Genotype and Trait Specific Responses to Rapamycin Intake in Drosophila melanogaster
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. D. melanogaster Husbandry
2.2. Life History and Stress Resistance Assays
2.2.1. Heat Stress Tolerance
2.2.2. Fecundity
2.2.3. Longevity
2.3. Statistical Analysis
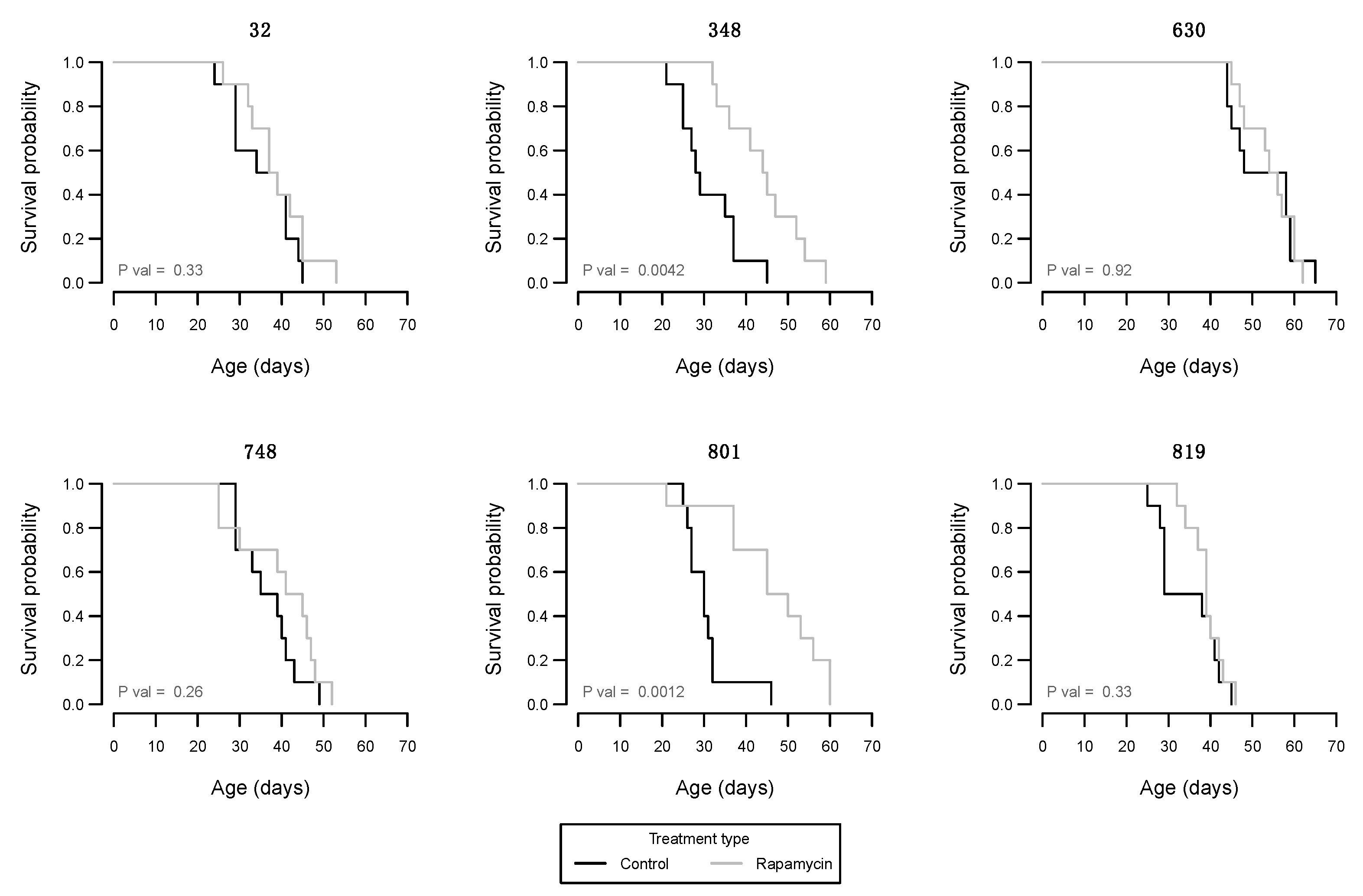
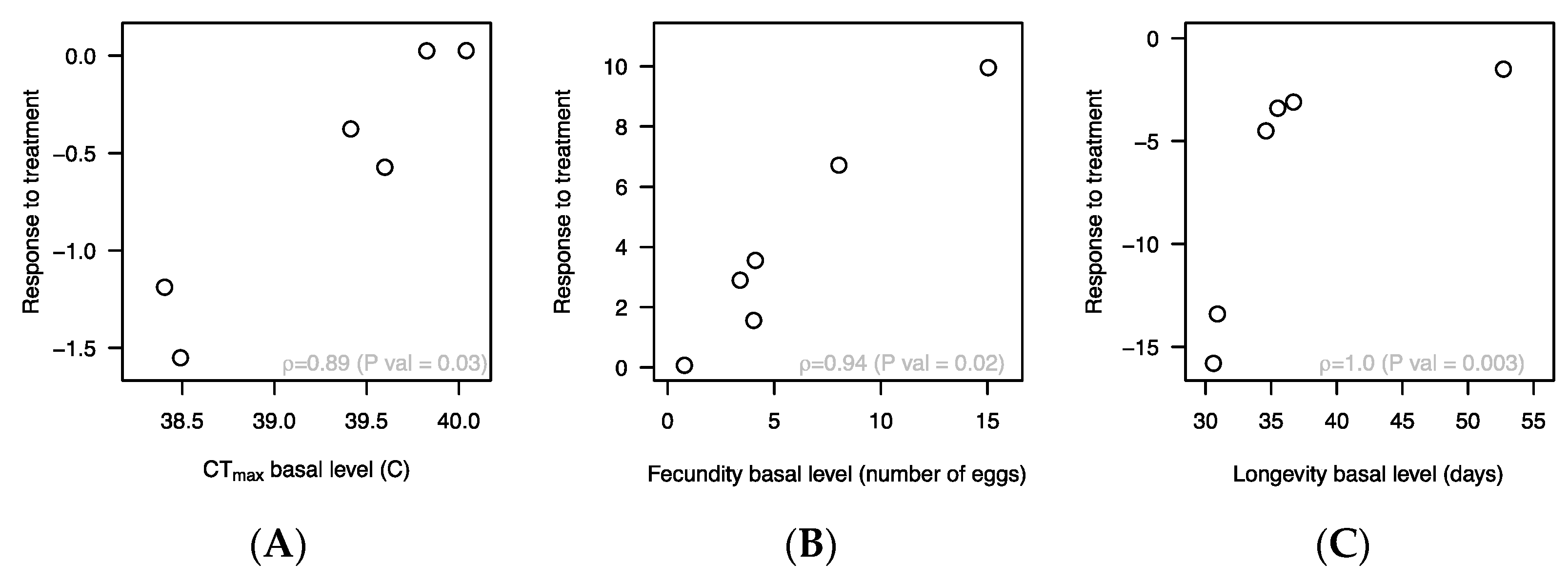
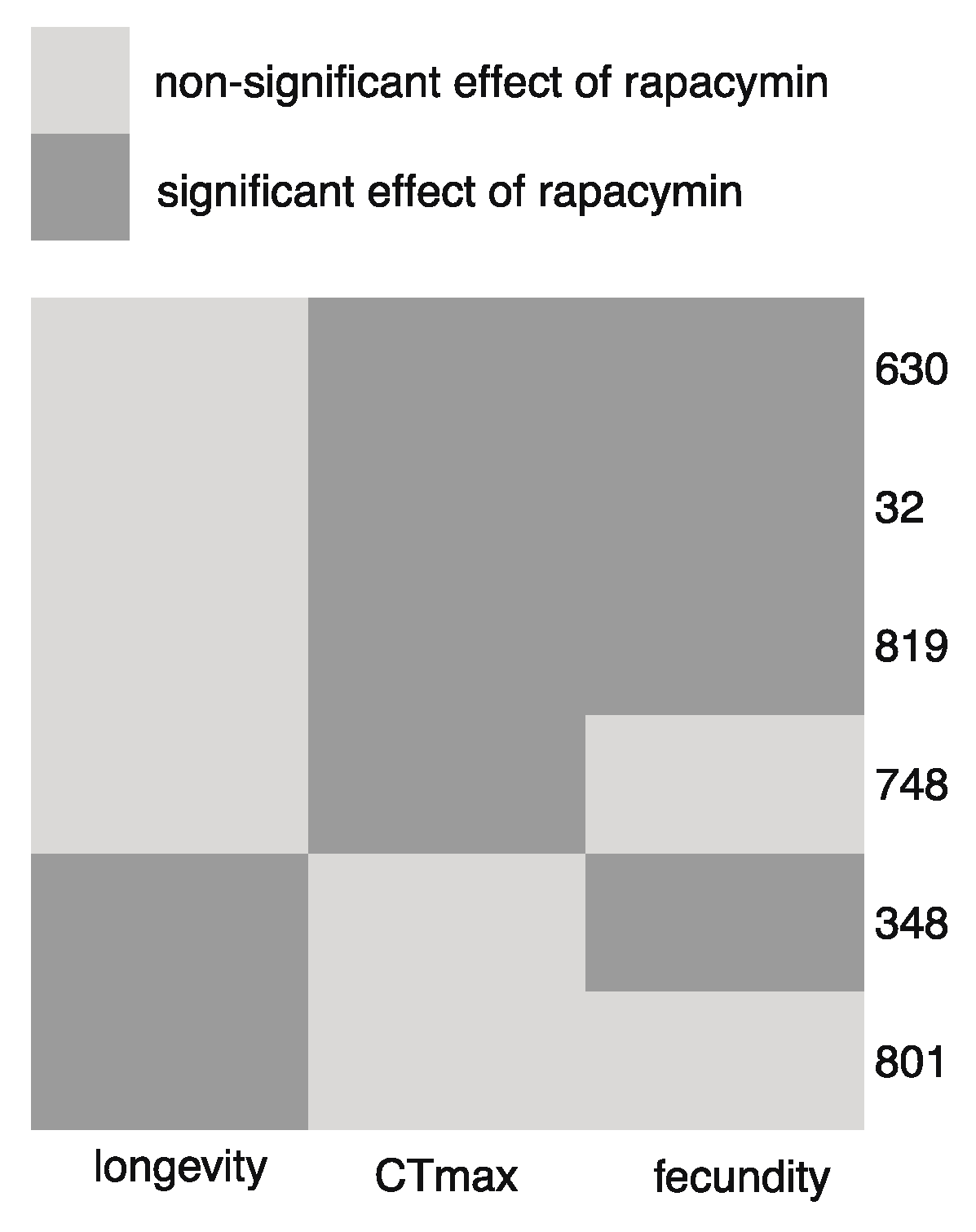
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rohani, N.; Eslahchi, C. Drug-drug interaction predicting by neural network using integrated similarity. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Malki, M.A.; Pearson, E.R. Drug–drug–gene interactions and adverse drug reactions. Pharm. J. 2020, 20, 355–366. [Google Scholar] [CrossRef]
- Veeren, J.C.; Weiss, M. Trends in emergency hospital admissions in England due to adverse drug reactions: 2008–2015. J. Pharm. Health Serv. Res. 2017, 8, 5–11. [Google Scholar] [CrossRef]
- Li, J.; Kim, S.G.; Blenis, J. Rapamycin: One drug, many effects. Cell Metab. 2014, 19, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.M.; Blenis, J. Molecular mechanisms of mTOR-mediated translational control. Nat. Rev. Mol. Cell Biol. 2009, 10, 307–318. [Google Scholar] [CrossRef]
- Saunders, R.N.; Metcalfe, M.S.; Nicholson, M.L. Rapamycin in transplantation: A review of the evidence. Kidney Int. 2001, 59, 3–16. [Google Scholar] [CrossRef]
- Serruys, P.W.; Regar, E.; Carter, A.J. Rapamycin eluting stent: The onset of a new era in interventional cardiology. Heart 2002, 87, 305–307. [Google Scholar] [CrossRef]
- Kennedy, B.K.; Pennypacker, J.K. Mammalian target of rapamycin: A target for (lung) diseases and aging. Ann. Am. Thorac. Soc. 2016, 13, S398–S401. [Google Scholar] [CrossRef]
- Kaeberlein, M. Resveratrol and rapamycin: Are they anti-aging drugs? BioEssays 2010, 32, 96–99. [Google Scholar] [CrossRef]
- Kapahi, P.; Zid, B.M.; Harper, T.; Koslover, D.; Sapin, V.; Benzer, S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 2004, 14, 1–6. [Google Scholar] [CrossRef]
- Bjedov, I.; Toivonen, J.M.; Kerr, F.; Slack, C.; Jacobson, J.; Foley, A.; Partridge, L. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 2010, 11, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Emran, S.; Yang, M.; He, X.; Zandveld, J.; Piper, M.D.W. Target of rapamycin signalling mediates the lifespan-extending effects of dietary restriction by essential amino acid alteration. Aging 2014, 6, 390–398. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.E.; Strong, R.; Sharp, Z.D.; Nelson, J.F.; Astle, C.M.; Flurkey, K.; Nadon, N.L.; Wilkinson, J.E.; Frenkel, K.; Carter, C.S.; et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 2009, 460, 392–395. [Google Scholar] [CrossRef] [PubMed]
- Fok, W.C.; Chen, Y.; Bokov, A.; Zhang, Y.; Salmon, A.B.; Diaz, V.; Javors, M.; Wood, W.H.; Zhang, Y.; Becker, K.G.; et al. Mice fed rapamycin have an increase in lifespan associated with major changes in the liver transcriptome. PLoS ONE 2014, 9. [Google Scholar] [CrossRef]
- Wilkinson, J.E.; Burmeister, L.; Brooks, S.V.; Chan, C.C.; Friedline, S.; Harrison, D.E.; Hejtmancik, J.F.; Nadon, N.; Strong, R.; Wood, L.K.; et al. Rapamycin slows aging in mice. Aging Cell 2012, 11, 675–682. [Google Scholar] [CrossRef]
- Ehninger, D.; Neff, F.; Xie, K. Longevity, aging and rapamycin. Cell. Mol. Life Sci. 2014, 71, 4325–4346. [Google Scholar] [CrossRef]
- Johnson, S.C.; Rabinovitch, P.S.; Kaeberlein, M. MTOR is a key modulator of ageing and age-related disease. Nature 2013, 493, 338–345. [Google Scholar] [CrossRef]
- Evans, W.E.; Mcleod, H.L. Pharmacogenomics—drug disposition, drug targets, and side effects. N. Engl. J. Med. 2003, 348, 538–549. [Google Scholar] [CrossRef]
- Rohde, P.D.; Kristensen, T.N. Untangling the genetic basis of drug response. Pharmacogenomics 2020, 21, 87–89. [Google Scholar] [CrossRef]
- Lee, S.H.; Min, K.J. Drosophila melanogaster as a model system in the study of pharmacological interventions in aging. Transl. Med. Aging 2019, 3, 98–103. [Google Scholar] [CrossRef]
- Su, T.T. Drug screening in Drosophila; why, when, and when not? WIREs Dev. Biol. 2019, 8, e346. [Google Scholar] [CrossRef] [PubMed]
- Jafari, M.; Long, A.D.; Mueller, L.D.; Rose, M.R. The pharmacology of ageing in Drosophila. Curr. Drug Targets 2006, 7, 1479–1483. [Google Scholar] [CrossRef] [PubMed]
- Pandey, U.B.; Nichols, C.D. Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol. Rev. 2011, 63, 411–436. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Massouras, A.; Inoue, Y.; Peiffer, J.; Ràmia, M.; Tarone, A.M.; Turlapati, L.; Zichner, T.; Zhu, D.; Lyman, R.F.; et al. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 2014, 24, 1193–1208. [Google Scholar] [CrossRef] [PubMed]
- Mackay, T.F.C.; Richards, S.; Stone, E.A.; Barbadilla, A.; Ayroles, J.F.; Zhu, D.; Casillas, S.; Han, Y.; Magwire, M.M.; Cridland, J.M.; et al. The Drosophila melanogaster Genetic Reference Panel. Nature 2012, 482, 173–178. [Google Scholar] [CrossRef]
- Mackay, T.F.C.; Huang, W. Charting the genotype–phenotype map: Lessons from the Drosophila melanogaster Genetic Reference Panel. Wiley Interdiscip. Rev. Dev. Biol. 2018, 7, 1–18. [Google Scholar] [CrossRef]
- Anholt, R.R.H.; Mackay, T.F.C. The road less traveled: From genotype to phenotype in flies and humans. Mamm. Genome 2018, 29, 5–23. [Google Scholar] [CrossRef]
- Rohde, P.D.; Jensen, I.R.; Sarup, P.M.; Ørsted, M.; Demontis, D.; Sørensen, P.; Kristensen, T.N. Genetic signatures of drug response variability in Drosophila melanogaster. Genetics 2019, 213, 633–650. [Google Scholar] [CrossRef]
- Kristensen, T.N.; Henningsen, A.K.; Aastrup, C.; Bech-Hansen, M.; Bjerre, L.B.H.; Carlsen, B.; Hagstrup, M.; Jensen, S.G.; Karlsen, P.; Kristensen, L.; et al. Fitness components of Drosophila melanogaster developed on a standard laboratory diet or a typical natural food source. Insect Sci. 2016, 23, 771–779. [Google Scholar] [CrossRef]
- Ja, W.W.; Carvalho, G.B.; Mak, E.M.; De La Rosa, N.N.; Fang, A.Y.; Liong, J.C.; Brummel, T.; Benzer, S. Prandiology of Drosophila and the CAFE assay. Proc. Natl. Acad. Sci. USA 2007, 104, 8253–8256. [Google Scholar] [CrossRef]
- Overgaard, J.; Kristensen, T.N.; Sørensen, J.G. Validity of thermal ramping assays used to assess thermal tolerance in arthropods. PLoS ONE 2012, 7, 1–7. [Google Scholar] [CrossRef] [PubMed]
- R Core Team R. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Bates, D.; Mächler, M.; Bolker, B.M.; Walker, S.C. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Therneau, T.M.; Grambsch, P.M. Modeling Survival Data: Extending the Cox Model; Springer: Berlin/Heidelberg, Germany, 2000; ISBN 0-387-98784-3. [Google Scholar]
- Fochler, S.; Morozova, T.V.; Davis, M.R.; Gearhart, A.W.; Huang, W.; Mackay, T.F.C.; Anholt, R.R.H. Genetics of alcohol consumption in Drosophila melanogaster. Genes Brain Behav. 2017, 16, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Battlay, P.; Schmidt, J.M.; Fournier-Level, A.; Robin, C. Genomic and transcriptomic associations identify a new insecticide resistance phenotype for the selective sweep at the Cyp6g1 locus of Drosophila melanogaster. G3 Genes Genomes Genet. 2016, 6, 2573–2581. [Google Scholar] [CrossRef]
- Montgomery, S.L.; Vorojeikina, D.; Huang, W.; Mackay, T.F.C.; Anholt, R.R.H.; Rand, M.D. Genome-wide association analysis of tolerance to methylmercury toxicity in Drosophila implicates myogenic and neuromuscular developmental pathways. PLoS ONE 2014, 9, e110375. [Google Scholar] [CrossRef]
- Guo, Z.; Yu, Q. Role of mTOR signaling in female reproduction. Front. Endocrinol. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Dou, X.; Sun, Y.; Li, J.; Zhang, J.; Hao, D.; Liu, W.; Wu, R.; Kong, F.; Peng, X.; Li, J. Short-term rapamycin treatment increases ovarian lifespan in young and middle-aged female mice. Aging Cell 2017, 16, 825–836. [Google Scholar] [CrossRef]
- Potter, S.; Sifers, J.; Yocom, E.; Blümich, S.L.E.; Potter, R.; Nadolski, J.; Harrison, D.A.; Cooper, R.L. Effects of inhibiting mTOR with rapamycin on behavior, development, neuromuscular physiology and cardiac function in larval Drosophila. Biol. Open 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Carey, J.R.; Harshman, L.G.; Liedo, P.; Müller, H.G.; Wang, J.L.; Zhang, Z. Longevity-fertility trade-offs in the tephritid fruit fly, Anastrepha ludens, across dietary-restriction gradients. Aging Cell 2008, 7, 470–477. [Google Scholar] [CrossRef]
- Chou, S.D.; Prince, T.; Gong, J.; Calderwood, S.K. mTOR is essential for the proteotoxic stress response, HSF1 activation and heat shock protein synthesis. PLoS ONE 2012, 7, 1–9. [Google Scholar] [CrossRef]
- Sørensen, J.G.; Kristensen, T.N.; Loeschcke, V. The evolutionary and ecological role of heat shock proteins. Ecol. Lett. 2003, 6, 1025–1037. [Google Scholar] [CrossRef]
- Sørensen, J.G.; Loeschcke, V.; Kristensen, T.N. Lessons from the use of genetically modified Drosophila melanogaster in ecological studies: Hsf mutant lines show highly trait-specific performance in field and laboratory thermal assays. Funct. Ecol. 2009, 23, 240–247. [Google Scholar] [CrossRef]
- Duncan, R.F. Rapamycin conditionally inhibits Hsp90 but not Hsp70 mRNA translation in Drosophila: Implications for the mechanisms of Hsp mRNA translation. Cell Stress Chaperones 2008, 13, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Welch, A.Z.; Gibney, P.A.; Botstein, D.; Koshland, D.E. TOR and RAS pathways regulate desiccation tolerance in Saccharomyces cerevisiae. Mol. Biol. Cell 2013, 24, 115–128. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, T.; Naito, H.; Kakigi, R.; Ichinoseki-Sekine, N.; Ogura, Y.; Sugiura, T.; Katamoto, S. Heat stress activates the Akt/mTOR signalling pathway in rat skeletal muscle. Acta Physiol. 2013, 207, 416–426. [Google Scholar] [CrossRef] [PubMed]
- Colinet, H.; Chertemps, T.; Boulogne, I.; Siaussat, D. Age-related decline of abiotic stress tolerance in young drosophila melanogaster adults. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2016, 71, 1574–1580. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vermeulen, C.J.; Loeschcke, V. Longevity and the stress response in Drosophila. Exp. Gerontol. 2007, 42, 153–159. [Google Scholar] [CrossRef]
- Kristensen, T.N.; Loeschcke, V.; Tan, Q.; Pertoldi, C.; Mengel-From, J. Sex and age specific reduction in stress resistance and mitochondrial DNA copy number in Drosophila melanogaster. Sci. Rep. 2019, 9, 1–7. [Google Scholar] [CrossRef]
- Grotewiel, M.S.; Martin, I.; Bhandari, P.; Cook-Wiens, E. Functional senescence in Drosophila melanogaster. Ageing Res. Rev. 2005, 4, 372–397. [Google Scholar] [CrossRef]
- Wang, A.; Mouser, J.; Pitt, J.; Promislow, D.; Kaeberlein, M. Rapamycin enhances survival in a Drosophila model of mitochondrial disease. Oncotarget 2016, 7, 80131–80139. [Google Scholar] [CrossRef]
- Harrison, B.; Tran, T.T.; Taylor, D.; Lee, S.D.; Min, K.J. Effect of rapamycin on lifespan in Drosophila. Geriatr. Gerontol. Int. 2010, 10, 110–112. [Google Scholar] [CrossRef] [PubMed]
- Cuesta, E.V.; Fan, F.; Rand, D.M. Rapamycin reduces Drosophila longevity under low nutrition. IOSR J. Pharm. 2014, 4, 43–51. [Google Scholar] [CrossRef]
- Dong, X.; Milholland, B.; Vijg, J. Evidence for a limit to human lifespan. Nature 2016, 538, 257–259. [Google Scholar] [CrossRef]
- Chandler, J.A.; Innocent, L.V.; Huang, I.L.; Yang, J.L.; Eisen, B.M.; Ludington, W.B. Chronic ethanol ingestion impairs Drosophila melanogaster health in a microbiome-dependent manner. BioRxiv 2018, 1–23. [Google Scholar] [CrossRef]
- Ivanov, D.K.; Escott-Price, V.; Ziehm, M.; Magwire, M.M.; Mackay, T.F.C.; Partridge, L.; Thornton, J.M. Longevity GWAS using the Drosophila Genetic Reference Panel. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2015, 70, 1470–1478. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Campbell, T.; Carbone, M.A.; Jones, W.E.; Unselt, D.; Anholt, R.R.H.; Mackay, T.F.C. Context-dependent genetic architecture of Drosophila life span. PLoS Biol. 2020, 18, 1–25. [Google Scholar] [CrossRef]
- McKenzie, J.A.; McKechnie, S.W. A comparative study of resource utilization in natural populations of Drosophila melanogaster and D. simulans. Oecologia 1979, 40, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, T.N.; Loeschcke, V.; Bilde, T.; Hoffmann, A.A.; Sgró, C.; Noreikiene, K.; Ondrésik, M.; Bechsgaard, J.S. No inbreeding depression for low temperature developmental acclimation across multiple Drosophila species. Evolution 2011, 65, 3195–3201. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 1–14. [Google Scholar] [CrossRef]
- Highfill, C.A.; Baker, B.M.; Stevens, S.D.; Anholt, R.R.H.; Mackay, T.F.C. Genetics of cocaine and methamphetamine consumption and preference in Drosophila melanogaster. PLoS Genet. 2019, 15, 1–24. [Google Scholar] [CrossRef]
- Baker, B.M.; Carbone, M.A.; Huang, W.; Anholt, R.R.H.; Mackay, T.F.C. Genetic basis of variation in cocaine and methamphetamine consumption in outbred populations of Drosophila melanogaster. BioRxiv Prepr. 2021. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rohde, P.D.; Bøcker, A.; Jensen, C.A.B.; Bergstrøm, A.L.; Madsen, M.I.J.; Christensen, S.L.; Villadsen, S.B.; Kristensen, T.N. Genotype and Trait Specific Responses to Rapamycin Intake in Drosophila melanogaster. Insects 2021, 12, 474. https://doi.org/10.3390/insects12050474
Rohde PD, Bøcker A, Jensen CAB, Bergstrøm AL, Madsen MIJ, Christensen SL, Villadsen SB, Kristensen TN. Genotype and Trait Specific Responses to Rapamycin Intake in Drosophila melanogaster. Insects. 2021; 12(5):474. https://doi.org/10.3390/insects12050474
Chicago/Turabian StyleRohde, Palle Duun, Asbjørn Bøcker, Caroline Amalie Bastholm Jensen, Anne Louise Bergstrøm, Morten Ib Juul Madsen, Sandra Læsø Christensen, Steffan Balling Villadsen, and Torsten Nygaard Kristensen. 2021. "Genotype and Trait Specific Responses to Rapamycin Intake in Drosophila melanogaster" Insects 12, no. 5: 474. https://doi.org/10.3390/insects12050474
APA StyleRohde, P. D., Bøcker, A., Jensen, C. A. B., Bergstrøm, A. L., Madsen, M. I. J., Christensen, S. L., Villadsen, S. B., & Kristensen, T. N. (2021). Genotype and Trait Specific Responses to Rapamycin Intake in Drosophila melanogaster. Insects, 12(5), 474. https://doi.org/10.3390/insects12050474





