dsRNA-Mediated Pest Management of Tuta absoluta Is Compatible with Its Biological Control Agent Nesidiocoris tenuis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing and Maintenance
2.2. Target Gene Selection and dsRNA Synthesis
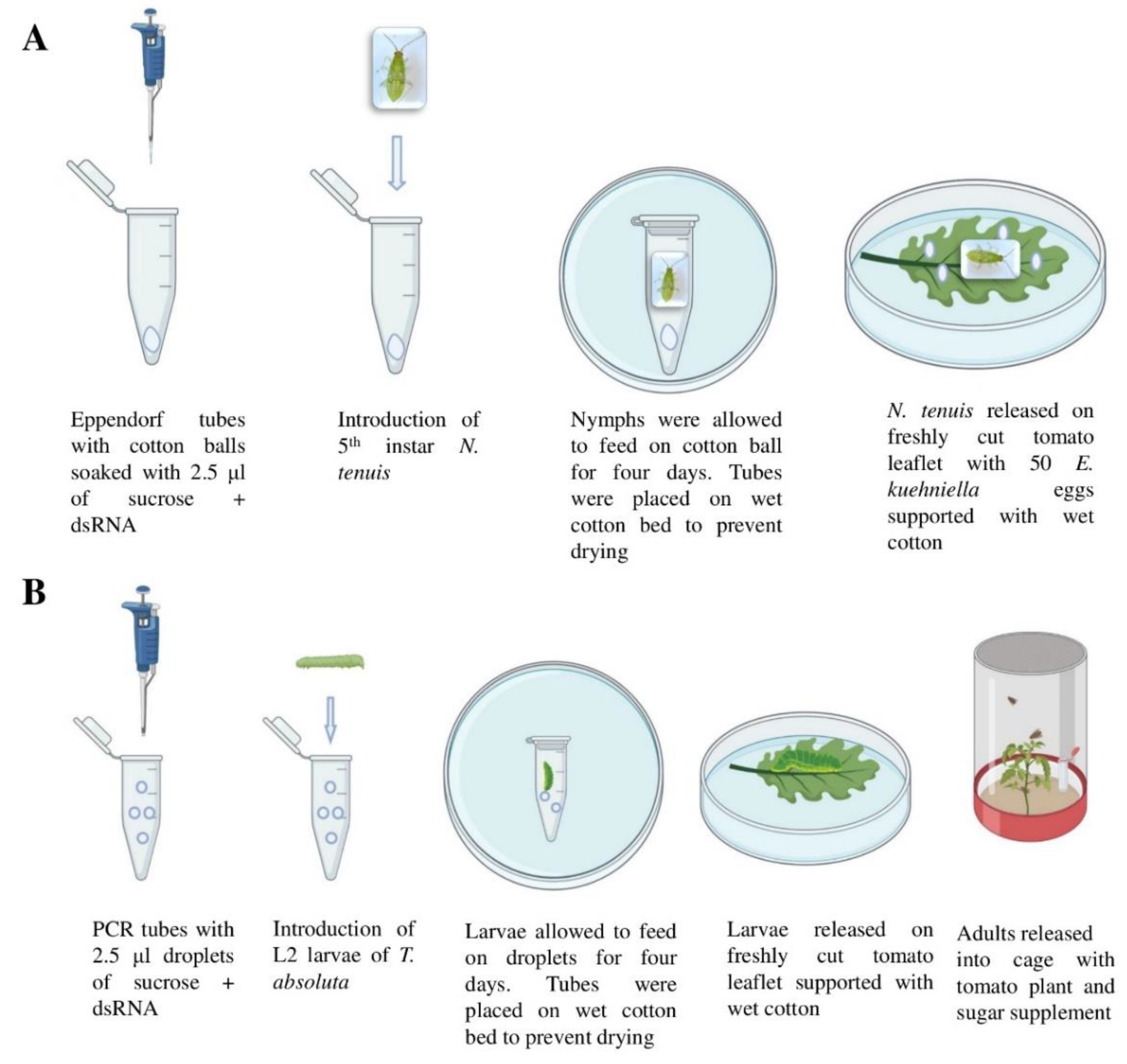
2.3. Oral Delivery of dsRNA to N. tenuis and T. absoluta
2.4. Gene Expression Analysis
2.5. Statistical Analysis of Data
3. Results
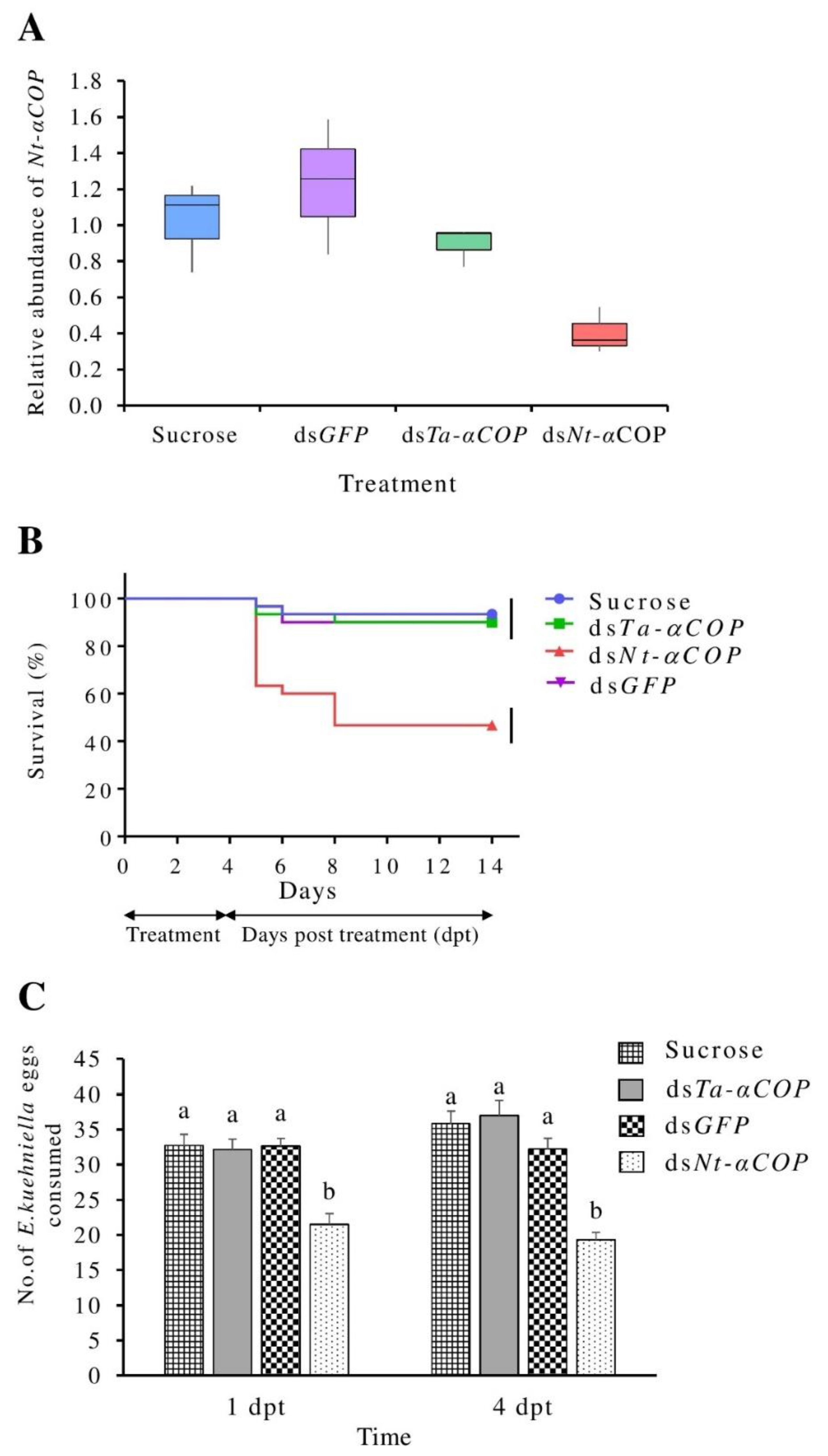
3.1. Effects of Oral Feeding of dsNt-αCOP in N. tenuis
3.2. No Significant Cross-Silencing Effects of dsTa-αCOP on N. tenuis
3.3. Oral Delivery of dsTa-αCOP to T. absoluta Can Cause Lethal to Sublethal Effects
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Garzia, T.G.; Siscaro, G.; Biondi, A.; Zappalà, L. Tuta absoluta, a South American pest of tomato now in the EPPO region: Biology, distribution and damage. EPPO Bull. 2012, 42, 205–210. [Google Scholar] [CrossRef]
- Desneux, N.; Wajnberg, E.; Wyckhuys, K.A.; Burgio, G.; Arpaia, S.; Narváez-Vasquez, C.A.; González-Cabrera, J.; Ruescas, D.C.; Tabone, E.; Frandon, J.; et al. Biological invasion of European tomato crops by Tuta absoluta: Ecology, geographic expansion and prospects for biological control. J. Pest. Sci. 2010, 83, 197–215. [Google Scholar] [CrossRef]
- Pérez-Hedo, M.; Suay, R.; Alonso, M.; Ruocco, M.; Giorgini, M.; Poncet, C.; Urbaneja, A. Resilience and robustness of IPM in protected horticulture in the face of potential invasive pests. Crop. Prot. 2017, 97, 119–127. [Google Scholar] [CrossRef]
- Urbaneja, A.; González-Cabrera, J.; Arnó, J.; Gabarra, R. Prospects for the biological control of Tuta absoluta in tomatoes of the Mediterranean basin. Pest. Manag. Sci. 2012, 68, 1215–1222. [Google Scholar] [CrossRef]
- Urbaneja, A.; Montón, H.; Mollá, O. Suitability of the tomato borer Tuta absoluta as prey for Macrolophus pygmaeus and Nesidiocoris tenuis. J. Appl. Entomol. 2009, 133, 292–296. [Google Scholar] [CrossRef]
- Mollá, O.; González-Cabrera, J.; Urbaneja, A. The combined use of Bacillus thuringiensis and Nesidiocoris tenuis against the tomato borer Tuta absoluta. BioControl 2011, 56, 883–891. [Google Scholar] [CrossRef]
- Schäfer, L.; Herz, A. Suitability of European Trichogramma species as biocontrol agents against the tomato leaf miner Tuta absoluta. Insects 2020, 11, 357. [Google Scholar] [CrossRef] [PubMed]
- Castañé, C.; Arnó, J.; Gabarra, R.; Alomar, O. Plant damage to vegetable crops by zoophytophagous mirid predators. BioControl 2011, 59, 22–29. [Google Scholar] [CrossRef]
- Pérez-Hedo, M.; Riahi, C.; Urbaneja, A. Use of zoophytophagous mirid bugs in horticultural crops: Current challenges and future perspectives. Pest. Manag. Sci. 2021, 77, 33–42. [Google Scholar] [CrossRef]
- Camargo, R.A.; Barbosa, G.O.; Possignolo, I.P.; Peres, L.E.; Lam, E.; Lima, J.E.; Figueira, A.; Marques-Souza, H. RNA interference as a gene silencing tool to control Tuta absoluta in tomato (Solanum lycopersicum). PeerJ 2016, 4, 2673. [Google Scholar] [CrossRef] [PubMed]
- Bento, F.M.; Marques, R.N.; Campana, F.B.; Demétrio, C.G.; Leandro, R.A.; Parra, J.R.P.; Figueira, A. Gene silencing by RNAi via oral delivery of dsRNA by bacteria in the South American tomato pinworm, Tuta absoluta. Pest. Manag. Sci. 2020, 76, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Tuschl, T. RNA interference and small interfering RNAs. Chembiochem 2001, 2, 239–245. [Google Scholar] [CrossRef]
- Dowling, D.; Pauli, T.; Donath, A.; Meusemann, K.; Podsiadlowski, L.; Petersen, M.; Peters, R.S.; Mayer, C.; Liu, S.; Zhou, X.; et al. Phylogenetic origin and diversification of RNAi pathway genes in insects. Genome Biol. Evol. 2016, 8, 3784–3793. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Kulkarni, M.M.; Booker, M.; Silver, S.J.; Friedman, A.; Hong, P.; Perrimon, N.; Mathey-Prevot, B. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat. Methods 2006, 3, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Khan, S.A.; Hasse, C.; Ruf, S.; Heckel, D.G.; Bock, R. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science 2015, 347, 991–994. [Google Scholar] [CrossRef]
- San Miguel, K.; Scott, J.G. The next generation of insecticides: dsRNA is stable as a foliar-applied insecticide. Pest. Manag. Sci. 2016, 72, 801–809. [Google Scholar] [CrossRef]
- Baum, J.A.; Bogaert, T.; Clinton, W.; Heck, G.R.; Feldmann, P.; Ilagan, O.; Johnson, S.; Plaetinck, G.; Munyikwa, T.; Pleau, M.; et al. Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 2007, 25, 1322–1326. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.B.; Cai, W.J.; Wang, J.W.; Hong, G.J.; Tao, X.Y.; Wang, L.J.; Huang, Y.P.; Chen, X.Y. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 2007, 25, 1307–1313. [Google Scholar] [CrossRef]
- Price, D.R.G.; Gatehouse, J.A. RNAi-mediated crop protection against insects. Trends Biotechnol. 2008, 26, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Dias, N.; Cagliari, D.; Kremer, F.S.; Rickes, L.N.; Nava, D.E.; Smagghe, G.; Zotti, M. The south American fruit fly: An important pest insect with RNAi-sensitive larval stages. Front. Physiol. 2019, 10, 794. [Google Scholar] [CrossRef]
- Tian, H.; Peng, H.; Yao, Q.; Chen, H.; Xie, Q.; Tang, B.; Zhang, W. Developmental control of a Lepidopteran pest Spodoptera exigua by ingestion of bacterial expressing dsRNA of a non-midgut gene. PLoS ONE 2009, 4, e6225. [Google Scholar] [CrossRef]
- Yu, N.; Christiaens, O.; Liu, J.; Niu, J.; Cappelle, K.; Caccia, S.; Huvenne, H.; Smagghe, G. Delivery of dsRNA for RNAi in insects: An overview and future directions. Insect Sci. 2013, 20, 4–14. [Google Scholar] [CrossRef]
- Chaitanya, B.N.; Asokan, R.; Sita, T.; Rebijith, K.B.; Ram Kumar, P.; Krishna Kumar, N.K. Silencing of JHEH and EcR genes of Plutella xylostella (Lepidoptera: Plutellidae) through double stranded RNA oral delivery. J. Asia Pac. Entomol. 2017, 20, 637–643. [Google Scholar] [CrossRef]
- Ganbaatar, O.; Cao, B.; Zhang, Y.; Bao, D.; Wuriyanghan, H. Knockdown of Mythimna separata chitinase genes via bacterial expression and oral delivery of RNAi effectors. BMC Biotechnol. 2017, 17, 9. [Google Scholar] [CrossRef] [PubMed]
- Rahmani, S.; Bandani, A.R. A gene silencing of V-ATPase subunit A interferes with survival and development of the tomato leafminer, Tuta absoluta. Arch. Insect Biochem. Physiol. 2021, 106, e21753. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, N.; Devos, Y.; Álvarez-Alfageme, F.; Lanzoni, A.; Waigmann, E. Risk assessment considerations for genetically modified RNAi plants: EFSA’s activities and perspective. Front. Plant Sci. 2020, 11, 445. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Yang, X.; Romeis, J.; Siegfried, B.D.; Zhou, X. Dietary RNAi toxicity assay exhibits differential responses to ingested dsRNAs among lady beetles. Pest. Manag. Sci. 2020, 76, 3606–3614. [Google Scholar] [CrossRef] [PubMed]
- Romeis, J.; Widmer, F. Assessing the risks of topically applied dsRNA-based products to non-target arthropods. Front. Plant Sci. 2020, 11, 679. [Google Scholar] [CrossRef] [PubMed]
- Taning, C.N.T.; Mezzetti, B.; Kleter, G.; Smagghe, G.; Baraldi, E. Does RNAi-based technology fit within EU sustainability goals? Trends Biotechnol. 2020, in press. [Google Scholar] [CrossRef]
- Castellanos, N.L.; Smagghe, G.; Sharma, R.; Oliveira, E.E.; Christiaens, O. Liposome encapsulation and EDTA formulation of dsRNA targeting essential genes increase oral RNAi-caused mortality in the Neotropical stink bug Euschistus heros. Pest. Manag. Sci. 2019, 75, 537–548. [Google Scholar] [CrossRef]
- Neumeier, J.; Meister, G. SiRNA specificity: RNAi mechanisms and strategies to reduce off-target effects. Front. Plant Sci. 2021, 11, 526455. [Google Scholar] [CrossRef] [PubMed]
- Taning, C.N.T.; Gui, S.H.; De Schutter, K.; Jahani, M.; Castellanos, N.L.; Christiaens, O.; Smagghe, G. A sequence complementarity-based approach for evaluating off-target transcript knockdown in Bombus terrestris, following ingestion of pest-specific dsRNA. J. Pest. Sci. 2021, 94, 487–503. [Google Scholar] [CrossRef]
- Yoo, B.C.; Kragler, F.; Varkonyi-Gasic, E.; Haywood, V.; Archer-Evans, S.; Lee, Y.M.; Lough, T.J.; Lucas, W.J. A systemic small RNA signaling system in plants. Plant Cell 2004, 16, 1979–2000. [Google Scholar] [CrossRef] [PubMed]
- Voloudakis, A.E.; Holeva, M.C.; Sarin, L.P.; Bamford, D.H.; Vargas, M.; Poranen, M.M.; Tenllado, F. Efficient double-stranded RNA production methods for utilization in plant virus control. Methods Mol. Biol. 2015, 1236, 255–274. [Google Scholar] [CrossRef] [PubMed]
- Taning, C.N.T.; Christiaens, O.; Berkvens, N.; Casteels, H.; Maes, M.; & Smagghe, G. Oral RNAi to control Drosophila suzukii: Laboratory testing against larval and adult stages. J. Pest. Sci. 2016, 89, 803–814. [Google Scholar] [CrossRef]
- Turner, C.T.; Davy, M.W.; MacDiarmid, R.M.; Plummer, K.M.; Birch, N.P.; Newcomb, R.D. RNA interference in the light brown apple moth, Epiphyas postvittana (Walker) induced by double-stranded RNA feeding. Insect Mol. Biol. 2006, 15, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Reyes, M.; Rocha, K.; Alarcón, L.; Siegwart, M.; Sauphanor, B. Metabolic mechanisms involved in the resistance of field populations of Tuta absoluta (Meyrick) (Lepidoptera: Gelechiidae) to spinosad. Pestic. Biochem. Physiol. 2012, 102, 45–50. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Sharma, R.; Christiaens, O.; Taning, C.N.T.; Smagghe, G. RNAi-mediated mortality in southern green stinkbug Nezara viridula by oral delivery of dsRNA. Pest. Manag. Sci. 2021, 77, 77–84. [Google Scholar] [CrossRef]
- Willow, J.; Sulg, S.; Taning, C.N.T.; Silva, A.I.; Christiaens, O.; Kaasik, R.; Prentice, K.; Lövei, G.L.; Smagghe, G.; Veromann, E. Targeting a coatomer protein complex-I gene via RNA interference results in effective lethality in the pollen beetle Brassicogethes aeneus. J. Pest. Sci. 2020, in press. [Google Scholar] [CrossRef]
- Prentice, K.; Christiaens, O.; Pertry, I.; Bailey, A.; Niblett, C.; Ghislain, M.; Gheysen, G.; Smagghe, G. RNAi-based gene silencing through dsRNA injection or ingestion against the African sweet potato weevil Cylas puncticollis (Coleoptera: Brentidae). Pest. Manag. Sci. 2017, 73, 44–52. [Google Scholar] [CrossRef]
- Li, H.; Guan, R.; Guo, H.; Miao, X. New insights into an RNAi approach for plant defence against piercing-sucking and stem-borer insect pests. Plant Cell Environ. 2015, 38, 2277–2285. [Google Scholar] [CrossRef] [PubMed]
- Mamta, B.; Rajam, M.V. RNAi technology: A new platform for crop pest control. Physiol. Mol. Biol. Plants 2017, 23, 487–501. [Google Scholar] [CrossRef]
- Christiaens, O.; Whyard, S.; Vélez, A.M.; Smagghe, G. Double-stranded RNA technology to control insect pests: Current status and challenges. Front. Plant Sci. 2020, 11, 451. [Google Scholar] [CrossRef] [PubMed]
- Mezzetti, B.; Smagghe, G.; Arpaia, S.; Christiaens, O.; Dietz-Pfeilstetter, A.; Jones, H.; Kostov, K.; Sabbadini, S.; Opsahl-Sorteberg, H.G.; Ventura, V.; et al. RNAi: What is its position in agriculture? J. Pest. Sci. 2020, 93, 1125–1130. [Google Scholar] [CrossRef]
- Arpaia, S.; Christiaens, O.; Giddings, K.; Jones, H.; Mezzetti, B.; Moronta-Barrios, F.; Perry, J.N.; Sweet, J.B.; Taning, C.N.T.; Smagghe, G.; et al. Biosafety of GM crop plants expressing dsRNA: Data requirements and EU regulatory considerations. Front. Plant Sci. 2020, 11, 940. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, S.; Smagghe, G.; Sweet, J.B. Biosafety of bee pollinators in GM agro-ecosystems: Current approach and further development in the EU. Pest. Manag. Sci. 2021, accepted. [Google Scholar] [CrossRef]
- Singh, I.K.; Singh, S.; Mogilicherla, K.; Shukla, J.N.; Palli, S.R. Comparative analysis of double-stranded RNA degradation and processing in insects. Sci. Rep. 2017, 7, 17059. [Google Scholar] [CrossRef]
- Grover, S.; Jindal, V.; Banta, G.; Taning, C.N.T.; Smagghe, G.; Christiaens, O. Potential of RNA interference in the study and management of the whitefly, Bemisia tabaci. Arch. Insect Biochem. Physiol. 2019, 100, e21522. [Google Scholar] [CrossRef] [PubMed]
- Tayler, A.; Heschuk, D.; Giesbrecht, D.; Park, J.Y.; Whyard, S. Efficiency of RNA interference is improved by knockdown of dsRNA nucleases in tephritid fruit flies. Open Biol. 2019, 9, 190198. [Google Scholar] [CrossRef] [PubMed]
- Soares, M.A.; Campos, M.R.; Passos, L.C.; Carvalho, G.A.; Haro, M.M.; Lavoir, A.V.; Biondi, A.; Zappalà, L.; Desneux, N. Botanical insecticide and natural enemies: A potential combination for pest management against Tuta absoluta. J. Pest. Sci. 2019, 92, 1433–1443. [Google Scholar] [CrossRef]

| Name | Type | Sequence (5′–3′) | Product Size (nt) | Target Species | Purpose |
|---|---|---|---|---|---|
| dsNt-αCOP (joined to T7 promoter) | F R | TAATACGACTCACTATAGGG CACACTGCCCCTGATCGTAT TAATACGACTCACTATAGGG GTCGAGTTTACGCAGGAAGC | 391 | N. tenuis | dsRNA production |
| qPCR-Nt-αCOP | F R | GGGAGGACTCGAAGAACATTT GATCGTGCCCTTCCAAGAC | 95 | N. tenuis | qPCR |
| qPCR-Nt-ATPB | F R | CATACGCCAAGGGAGGTAAA CTGGGTGAAACGGAAAATGT | 356 | N. tenuis | qPCR |
| dsTa-αCOP (joined to T7 promoter) | F R | TAATACGACTCACTATAGGG CCGTTTTCATCACAGGTCT TAATACGACTCACTATAGGG CGGTCATGGCCACTAAGAAT | 505 | T. absoluta | dsRNA production |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarmah, N.; Kaldis, A.; Taning, C.N.T.; Perdikis, D.; Smagghe, G.; Voloudakis, A. dsRNA-Mediated Pest Management of Tuta absoluta Is Compatible with Its Biological Control Agent Nesidiocoris tenuis. Insects 2021, 12, 274. https://doi.org/10.3390/insects12040274
Sarmah N, Kaldis A, Taning CNT, Perdikis D, Smagghe G, Voloudakis A. dsRNA-Mediated Pest Management of Tuta absoluta Is Compatible with Its Biological Control Agent Nesidiocoris tenuis. Insects. 2021; 12(4):274. https://doi.org/10.3390/insects12040274
Chicago/Turabian StyleSarmah, Nomi, Athanasios Kaldis, Clauvis Nji Tizi Taning, Dionysios Perdikis, Guy Smagghe, and Andreas Voloudakis. 2021. "dsRNA-Mediated Pest Management of Tuta absoluta Is Compatible with Its Biological Control Agent Nesidiocoris tenuis" Insects 12, no. 4: 274. https://doi.org/10.3390/insects12040274
APA StyleSarmah, N., Kaldis, A., Taning, C. N. T., Perdikis, D., Smagghe, G., & Voloudakis, A. (2021). dsRNA-Mediated Pest Management of Tuta absoluta Is Compatible with Its Biological Control Agent Nesidiocoris tenuis. Insects, 12(4), 274. https://doi.org/10.3390/insects12040274








