Vectors of Dutch Elm Disease in Northern Europe
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
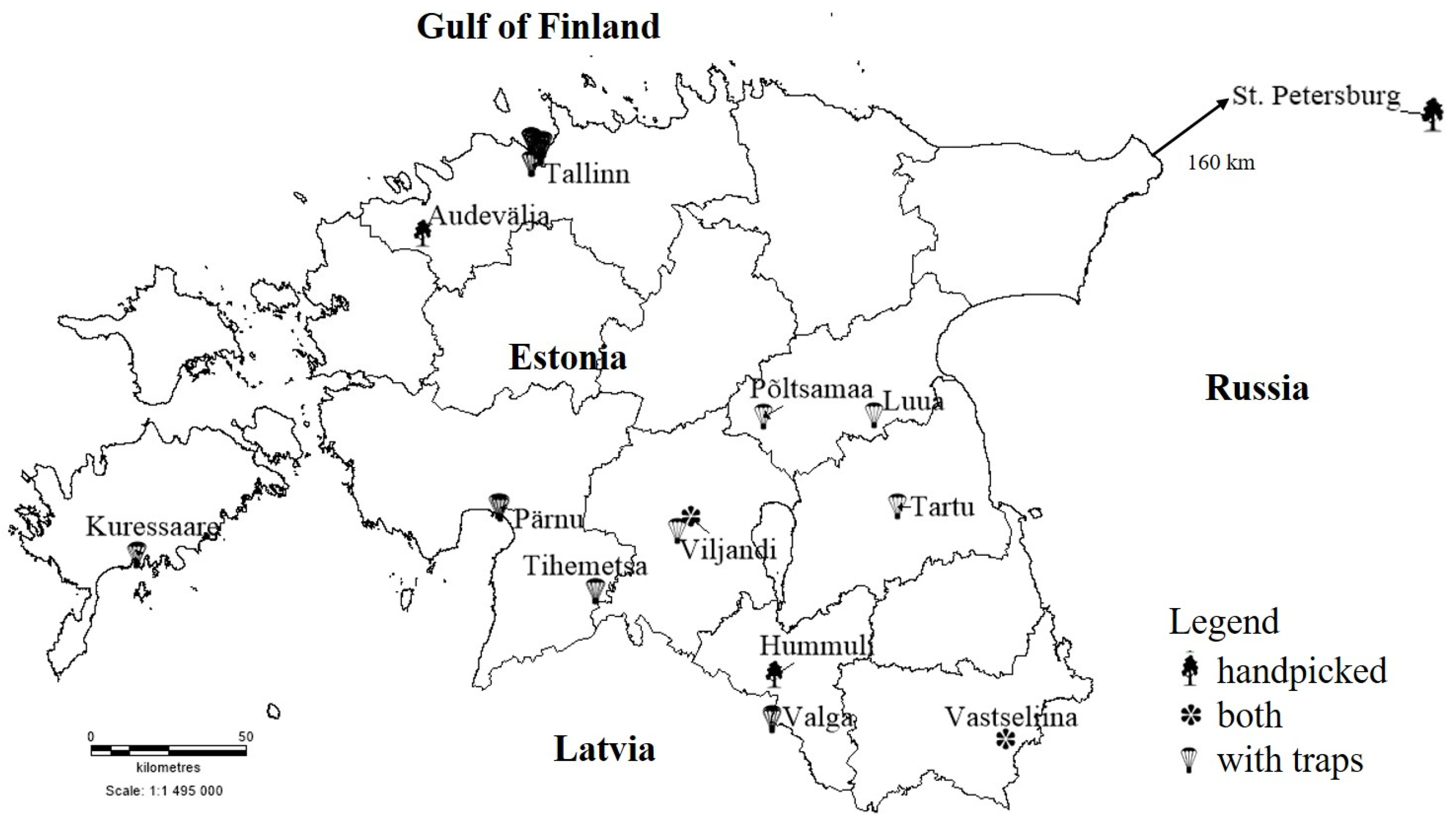
2.1. Study Sites and Sampling
2.2. Beetles’ Identification
2.3. Molecular Analyses
2.4. Bioinformatics and Statistical Analysis
3. Results
3.1. Collected Beetle Species
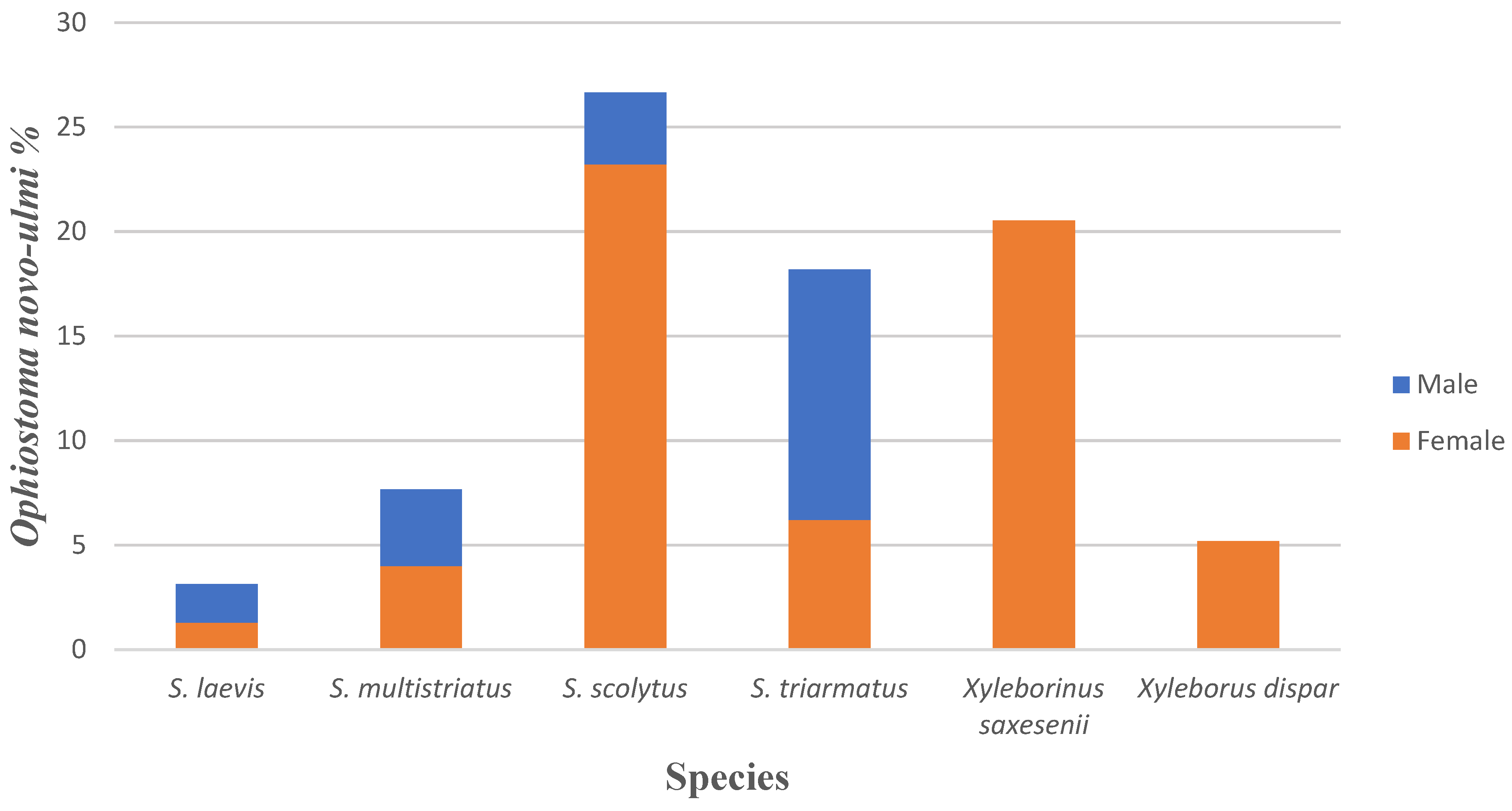
3.2. Ophiostoma novo-ulmi on Vector Beetles
4. Discussion
4.1. Beetles as Vectors of O. novo-ulmi and Pathogen Detection
4.2. Spread of Vectors for DED in Estonia and Northwest Russia
4.3. The Traps and Alcohol as a Baiting Compound
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kukk, T.; Kull, T. Eesti Taimede Levikuatlas (Atlas of the Estonian Flora); Eesti Maaülikooli Põllumajandus- ja Keskkonnainstituut; Institute of Agricultural and Environmental Sciences of the Estonian University of Life Sciences: Tartu, Estonia, 2005; 528p. (In Estonian) [Google Scholar]
- Caudullo, G.; De Rigo, D. Ulmus—elms in Europe: Distribution, habitat, usage and threats. In European Atlas of Forest Tree Species; San-Miguel-Ayanz, J., de Rigo, D., Caudullo, G., Durrant, H.T., Mauri, A., Eds.; Publications Office of the EU: Luxembourg, 2016; p. e01bd40. [Google Scholar]
- Saarse, L.; Veski, S. Spread of broad-leaved trees in Estonia. Proc. Est. Acad. Sci. Geol. 2001, 50, 51. Available online: https://www.researchgate.net/publication/252516467_Spread_of_broad-leaved_trees_in_Estonia (accessed on 6 April 2021).
- Santini, A.; Faccoli, M. Dutch elm disease and elm bark beetles: A century of association. iForest Biogeosci. For. 2015, 8, 126–134. [Google Scholar] [CrossRef] [Green Version]
- Smith, S.M.; Hulcr, J. Scolytus and other Economically Important Bark and Ambrosia Beetles. Bark Beetles 2015, 495–531. [Google Scholar] [CrossRef]
- Wingfield, M.J.; Garnas, J.R.; Hajek, A.; Hurley, B.P.; De Beer, Z.W.; Taerum, S.J. Novel and co-evolved associations between insects and microorganisms as drivers of forest pestilence. Biol. Invasions 2016, 18, 1045–1056. [Google Scholar] [CrossRef]
- Martín, J.A.; Sobrino-Plata, J.; Rodríguez-Calcerrada, J.; Collada, C.; Gil, L. Breeding and scientific advances in the fight against Dutch elm disease: Will they allow the use of elms in forest restoration? New For. 2019, 50, 183–215. [Google Scholar] [CrossRef] [Green Version]
- Brasier, C.M. Intercontinental Spread and Continuing Evolution of the Dutch Elm Disease Pathogens. In The Elms; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2000; pp. 61–72. [Google Scholar]
- Clinton, G.P.; McCormick, F.A. Dutch Elm Disease. Graphium Ulmi; Connecticut Agricultural Experimental Station, Bulletin 386: New Haven, CT, USA, 1936; pp. 701–752. [Google Scholar]
- Stipes, R.J.; Campana, R.J. Compendium of Elm Diseases; Stipes, R.J., Campana, R.J., Eds.; American Phytopathological Society: Saint Paul, MN, USA, 1981. [Google Scholar]
- Peace, T.R. The status and development of Elm Disease in Britain. Comm Bull. 1960, 33, 44. [Google Scholar]
- Gibbs, J.N. Intercontinental Epidemiology of Dutch Elm Disease. Annu. Rev. Phytopathol. 1978, 16, 287–307. [Google Scholar] [CrossRef]
- Brasier, C.; Buck, K. Rapid Evolutionary Changes in a Globally Invading Fungal Pathogen (Dutch Elm Disease). Biol. Invasions 2001, 3, 223–233. [Google Scholar] [CrossRef]
- Kirisits, T. Dutch elm disease and other Ophiostoma diseases. Infect. For. Dis. 2013, 2013, 256–282. [Google Scholar] [CrossRef]
- Brasier, C.M. Rapid Evolution of Introduced Plant Pathogens via Interspecific Hybridization is leading to rapid evolution of Dutch elm disease and other fungal plant pathogens. Bioscience 2001, 51, 123–133. [Google Scholar] [CrossRef]
- Gärdenfors, U. The 2010 Red List of Swedish Species. Available online: https://www.artdatabanken.se/globalassets/ew/subw/artd/2.-var-verksamhet/publikationer/4.-rodlista-2010/274614_inlaga_liten_sid-del1-1-199.pdf (accessed on 27 November 2019).
- Jürisoo, L.; Selikhovkin, A.V.; Padari, A.; Shevchenko, S.V.; Shcherbakova, L.N.; Popovichev, B.G.; Drenkhan, R. The extensive damages of elms by Dutch elm disease agents and their hybrids in north-western Russia. Urban For. Urban Green. 2021. submitted. [Google Scholar]
- Matisone, I.; Kenigsvalde, K.; Zaļuma, A.; Burņeviča, N.; Šņepste, I.; Matisons, R.; Gaitnieks, T. First report on the Dutch elm disease pathogen Ophiostoma novo-ulmi from Latvia. For. Pathol. 2020, e12601. [Google Scholar] [CrossRef]
- Motiejūnaitė, J.; Kutorga, E.; Kasparavičius, J.; Lygis, V.; Norkutė, G.; Jurga, M.; Ernestas, K.; Jonas, K.; Vaidotas, L.; Goda, N. New records from Lithuania of fungi alien to Europe. Mycotaxon 2016, 131, 49–60. [Google Scholar] [CrossRef]
- Jürisoo, L.; Padari, A.; Drenkhan, R. Jalakasurma levikust ja ohtlikkusest Eestis (Spread and riskiness of Dutch elm disease in Estonia). For. Stud. Metsanduslikud Uurim. 2021. submitted. (In Estonian) [Google Scholar]
- Jürisoo, L.; Adamson, K.; Padari, A.; Drenkhan, R. Health of elms and Dutch elm disease in Estonia. Eur. J. Plant Pathol. 2019, 154, 823–841. [Google Scholar] [CrossRef]
- Jankowiak, R.; Strzałka, B.; Bilański, P.; Kacprzyk, M.; Wieczorek, P.; Linnakoski, R. Ophiostomatoid fungi associated with hardwood-infesting bark and ambrosia beetles in Poland: Taxonomic diversity and vector specificity. Fungal Ecol. 2019, 39, 152–167. [Google Scholar] [CrossRef]
- Pajares, J.A.; García, S.; Díez, J.J.; Martín, D.; García-Vallejo, M.C. Feeding Responses by Scolytus scolytus to Twig Bark Extracts from Elms. Investig. Agraria Sist. Recur. For. 2004, 13, 217–225. [Google Scholar]
- Kirisits, T. Fungal Associates of European Bark Beetles with Special Emphasis on the Ophiostomatoid Fungi. In Bark and Wood Boring Insects in Living Trees in Europe, a Synthesis; Metzler, J.B., Ed.; Springer: Berlin, Germany, 2007; Volume 2007, pp. 181–236. [Google Scholar]
- Linnakoski, R. Bark beetle-associated fungi in Fennoscandia with special emphasis on species of Ophiostoma and Grosmannia. Diss. For. 2011, 2011. [Google Scholar] [CrossRef] [Green Version]
- Aas, T.; Solheim, H.; Jankowiak, R.; Bilański, P.; Hausner, G. Four new Ophiostoma species associated with hardwood-infesting bark beetles in Norway and Poland. Fungal Biol. 2018, 122, 1142–1158. [Google Scholar] [CrossRef]
- Mayers, C.G.; McNew, D.L.; Harrington, T.C.; Roeper, R.A.; Fraedrich, S.W.; Biedermann, P.H.; Castrillo, L.A.; Reed, S.E. Three genera in the Ceratocystidaceae are the respective symbionts of three independent lineages of ambrosia beetles with large, complex mycangia. Fungal Biol. 2015, 119, 1075–1092. [Google Scholar] [CrossRef] [Green Version]
- Heliövaara, K.; Peltonen, M. Bark Beetles in a Changing Environment. Ecol. Bull. 1999, 48–53. [Google Scholar] [CrossRef]
- Ohmart, C.P. Why are There so Few Tree-Killing Bark Beetles Associated with Angiosperms? Oikos 1989, 54, 242. [Google Scholar] [CrossRef]
- Webber, J.F. Relative effectiveness of Scolytus scolytus, S. multistriatus and S. kirschi as vectors of Dutch elm disease. For. Pathol. 1990, 20, 184–192. [Google Scholar] [CrossRef]
- Sherif, S.; Jones, A.; Shukla, M.; Saxena, P. Establishment of invasive and non-invasive reporter systems to investigate American elm–Ophiostoma novo-ulmi interactions. Fungal Genet. Biol. 2014, 71, 32–41. [Google Scholar] [CrossRef]
- Menkis, A.; Östbrant, I.-L.; Davydenko, K.; Bakys, R.; Balalaikins, M.; Vasaitis, R. Scolytus multistriatus associated with Dutch elm disease on the island of Gotland: Phenology and communities of vectored fungi. Mycol. Prog. 2016, 15, 1–8. [Google Scholar] [CrossRef]
- Anderbrant, O.; Schlyter, F. Ecology of the Dutch Elm Disease Vectors Scolytus laevis and S. scolytus (Coleoptera: Scolytidae) in Southern Sweden. J. Appl. Ecol. 1987, 24, 539. [Google Scholar] [CrossRef]
- Anderbrant, O.; Yuvaraj, J.K.; Martin, J.A.; Rodriguez-Gil, J.L.; Witzell, J. Feeding by Scolytus bark beetles to test for differently susceptible elm varieties. J. Appl. Entomol. 2017, 141, 417–420. [Google Scholar] [CrossRef]
- Tarigan, M.; Roux, J.; Van Wyk, M.; Tjahjono, B.; Wingfield, M. A new wilt and die-back disease of Acacia mangium associated with Ceratocystis manginecans and C. acaciivora sp. nov. in Indonesia. South. Afr. J. Bot. 2011, 77, 292–304. [Google Scholar] [CrossRef] [Green Version]
- Webber, J.F. Experimental studies on factors influencing the transmission of Dutch elm disease. Investig. Agrar. Sist. Recur. 2004, 13, 197. [Google Scholar]
- Lindelöw, Å. Introduced or Overlooked? New Bark Beetle Species in Sweden (Coleoptera; Curculionidae). Forstsch. Aktuell. 2012, 55, 28. Available online: https://bfw.ac.at/400/pdf/fsaktuell_55_10.pdf (accessed on 24 May 2019).
- Waller, M. Drought, disease, defoliation and death: Forest pathogens as agents of past vegetation change. J. Quat. Sci. 2013, 28, 336–342. [Google Scholar] [CrossRef]
- Hulcr, J.; Stelinski, L.L. The Ambrosia Symbiosis: From Evolutionary Ecology to Practical Management. Annu. Rev. Entomol. 2017, 62, 285–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkendall, L.R.; Biedermann, P.H.; Jordal, B.H. Evolution and Diversity of Bark and Ambrosia Beetles. Bark Beetles 2015, 2015, 85–156. [Google Scholar] [CrossRef]
- Bernier, L.; Aoun, M.; Bouvet, G.; Comeau, A.; Dufour, J.; Naruzawa, E.; Nigg, M.; Plourde, K. Genomics of the Dutch elm disease pathosystem: Are we there yet? iForest Biogeosci. For. 2015, 8, 149–157. [Google Scholar] [CrossRef] [Green Version]
- Moser, J.C.; Konrad, H.; Blomquist, S.R.; Kirisits, T. Do mites phoretic on elm bark beetles contribute to the transmission of Dutch elm disease? Naturwissenschaften 2009, 97, 219–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sherif, S.M.; Erland, L.A.; Shukla, M.R.; Saxena, P.K. Bark and wood tissues of American elm exhibit distinct responses to Dutch elm disease. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Koch, K. Die Kaefer Mitteleuropas, 3rd ed.; Goecke & Evers: Krefeld, Germany, 1992; 370p. [Google Scholar]
- Nikulina, T.; Mandelshtam, M.; Petrov, A.; Nazarenko, V.; Yunakov, N. A survey of the weevils of Ukraine. Bark and ambrosia beetles (Coleoptera: Curculionidae: Platypodinae and Scolytinae). Zootaxa 2015, 3912, 1–61. [Google Scholar] [CrossRef] [Green Version]
- Yanovskiy, V.M. Annotirovannyy spisok koroyedov (Scolytidae) Severnoy Azii (Annotated list of bark beetles (Scolytidae) of North Asia). Entomol. Rev. 1999, 78, 327–362. (In Russian) [Google Scholar]
- Beaver, R.A. Notes on the fauna associated with elm bark beetles in Wytham Wood, Berks—I Coleoptera. Entomol. Mon. Mag. 1966, 102, 163–170. [Google Scholar]
- Borowski, J.; Mazur, S. Beetles (Coleoptera) of the Rogów Region—Part IV—Clown Beetles (Histeridae) and False Clown Beetles (Sphaeritidae). Int. Lett. Nat. Sci. 2015, 37, 10–17. [Google Scholar] [CrossRef] [Green Version]
- Troukens, W. Paromalus parallelepipedus (Coleoptera: Histeridae) aan de westrand van Brussel (Paromalus parallelepipedus (Coleoptera: Histeridae) at the westside of Brussels). Phegea 2015, 43. Available online: www.faunaeur.org (accessed on 21 January 2021). (In Dutch).
- Vose, R.S.; Easterling, D.R.; Gleason, B. Maximum and minimum temperature trends for the globe: An update through Geophys. Res. Lett. 2005, 32, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Bentz, B.J.; Jönsson, A.M. Modeling Bark Beetle Responses to Climate Change. Bark Beetles 2015, 2015, 533–553. [Google Scholar] [CrossRef]
- Selikhovkin, A.V.; Drenkhan, R.; Mandelshtam, M.Y.; Musolin, D.L. Invasions of insect pests and fungal pathogens of woody plants into the northwestern part of European Russia. Vestnik St. Petersburg Univ. Earth Sci. 2020, 65, 263–283. [Google Scholar] [CrossRef]
- Sturrock, R.N.; Frankel, S.J.; Brown, A.V.; Hennon, P.E.; Kliejunas, J.T.; Lewis, K.J.; Worrall, J.J.; Woods, A.J. Climate change and forest diseases. Plant Pathol. 2011, 60, 133–149. [Google Scholar] [CrossRef]
- Kullman, L. Changes in alpine plant cover—effects of climate warming. Sven. Bot. Tidskr. 2003, 97, 210. [Google Scholar]
- Drobyshev, I.V. Effect of natural disturbances on the abundance of Norway spruce (Picea abies (L.) Karst.) regeneration in nemoral forests of the southern boreal zone. For. Ecol. Manag. 2001, 140, 151–161. [Google Scholar] [CrossRef]
- Roloff, A.; Korn, S.; Gillner, S. The Climate-Species-Matrix to select tree species for urban habitats considering climate change. Urban For. Urban Green. 2009, 8, 295–308. [Google Scholar] [CrossRef]
- Hanso, M.; Drenkhan, R. Metsa-ja linnapuud ilmastiku äärmuste vaevas (Trees in forests and towns are suffering from the extreme weather conditions). Eesti Lood Est. Nat. 2007, 58, 6–13. (In Estonian) [Google Scholar]
- Hanso, M.; Drenkhan, R. Simple visualization of climate change for improving the public perception in forest pathology/Kliimamuutuste visualiseerimise lihtne viis seoste paremaks tajumiseks metsapatoloogias. For. Stud. 2013, 58, 37–45. [Google Scholar] [CrossRef] [Green Version]
- Adamson, K.; Drenkhan, R.; Hanso, M. Invasive brown spot needle blight caused by Lecanosticta acicola in Estonia. Scand. J. For. Res. 2015, 30, 587–593. [Google Scholar] [CrossRef]
- Drenkhan, R.; Riit, T.; Adamson, K.; Hanso, M. The earliest samples of Hymenoscyphus albidus vs. H. fraxineus in Estonian mycological herbaria. Mycol. Prog. 2016, 15, 835–844. [Google Scholar] [CrossRef]
- Hanso, M.; Drenkhan, R. Diplodia pinea is a new pathogen on Austrian pine (Pinus nigra) in Estonia. Plant Pathol. 2009, 58, 797. [Google Scholar] [CrossRef]
- Hanso, M.; Drenkhan, R. Lophodermium needle cast, insect defoliation and growth responses of young Scots pines in Estonia. For. Pathol. 2011, 42, 124–135. [Google Scholar] [CrossRef]
- Lutter, R.; Drenkhan, R.; Tullus, A.; Jürimaa, K.; Tullus, T.; Tullus, H. First record of Entoleuca mammata in hybrid aspen plantations in hemiboreal Estonia and stand–environmental factors affecting its prevalence. Eur. J. For. Res. 2019, 138, 263–274. [Google Scholar] [CrossRef]
- Braschler, B.; Hill, J.K. Role of larval host plants in the climate-driven range expansion of the butterfly Polygonia c-album. J. Anim. Ecol. 2007, 76, 415–423. [Google Scholar] [CrossRef]
- Hemery, G.E.; Clark, J.R.; Aldinger, E.; Claessens, H.; Malvolti, M.E.; O’Connor, E.; Raftoyannis, Y.; Savill, P.S.; Brus, R. Growing scattered broadleaved tree species in Europe in a changing climate: A review of risks and opportunities. Forestry 2009, 83, 65–81. [Google Scholar] [CrossRef] [Green Version]
- Cudmore, T.J.; Björklund, N.; Carroll, A.L.; Lindgren, B.S. Climate change and range expansion of an aggressive bark beetle: Evidence of higher beetle reproduction in naïve host tree populations. J. Appl. Ecol. 2010, 47, 1036–1043. [Google Scholar] [CrossRef] [Green Version]
- Lekander, B.; Bejer-Petersen, B.; Kangas, E.; Bakke, A. The distribution of bark beetles in the Nordic countries. Acta. Entomol. Fenn. 1977, 32. Available online: https://www.cabdirect.org/cabdirect/abstract/19770548371 (accessed on 15 January 2021).
- Caulton, E.; Aitken, W.; Rashid, N. Aerobiological aspects of elm (Ulmus spp.) in South-East Scotland in relation to elm decline from Dutch Elm disease (1976–1996). Aerobiologia 1998, 14, 147–153. [Google Scholar] [CrossRef]
- Brand, J.M.; Young, J.C.; Silverstein, R.M. Insect Pheromones: A Critical Review of Recent Advances in Their Chemistry, Biology, and Application. In Fortschritte der Chemie Organischer Naturstoffe Progress in the Chemistry of Organic Natural Products; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 1979; Volume 37, pp. 1–190. [Google Scholar]
- Cuthbert, R.A.; Peacock, J.W. Attraction of Scolytus multistriatus to Pheromone-baited Traps at Different Heights. Environ. Entomol. 1975, 4, 889–890. [Google Scholar] [CrossRef]
- Neumann, F.G.; Minko, G. Studies on the introduced smaller European elm bark beetle, Scolytus multistriatus, a potential vector of Dutch elm disease in Victoria. Aust. For. 1985, 48, 116–126. [Google Scholar] [CrossRef]
- Schedl, K.E. Familie: Scolytidae (Borken- und Ambrosiakäfer). In Die Käfer Mitteleuropas; Freude, H., Harde, K.W., Lohse, G.A., Eds.; Goecke & Evers: Krefeld, Germany, 1981; pp. 34–99. [Google Scholar]
- Pfeffer, A. Familie: Scolytidae. In Die Käfer Mitteleuropas, Supplementband Mit Katalogteil; Lohse, G.A., Lucht, W.H., Eds.; Goecke & Evers: Krefeld, Germany, 1994; pp. 153–180. [Google Scholar]
- Pfeffer, A. Zentral- und Westpaläarktische Borken- und Kernkäfer (Coleoptera: Scolytidae, Platypodidae); Pro Entomologia, c/o Naturhistorisches Museum: Basel, Switzerland, 1995; 370p. [Google Scholar]
- Petrov, A.V.; Mandelshtam, M.Y.; Beaver, R.A. A key to species of the tribe Scolytini Latreille, 1804 (Coleoptera: Curculionidae: Scolytinae) from Russia and adjacent countries. REJ 2019, 28, 286–302. [Google Scholar] [CrossRef] [Green Version]
- Voolma, K.; Õunap, H.; Süda, I. Eesti Ürasklaste (Coleoptera, Scolytidae) Määraja (A Key of Estonian Bark Beetles (Coleoptera, Scolytidae)); Eesti Loodusfoto: Tartu, Estonia, 1997; 43p. (In Estonian) [Google Scholar]
- Drenkhan, T.; Voolma, K.; Adamson, K.; Sibul, I.; Drenkhan, R. The large pine weevil Hylobius abietis (L.) as a potential vector of the pathogenic fungus Diplodia sapinea (Fr.) Fuckel. Agric. For. Entomol. 2017, 19, 4–9. [Google Scholar] [CrossRef] [Green Version]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.L.C.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [Green Version]
- Tedersoo, L.; Anslan, S. Towards PacBio-based pan-eukaryote metabarcoding using full-length ITS sequences. Environ. Microbiol. Rep. 2019, 11, 659–668. [Google Scholar] [CrossRef]
- Loit, K.; Adamson, K.; Bahram, M.; Puusepp, R.; Anslan, S.; Kiiker, R.; Drenkhan, R.; Tedersoo, L. Relative performance of Oxford Nanopore MinION vs. Pacific Biosciences Sequel third-generation sequencing platforms in identification of agricultural and forest pathogens. Appl. Environ. Microbiol. 2019, 85, e01368-19. [Google Scholar] [CrossRef]
- Tedersoo, L.; Drenkhan, R.; Anslan, S.; Morales-Rodriguez, C.; Cleary, M. High-throughput identification and diagnostics of pathogens and pests: Overview and practical recommendations. Mol. Ecol. Resour. 2018, 19, 47–76. [Google Scholar] [CrossRef] [Green Version]
- Agan, A.; Drenkhan, R.; Adamson, K.; Tedersoo, L.; Solheim, H.; Børja, I.; Matsiakh, I.; Timmermann, V.; Nagy, N.E.; Hietala, A.M. The Relationship between Fungal Diversity and Invasibility of a Foliar Niche—The Case of Ash Dieback. J. Fungi 2020, 6, 150. [Google Scholar] [CrossRef]
- Tedersoo, L.; Tooming-Klunderud, A.; Anslan, S. PacBio metabarcoding of Fungi and other eukaryotes: Errors, biases and perspectives. New Phytol. 2018, 217, 1370–1385. [Google Scholar] [CrossRef] [Green Version]
- Anslan, S.; Bahram, M.; Hiiesalu, I.; Tedersoo, L. PipeCraft: Flexible open-source toolkit for bioinformatics analysis of custom high-throughput amplicon sequencing data. Mol. Ecol. Resour. 2017, 17, e234–e240. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bengtsson-Palme, J.; Ryberg, M.; Hartmann, M.; Branco, S.; Wang, Z.; Godhe, A.; De Wit, P.J.G.M.; Sánchez-García, M.; Ebersberger, I.; De Sousa, F.; et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013, 4, 914–919. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kühnholz, S.; Borden, J.H.; Uzunovic, A. Secondary Ambrosia Beetles in Apparently Healthy Trees: Adaptations, Potential Causes and Suggested Research. Integr. Pest Manag. Rev. 2001, 6, 209–219. [Google Scholar] [CrossRef]
- Speranza, S.; Bucini, D.; Paparatti, B. New observation on biology of european shot-hole borer [xyleborus dispar (f.)] on hazel in northern latium (central italy). Acta Hortic. 2009, 845, 539–542. [Google Scholar] [CrossRef]
- Deyrup, M.; Atkinson, T. Comparative Biology of Temperate and Subtropical Bark and Ambrosia Beetles (Coleoptera: Scoly-tidae, Platypodidae) in Indiana and Florida. Gt Lakes Entomol. 1987, 20, 59. [Google Scholar]
- Steininger, M.S.; Hulcr, J.; Igut, M.; Lucky, A.; Sigut, M. Simple and Efficient Trap for Bark and Ambrosia Beetles (Coleoptera: Curculionidae) to Facilitate Invasive Species Monitoring and Citizen Involvement. J. Econ. Entomol. 2015, 108, 1115–1123. [Google Scholar] [CrossRef] [Green Version]
- Rabaglia, R.J.; Dole, S.A.; Cognato, A.I. Review of American Xyleborina (Coleoptera: Curculionidae: Scolytinae) Occurring North of Mexico, with an Illustrated Key. Ann. Entomol. Soc. Am. 2006, 99, 1034–1056. [Google Scholar] [CrossRef]
- Chang, R.; Duong, T.A.; Taerum, S.J.; Wingfield, M.J.; Zhou, X.; De Beer, Z.W. Ophiostomatoid fungi associated with conifer-infesting beetles and their phoretic mites in Yunnan, China. MycoKeys 2017, 28, 19–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brasier, C.M.; Kirk, S.A. Designation of the EAN and NAN races of Ophiostoma novo-ulmi as subspecies. Mycol. Res. 2001, 105, 547–554. [Google Scholar] [CrossRef]
- Mandelshtam, M.Y.; Popovichev, B.G. Annotated list of bark beetle species (Coleoptera, Scoytidae) of the Leningrad Region. Entomol. Rev. 2000, 80, 887–903. [Google Scholar]
- Mandelshtam, M.Y.; Selikhovkin, A.V. Bark and Ambrosia Beetles (Coleoptera, Curculionidae: Scolytinae) of Northwest Russia: History of the Study, Composition and Genesis of the Fauna. Entomol. Rev. 2020, 100, 1–27. [Google Scholar] [CrossRef]
- Zolk, K. Kodumaa ürasklased (Ipidae) ühes lühikese ülevaatega nende bionoomiast ja levimisest Eestis (Native bark beetles (Ipidae) together with a brief overview of their bionomy and distribution in Estonia). Eesti Metsanduse Aastaraam. Est. Yearb. 1932, 6, 127–176. (In Estonian) [Google Scholar]
- Voolma, K.; Süda, I. Rare species of Cerambycidae and Scolytidae (Coleoptera) in Estonia. In Proceedings of the XXIV Nordic Congress of Entomology, Tartu, Estonia, 8–11 August 1997; pp. 187–192. [Google Scholar]
- Shcherbakova, L.N.; Mandelshtam, M.Y. Vyazy Sankt Peterburga: Posle tret’yego zvonka (Elms of Saint Petersburg: After the third ring). In Proceedings of the International Conference the Kataev Memorial Readings—VIII, St. Peterburg, Russia, 18–20 November 2014; Musolin, D.L., Selikhovkin, A.V., Eds.; Saint Petersburg State Forest Technical University: Saint Petersburg, Russia, 2014. (In Russian). [Google Scholar]
- Kristian, J. Mõnest uuest üraskiliigist Eestis (About some new beetle species in Estonia). Eesti Mets Estonian For. 1937, 17, 399. (In Estonian) [Google Scholar]
- Leius, K. Täiendavaid andmeid ürasklaste (Ipidae) esinemise kohta Eestis (Additional data on the occurrence of bark beetles (Ipidae) in Estonia). Eesti Metsanduse Aastaraam. Est. Yearb. 1939, 9, 318–328. (In Estonian) [Google Scholar]
- Voolma, K.; Õunap, H.; Süda, I. Eesti Putukate Levikuatlas, 2: Ürasklased—Scolytidae (Distribution Maps of Estonian Insects, 2: Scolytidae); Eesti Loodusfoto: Tartu, Estonia, 2000; 84p. (In Estonian) [Google Scholar]
- Voolma, K.; Mandelshtam, M.; Shcherbakov, A.; Yakovlev, E.; Õunap, H.; Süda, I.; Popovichev, B.; Sharapa, T.; Galasjeva, T.; Khairetdinov, R.; et al. Distribution and spread of bark beetles (Coleoptera: Scolytidae) around the Gulf of Finland: A comparative study with notes on rare species of Estonia, Finland and North-Western Russia. Entomol. Fenn. 2004, 15, 198–210. [Google Scholar] [CrossRef]
- Voolma, K.; Süda, I.; Õunap, H. New records of bark beetles (Coleoptera, Scolytidae) from Estonia. Proc. Est. Acad. Sci. Biol. Ecol. 1998, 47, 73–78. [Google Scholar]
- Süda, I. Jalaka-maltsaürask (Scolytus triarmatus (Eggers, 1912))—uus üraskiliik Baltikumis Scolytus triarmatus (Eggers, 1912)—new bark beetle in the Baltics. Metsanduslikud Uurim. 2006, 44, 112–117. (In Estonian) [Google Scholar]
- Mandelshtam, M.Y.; Khayretdinov, R.R. Additions to the list of species of bark beetles (Coleoptera, Curculionidae: Scolytinae) of the Leningrad Region (Dopolneniya k spisku vidov koroyedov (Coleoptera, Curculionidae: Scolytinae) Leningradskoy oblasti. Entomol. Rev. 2017, 86, 512–521. (In Russian) [Google Scholar]
- Petrov, A.V.; Nikitsky, N.B. Scolytid fauna (Coleoptera, Scolytidae) of Moscow Province. Entomol. Obozr. 2001, 80, 353. [Google Scholar]
- Vlasov, D.V.; Mandelshtam, M.Y. Elm bark beetles of the genus Scolytus Geoffroy, 1762 (Coleoptera: Scolytidae) as new serious pests in the parks of Yaroslavl and Saint Petersburg. In Proceedings of the Fitosanitarnoe Ozdorovlenie Ekosistem. Materialy Vtorogo Vserossiiskogo s”Ezda Po Zashchite Rastenii: V 2 Tomakh (Phytosanitary Improvement of Ecosystems: Proceedings of the 2nd All-Russian Plant Protection Congress in Two Volumes, St. Petersburg, FL, USA, 9–11 September 2005; Volume 1, p. 262. (In Russian). [Google Scholar]
- Ganley, R.J.; Bulman, L.S. Dutch elm disease in New Zealand: Impacts from eradication and management programmes. Plant Pathol. 2016, 65, 1047–1055. [Google Scholar] [CrossRef]
- Süda, I. Metsamardikate (Coleoptera) uued liigid Eestis (New woodland beetle species (Coleoptera) in Estonian fauna). For. Stud. 2009, 50, 98–114. (In Estonian) [Google Scholar] [CrossRef] [Green Version]
- Biedermann, P.H. Observations on sex ratio and behavior of males in Xyleborinus saxesenii Ratzeburg (Scolytinae, Coleoptera). ZooKeys 2010, 56, 253–267. [Google Scholar] [CrossRef]
- Salmane, I.; Ciematnieks, R.; Ozoliņa-Pole, L.; Ralle, B.; Ievinsh, G. Investigation of European shot-hole borer, Xyleborus dispar (Coleoptera, Scolytidae), in apple orchards of Latvia. In Proceedings of the International Scientific and Practical Conference, Rezekne, Latvia, 18–20 June 2015; Volume 2, pp. 256–260. [Google Scholar]
- Süda, I. Metsamardikate (Coleoptera) uued liigid Eestis. 2 (New woodland beetle species (Coleoptera) in Estonian fauna. 2). For. Stud. 2016, 64, 51–69. (In Estonian) [Google Scholar] [CrossRef] [Green Version]
- Süda, I. Mardikauurimisest viimastel aastakümnetel (1990–2009) (Coleoptera studies in recent decades (1990−2009)). In Year-Book of the Estonian Naturalists’ Society, 86th ed.; Kull, T., Liira, J., Sammul, M., Eds.; Eesti Looduseuurijate Seltsi Aastaraamat; Eesti Looduseuurijate Selts: Tartu, Estonia, 2011; pp. 225–229. (In Estonian) [Google Scholar]
- Helland, I.S.; Hoff, J.M.; Anderbrant, O. Attraction of bark beetles (Coleoptera: Scolytidae) to a pheromone trap. J. Chem. Ecol. 1984, 10, 723–752. [Google Scholar] [CrossRef]
- Bouget, C.; Brustel, H.; Brin, A.; Valladares, L. Evaluation of window flight traps for effectiveness at monitoring dead wood-associated beetles: The effect of ethanol lure under contrasting environmental conditions. Agric. For. Entomol. 2009, 11, 143–152. [Google Scholar] [CrossRef] [Green Version]
- Wegensteiner, R.; Wermelinger, B.; Herrmann, M. Natural Enemies of Bark Beetles: Predators, Parasitoids, Pathogens, and Nematodes. In Bark Beetles; Academic Press: Cambridge, MA, USA, 2015; pp. 247–304. [Google Scholar] [CrossRef]
| Species of Beetles | Country | Traps | Handpicked | Total | ||
|---|---|---|---|---|---|---|
| Sex | Sex | |||||
| Male | Female | Male | Female | |||
| Scolytus multistriatus | Estonia | 3 | 3 | - | - | 6 |
| Russia | - | - | 5 | - | 5 | |
| Scolytus triarmatus | Estonia | 4 | 2 | 66 | 114 | 186 |
| Scolytus laevis | Estonia | - | - | 21 | 18 | 39 |
| Scolytus scolytus | Russia | - | - | 2 | 2 | 4 |
| Scolytus pygmaeus | Russia | - | - | - | 1 | 1 |
| Xyleborinus saxesenii | Estonia | - | 10 | - | - | 10 |
| Xyleborus dispar | Estonia | - | 10 | - | - | 10 |
| Total | 32 | 229 | 261 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jürisoo, L.; Süda, I.; Agan, A.; Drenkhan, R. Vectors of Dutch Elm Disease in Northern Europe. Insects 2021, 12, 393. https://doi.org/10.3390/insects12050393
Jürisoo L, Süda I, Agan A, Drenkhan R. Vectors of Dutch Elm Disease in Northern Europe. Insects. 2021; 12(5):393. https://doi.org/10.3390/insects12050393
Chicago/Turabian StyleJürisoo, Liina, Ilmar Süda, Ahto Agan, and Rein Drenkhan. 2021. "Vectors of Dutch Elm Disease in Northern Europe" Insects 12, no. 5: 393. https://doi.org/10.3390/insects12050393






