsterdam, The Netherlands, 1996; p. 254. [Google Scholar]
- Ren, S.Z. Two New Species of Harpactorinae from South China. J. Tianjin Agric. Coll. 2001, 8, 1–4. [Google Scholar]
- Planas, E.; Ribera, C. Description of six new species of Loxosceles (Araneae: Sicariidae) endemic to the Canary Islands and the utility of DNA barcoding for their fast and accurate identification. Zool. J. Linn. Soc. Lond. 2015, 1, 47–73. [Google Scholar] [CrossRef] [Green Version]
- Zittra, C.; Wöss, G.; Van der Vloet, L.; Bakran-Lebl, K.; Barogh, B.S.; Sehnal, P.; Fuehrer, H.P. Barcoding of the genus Culicoides (Diptera: Ceratopogonidae) in Austria—An update of the species inventory including the first records of three species in Austria. Pathogens 2020, 9, 406. [Google Scholar] [CrossRef]
- Kim, S.; Lee, Y.; Mutanen, M.; Seung, J.; Lee, S. High functionality of DNA barcodes and revealed cases of cryptic diversity in Korean curved-horn moths (Lepidoptera: Gelechioidea). Sci. Rep 2020, 10, 6208. [Google Scholar] [CrossRef] [Green Version]
- Lavery, S.D.; Farhadi, A.; Farahmand, H.; Chan, T.Y.; Azhdehakoshpour, A.; Thakur, V.; Jeffs, A.G. Evolutionary divergence of geographic subspecies within the scalloped spiny lobster Panulirus homarus (Linnaeus, 1758). PLoS ONE 2014, 9, e97247. [Google Scholar] [CrossRef] [Green Version]
- Khadem, M. Deep Interisland Genetic Divergence in the Macaronesian Endemic Mosquito Ochlerotatus eatoni (Diptera: Culicidae), Indication of Cryptic Species. J. Med. Entomol. 2015, 52, 1175–1180. [Google Scholar] [CrossRef]
- Hebert, P.D.N.; Ratnasingham, S.; deWaard, J.R. Barcoding animal life: Cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B Biol. Sci. 2003, 270, S96–S99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Jung, S. COI barcoding of plant bugs (Insecta: Hemiptera: Miridae). PeerJ 2018, 6, e6070. [Google Scholar] [CrossRef]
- Jung, S.; Duwal, R.K.; Lee, S. COI barcoding of true bugs (Insecta, Heteroptera). Mol. Ecol. Resour. 2011, 11, 266–270. [Google Scholar] [CrossRef]
- Park, D.S.; Foottit, R.; Maw, E.; Hebert, P.D.N. Barcoding Bugs: DNA-Based Identification of the True Bugs (Insecta: Hemiptera: Heteroptera). PLoS ONE 2011, 6, e18749. [Google Scholar] [CrossRef]
- Rubén, G.M.; Eva, M.N.; Yolanda, J.R.; Paloma, M.P.; Ghanavi, H.R.; Mario, G.P. Speciation patterns in the Forficula auricularia species complex: Cryptic and not so cryptic taxa across the western Palaearctic region. Zool. J. Linn. Soc Lond. 2020, 20, 1–36. [Google Scholar] [CrossRef]
- Jacobina, U.P.; Torres, R.A.; De Mello Affonso, P.R.A.; Dos Santos, E.V.; Calado, L.L.; De Araújo Bitencourt, J. DNA barcoding reveals cryptic diversity and peculiar phylogeographic patterns in mojarras (Perciformes: Gerreidae) from the Caribbean and South-western Atlantic. J. Mar. Biol. Assoc. 2020, 100, 277–283. [Google Scholar] [CrossRef]
- Qin, L.; Wang, J.; Bing, X.L.; Liu, S.S. Identification of nine cryptic species of Bemisia tabaci (Hemiptera: Aleyrodidae) from China by using the mtCOI PCR-RFLP technique. Acta Entomol. Sin. 2013, 56, 186–194. [Google Scholar]
- Weis, A.; Meyer, R.; Dietz, L.; Doemel, J.S.; Leese, F.; Melzer, R.R. Pallenopsis patagonica (Hoek, 1881)—A species complex revealed by morphology and DNA barcoding, with description of a new species of Pallenopsis Wilson, 1881. Zool. J. Linn. Soc. Lond. 2014, 170, 110–131. [Google Scholar] [CrossRef]
- Elderkin, C.L.; Clewing, C.; Ndeo, O.W.; Albrecht, C. Molecular phylogeny and DNA barcoding confirm cryptic species in the African freshwater Oyster Etheria elliptica Lamarck, 1807 (Bivalvia: Etheriidae). Biol. J. Linn. Soc. 2016, 118, 369–381. [Google Scholar] [CrossRef] [Green Version]
- Pfeiler, E.; Nazario-Yepiz, N.O. DNA-based taxonomy and potential suppression of long-established names: The case of Telegonus fulgerator (Lepidoptera: Hesperiidae). Syst. Biodivers. 2020, 1–9. [Google Scholar] [CrossRef]
- Lee, Y.; Lee, W.; Lee, S.; Kim, H. A cryptic species of Aphis gossypii (Hemiptera: Aphididae) complex revealed by genetic divergence and different host plant association. B. Entomol. Res. 2015, 105, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Lent, H.; Wygodzinsky, P. Revision of the Triatominae (Hemiptera, Reduviidae) and Their Significance as Vectors of Chagas’ Disease. B. Am. Mus. of Nat. Hist. 1979, 163, 125–520. Available online: http://fossilinsects.net/node/10958 (accessed on 25 November 2013).
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Mol. Biol. Evol. 2016, 34, 772–773. [Google Scholar] [CrossRef] [Green Version]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef]
- Bouckaert, R.; Heled, J.; Kühnert, D.; Vaughan, T.; Wu, C.H.; Xie, D.; Suchard, M.A.; Rambaut, A.; Drummond, A.J. BEAST 2: A Software Platform for Bayesian Evolutionary Analysis. PLoS Comput. Biol. 2014, 10, e1003537. [Google Scholar] [CrossRef] [Green Version]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papadopoulou, A.; Anastasiou, I.; Vogler, A.P. Revisiting the insect Mitochondrial molecular clock: The mid-Aegean trench calibration. Mol. Biol. Evol. 2010, 27, 1659–1672. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [Green Version]
- Huang, K.R.; Shang, H.; Zhou, Q.; Yan, Y.H.; Chen, S.; Li, G. The life history and biological characteristics of Sclomina erinacea. J. Environ. Entomol. 2017, 39, 1350–1355. [Google Scholar]
- Hwang, W.S.; Weirauch, C. Evolutionary history of assassin bugs (Insecta: Hemiptera: Reduviidae): Insights from divergence dating and ancestral state reconstruction. PLoS ONE 2012, 7, e45523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoorn, C.; Mosbrugger, V.; Mulch, A.; Antonelli, A. Biodiversity from mountain building. Nat. Geosci. 2013, 6, 154. [Google Scholar] [CrossRef]
- Forero, D.; Choe, D.H.; Weirauch, C. Resin gathering in neotropical resin bugs (Insecta: Hemiptera: Reduviidae): Functional and comparative morphology. J. Morphol. 2011, 272, 204–229. [Google Scholar] [CrossRef]
- Forero, D.; Weirauch, C. Comparative genitalic morphology in the New World resin bugs Apiomerini (Hemiptera, Heteroptera, Reduviidae, Harpactorinae). Deut. Entomol. Z. 2012, 59, 5–41. [Google Scholar] [CrossRef]
- Garrouste, R.; Hugel, S.; Jacquelin, L.; Rostan, P.; Steyer, J.S.; Desutter-Grandcolas, L.; Nel, A. Insect mimicry of plants dates back to the permian. Nat. Commun. 2016, 7, 13735. [Google Scholar] [CrossRef]
- Goodrich, K.R.; Jürgens, A. Pollination systems involving floral mimicry of fruit: Aspects of their ecology and evolution. New Phytol. 2018, 1, 217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Chen, Q.; Chen, T.; Tang, H.; Liu, L.; Wang, X. Phylogenetic insights into Chinese Rubus (Rosaceae) from Multiple Chloroplast and Nuclear DNAs. Front. Plant Sci. 2016, 7, 968. [Google Scholar] [CrossRef] [Green Version]










Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, P.; Du, Z.; Zhao, Q.; Li, D.; Shao, X.; Li, H.; Cai, W. Integrative Taxonomy of the Spinous Assassin Bug Genus Sclomina (Heteroptera: Reduviidae: Harpactorinae) Reveals Three Cryptic Species Based on DNA Barcoding and Morphological Evidence. Insects 2021, 12, 251. https://doi.org/10.3390/insects12030251
Zhao P, Du Z, Zhao Q, Li D, Shao X, Li H, Cai W. Integrative Taxonomy of the Spinous Assassin Bug Genus Sclomina (Heteroptera: Reduviidae: Harpactorinae) Reveals Three Cryptic Species Based on DNA Barcoding and Morphological Evidence. Insects. 2021; 12(3):251. https://doi.org/10.3390/insects12030251
Chicago/Turabian StyleZhao, Ping, Zhenyong Du, Qian Zhao, Donghai Li, Xiaolan Shao, Hu Li, and Wanzhi Cai. 2021. "Integrative Taxonomy of the Spinous Assassin Bug Genus Sclomina (Heteroptera: Reduviidae: Harpactorinae) Reveals Three Cryptic Species Based on DNA Barcoding and Morphological Evidence" Insects 12, no. 3: 251. https://doi.org/10.3390/insects12030251
APA StyleZhao, P., Du, Z., Zhao, Q., Li, D., Shao, X., Li, H., & Cai, W. (2021). Integrative Taxonomy of the Spinous Assassin Bug Genus Sclomina (Heteroptera: Reduviidae: Harpactorinae) Reveals Three Cryptic Species Based on DNA Barcoding and Morphological Evidence. Insects, 12(3), 251. https://doi.org/10.3390/insects12030251