Contribution of Epigenetic Mechanisms in the Regulation of Environmentally-Induced Polyphenism in Insects
Abstract
:Simple Summary
Abstract
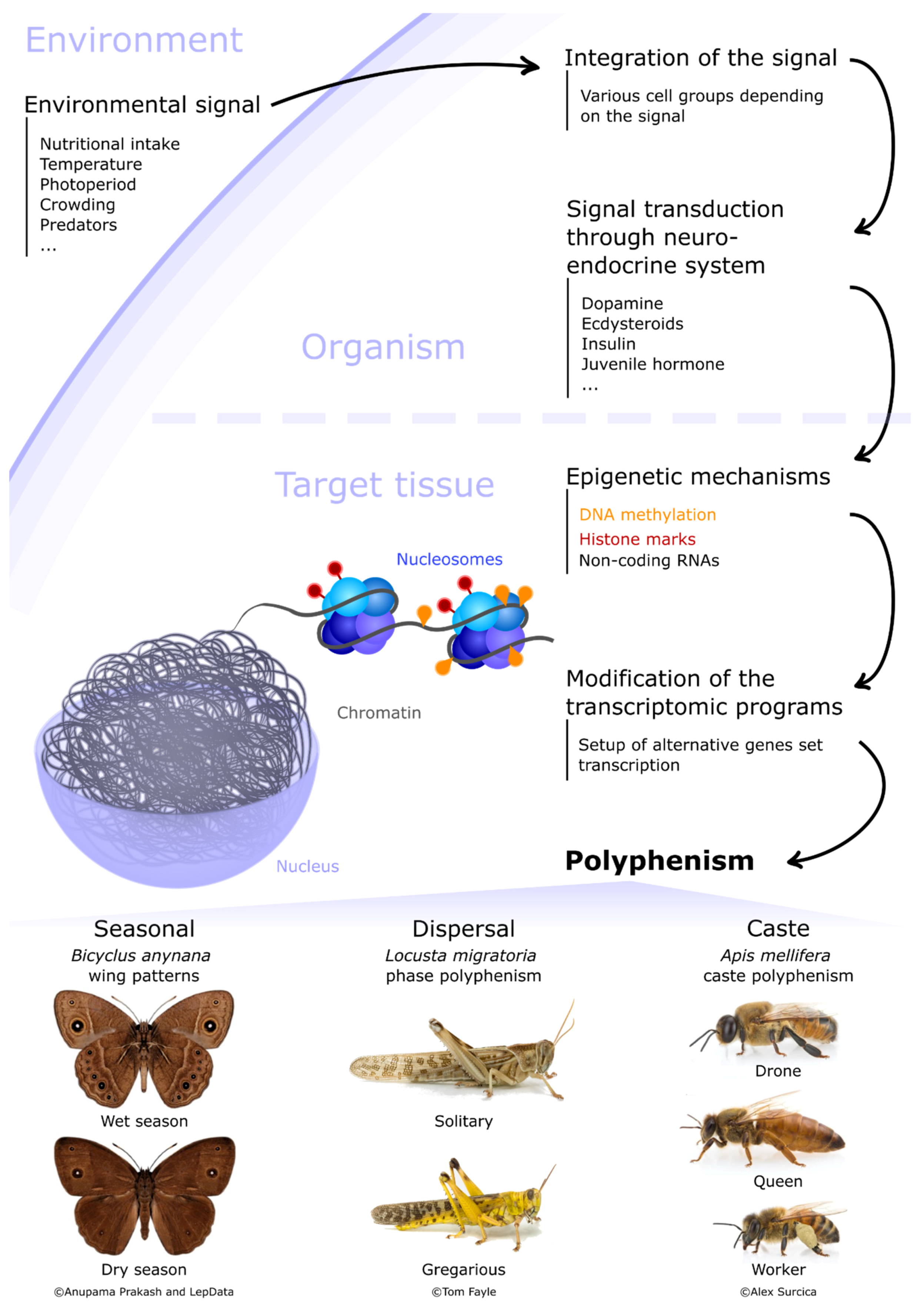
1. Introduction
2. Seasonal Polyphenism
2.1. Wing Patterning Polyphenism in Butterflies
2.2. Reproductive Polyphenism in Aphids
3. Dispersal Polyphenism
3.1. Phase Polyphenism in Locusts
3.2. Wing Polyphenism in Aphids
4. Caste Polyphenism
4.1. Nutrient Sensing and Neuro-Endocrine Signalling as Key Triggers of Caste Differentiation
4.2. DNA Methylation Patterns Changes during Caste Polyphenism
4.3. Contribution of Transcription Factors and Chromatin Remodelling Events in Caste Polyphenism
4.4. Non-Coding RNAs and Caste Polyphenism
4.5. Conclusions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Fordyce, J.A. The evolutionary consequences of ecological interactions mediated through phenotypic plasticity. J. Exp. Biol. 2006, 209, 2377–2383. [Google Scholar] [CrossRef] [Green Version]
- Via, S.; Gomulkiewicz, R.; De Jong, G.; Scheiner, S.; Schlichting, C.D.; Van Tienderen, P.H. Adaptive phenotypic plasticity: Consensus and controversy. Trends Ecol. Evol. 1995, 10, 212–217. [Google Scholar] [CrossRef]
- Mayr, E. Animal Species and Evolution; Harvard University Press: Cambridge, MA, USA; Oxford University Press: London, UK, 1963; 797p. [Google Scholar]
- Simpson, S.J.; Sword, G.; Lo, N. Polyphenism in Insects. Curr. Biol. 2011, 21, R738–R749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roessingh, P.; Bouaïchi, A.; Simpson, S.J. Effects of sensory stimuli on the behavioural phase state of the desert locust, Schistocerca gregaria. J. Insect Physiol. 1998, 44, 883–893. [Google Scholar] [CrossRef] [Green Version]
- Braendle, C.; Davis, G.K.; Brisson, J.; Stern, D. Wing dimorphism in aphids. Heredity 2006, 97, 192–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richard, G.; Le Trionnaire, G.; Danchin, E.; Sentis, A. Epigenetics and insect polyphenism: Mechanisms and climate change impacts. Curr. Opin. Insect Sci. 2019, 35, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Glastad, K.; Hunt, B.G.; Goodisman, M.A. Epigenetics in Insects: Genome Regulation and the Generation of Phenotypic Diversity. Annu. Rev. Entomol. 2019, 64, 185–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bannister, A.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Klose, R.J.; Bird, A.P. Genomic DNA methylation: The mark and its mediators. Trends Biochem. Sci. 2006, 31, 89–97. [Google Scholar] [CrossRef]
- Shi, D.-Q.; Ali, I.; Tang, J.; Yang, W.-C. New Insights into 5hmC DNA modification: Generation, distribution and function. Front. Genet. 2017, 8, 100. [Google Scholar] [CrossRef]
- Wion, D.; Casadesús, J. N6-methyl-adenine: An epigenetic signal for DNA-protein interactions. Nat. Rev. Microbiol. 2006, 4, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Kriaucionis, S.; Heintz, N. The Nuclear DNA Base 5-Hydroxymethylcytosine Is Present in Purkinje Neurons and the Brain. Science 2009, 324, 929–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Y.-F.; Li, B.-Z.; Li, Z.; Liu, P.; Wang, Y.; Tang, Q.; Ding, J.; Jia, Y.; Chen, Z.; Li, L.; et al. Tet-Mediated Formation of 5-Carboxylcytosine and Its Excision by TDG in Mammalian DNA. Science 2011, 333, 1303–1307. [Google Scholar] [CrossRef] [Green Version]
- Ito, S.; Shen, L.; Dai, Q.; Wu, S.C.; Collins, L.B.; Swenberg, J.A.; He, C.; Zhang, Y. Tet Proteins Can Convert 5-Methylcytosine to 5-Formylcytosine and 5-Carboxylcytosine. Science 2011, 333, 1300–1303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfaffeneder, T.; Hackner, B.; Truß, M.; Münzel, M.; Müller, M.; Deiml, C.A.; Hagemeier, C.; Carell, T. The Discovery of 5-Formylcytosine in Embryonic Stem Cell DNA. Angew. Chem. 2011, 123, 7146–7150. [Google Scholar] [CrossRef]
- Fu, Y.; He, C. Nucleic acid modifications with epigenetic significance. Curr. Opin. Chem. Biol. 2012, 16, 516–524. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Zhu, Y.; Luo, G.-Z.; Wang, X.; Yue, Y.; Wang, X.; Zong, X.; Chen, K.; Yin, H.; Fu, Y.; et al. Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig. Nat. Commun. 2016, 7, 13052. [Google Scholar] [CrossRef]
- Kim, V.N.; Han, J.; Siomi, M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009, 10, 126–139. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Ollikainen, N.; Guttman, M. Long non-coding RNAs: Spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 2016, 17, 756–770. [Google Scholar] [CrossRef] [Green Version]
- Bonev, B.; Cavalli, G. Organization and function of the 3D genome. Nat. Rev. Genet. 2016, 17, 661–678. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; Van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uriel, Y.; Abram, P.K.; Gries, G. Parasitoid pressure does not elicit defensive polyphenism in the green peach aphid. Ecol. Entomol. 2021, 46, 668–676. [Google Scholar] [CrossRef]
- Brakefield, P.M.; Frankino, W.A. Polyphenisms in Lepidoptera: Multidisciplinary approaches to studies of evolution and development. In Phenotypic Plast Insects Mech Consequences; Science Publishers: New York, NY, USA, 2009; pp. 337–368. [Google Scholar]
- Oostra, V.; de Jong, M.; Invergo, B.M.; Kesbeke, F.; Wende, F.; Brakefield, P.M.; Zwaan, B.J. Translating environmental gradients into discontinuous reaction norms via hormone signalling in a polyphenic butterfly. Proc. R. Soc. B Biol. Sci. 2010, 278, 789–797. [Google Scholar] [CrossRef] [Green Version]
- Brakefield, P.M.; Gates, J.; Keys, D.; Kesbeke, F.; Wijngaarden, P.J.; Monteiro, A.; French, V.; Carroll, S.B. Development, plasticity and evolution of butterfly eyespot patterns. Nature 1996, 384, 236–242. [Google Scholar] [CrossRef]
- Brakefield, P.M.; Koch, K. The Regulation of Phenotypic Plasticity of Eyespots in the Butterfly Bicyclus anynana. Am. Nat. 1998, 152, 853. [Google Scholar] [CrossRef]
- Tagu, D.; Sabater-Muñoz, B.; Simon, J.-C. Deciphering reproductive polyphenism in aphids. Invertebr. Reprod. Dev. 2005, 48, 71–80. [Google Scholar] [CrossRef]
- Le Trionnaire, G.; Wucher, V.; Tagu, D. Genome expression control during the photoperiodic response of aphids. Physiol. Entomol. 2013, 38, 117–125. [Google Scholar] [CrossRef]
- Steel, C.G.; Lees, A.D. The role of neurosecretion in the photoperiodic control of polymorphism in the aphid Megoura viciae. J. Exp. Biol. 1977, 67, 117–135. [Google Scholar] [CrossRef]
- Hardie, J.; Lees, A.D. The induction of normal and teratoid viviparae by a juvenile hormone and kinoprene in two species of aphids. Physiol. Entomol. 1985, 10, 65–74. [Google Scholar] [CrossRef]
- Gao, N.; von Schantz, M.; Foster, R.G.; Hardie, J. The putative brain photoperiodic photoreceptors in the vetch aphid, Megoura viciae. J. Insect Physiol. 1999, 45, 1011–1019. [Google Scholar] [CrossRef]
- Collantes-Alegre, J.M.; Mattenberger, F.; Barberà, M.; Martínez-Torres, D. Characterisation, analysis of expression and localisation of the opsin gene repertoire from the perspective of photoperiodism in the aphid Acyrthosiphon pisum. J. Insect Physiol. 2018, 104, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Cortes, T.; Ortiz-Rivas, B.; Martínez-Torres, D.; Cortes, T.; Ortiz-Rivas, B.; Martínez-Torres, D. Identification and characterization of circadian clock genes in the pea aphid Acyrthosiphon pisum. Insect Mol. Biol. 2010, 19, 123–139. [Google Scholar] [CrossRef]
- Barberà, M.; Collantes-Alegre, J.M.; Martínez-Torres, D. Characterisation, analysis of expression and localisation of circadian clock genes from the perspective of photoperiodism in the aphid Acyrthosiphon pisum. Insect Biochem. Mol. Biol. 2017, 83, 54–67. [Google Scholar] [CrossRef] [Green Version]
- Barberà, M.; Cañas-Cañas, R.; Martínez-Torres, D. Insulin-like peptides involved in photoperiodism in the aphid Acyrthosiphon pisum. Insect Biochem. Mol. Biol. 2019, 112, 103185. [Google Scholar] [CrossRef] [PubMed]
- Le Trionnaire, G.; Francis, F.; Jaubert-Possamai, S.; Bonhomme, J.; De Pauw, E.; Gauthier, J.-P.; Haubruge, E.; Legeai, F.; Prunier-Leterme, N.; Simon, J.-C.; et al. Transcriptomic and proteomic analyses of seasonal photoperiodism in the pea aphid. BMC Genom. 2009, 10, 456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gallot, A.; Rispe, C.; Leterme, N.; Gauthier, J.-P.; Jaubert-Possamai, S.; Tagu, D. Cuticular proteins and seasonal photoperiodism in aphids. Insect Biochem. Mol. Biol. 2010, 40, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Trionnaire, G.L.; Hudaverdian, S.; Richard, G.; Tanguy, S.; Gleonnec, F.; Prunier-Leterme, N.; Gauthier, J.-P.; Tagu, D. Dopamine pathway characterization during the reproductive mode switch in the pea aphid. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Walsh, T.K.; Brisson, J.; Robertson, H.M.; Gordon, K.; Jaubert-Possamai, S.; Tagu, D.; Edwards, O.R. A functional DNA methylation system in the pea aphid, Acyrthosiphon pisum. Insect Mol. Biol. 2010, 19, 215–228. [Google Scholar] [CrossRef]
- Rider, S.D., Jr.; Srinivasan, D.G.; Hilgarth, R.S. Chromatin-remodelling proteins of the pea aphid, Acyrthosiphon pisum (Harris). Insect Mol. Biol. 2010, 19, 201–214. [Google Scholar] [CrossRef]
- Gallot, A.; Shigenobu, S.; Hashiyama, T.; Jaubert-Possamai, S.; Tagu, D. Sexual and asexual oogenesis require the expression of unique and shared sets of genes in the insect Acyrthosiphon pisum. BMC Genom. 2012, 13, 76. [Google Scholar] [CrossRef] [Green Version]
- Richard, G.; Legeai, F.; Prunier-Leterme, N.; Bretaudeau, A.; Tagu, D.; Jaquiéry, J.; Le Trionnaire, G. Dosage compensation and sex-specific epigenetic landscape of the X chromosome in the pea aphid. Epigenet. Chromatin 2017, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Mathers, T.C.; Mugford, S.T.; Percival-Alwyn, L.; Chen, Y.; Kaithakottil, G.; Swarbreck, D.; Hogenhout, S.A.; Van Oosterhout, C. Sex-specific changes in the aphid DNA methylation landscape. Mol. Ecol. 2019, 28, 4228–4241. [Google Scholar] [CrossRef] [Green Version]
- Harrison, R.G. Dispersal Polymorphisms in Insects. Annu. Rev. Ecol. Syst. 1980, 11, 95–118. [Google Scholar] [CrossRef]
- Ernst, U.; Van Hiel, B.; Depuydt, G.; Boerjan, B.; De Loof, A.; Schoofs, L. Epigenetics and locust life phase transitions. J. Exp. Biol. 2015, 218, 88–99. [Google Scholar] [CrossRef] [Green Version]
- Burrows, M.; Rogers, S.M.; Ott, S.R. Epigenetic remodelling of brain, body and behaviour during phase change in locusts. Neural Syst. Circuits 2011, 1, 11. [Google Scholar] [CrossRef] [Green Version]
- Rogers, S.M.; Matheson, T.; Sasaki, K.; Kendrick, K.; Simpson, S.J.; Burrows, M. Substantial changes in central nervous system neurotransmitters and neuromodulators accompany phase change in the locust. J. Exp. Biol. 2004, 207 Pt 20, 3603–3617. [Google Scholar] [CrossRef] [Green Version]
- Ott, S.R.; Verlinden, H.; Rogers, S.M.; Brighton, C.H.; Quah, P.S.; Vleugels, R.K.; Verdonck, R.; Broeck, J.V. Critical role for protein kinase A in the acquisition of gregarious behavior in the desert locust. Proc. Natl. Acad. Sci. USA 2012, 109, E381–E387. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Guo, W.; Guo, X.; Wang, X.; Kang, L. Modulation of behavioral phase changes of the migratory locust by the catecholamine metabolic pathway. Proc. Natl. Acad. Sci. USA 2011, 108, 3882–3887. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, S. Corazonin and locust phase polyphenism. Appl. Entomol. Zool. 2006, 41, 179–193. [Google Scholar] [CrossRef] [Green Version]
- Kang, L.; Chen, X.; Zhou, Y.; Liu, B.; Zheng, W.; Li, R.; Wang, J.; Yu, J. The analysis of large-scale gene expression correlated to the phase changes of the migratory locust. Proc. Natl. Acad. Sci. USA 2004, 101, 17611–17615. [Google Scholar] [CrossRef] [Green Version]
- Badisco, L.; Ott, S.R.; Rogers, S.M.; Matheson, T.; Knapen, D.; Vergauwen, L.; Verlinden, H.; Marchal, E.; Sheehy, M.R.J.; Burrows, M.; et al. Microarray-based transcriptomic analysis of differences between long-term gregarious and solitarious desert locusts. PLoS ONE 2011, 6, e28110. [Google Scholar]
- Chen, S.; Yang, P.; Jiang, F.; Wei, Y.; Ma, Z.; Kang, L. De Novo Analysis of Transcriptome Dynamics in the Migratory Locust during the Development of Phase Traits. PLoS ONE 2010, 5, e15633. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wang, X.; Ma, Z.; Xue, L.; Han, J.; Yu, D.; Kang, L. CSP and Takeout Genes Modulate the Switch between Attraction and Repulsion during Behavioral Phase Change in the Migratory Locust. PLoS Genet. 2011, 7, e1001291. [Google Scholar] [CrossRef] [PubMed]
- Takayama, S.; Dhahbi, J.; Roberts, A.; Mao, G.; Heo, S.-J.; Pachter, L.; Martin, D.I.K.; Boffelli, D. Genome methylation in Drosophila melanogaster is found at specific short motifs and is independent of DNMT2 activity. Genome Res. 2014, 24, 821–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falckenhayn, C.; Boerjan, B.; Raddatz, G.; Frohme, M.; Schoofs, L.; Lyko, F. Characterization of genome methylation patterns in the desert locust Schistocerca gregaria. J. Exp. Biol. 2013, 216, 1501–1515. [Google Scholar] [CrossRef] [Green Version]
- Robinson, K.L.; Tohidi-Esfahani, D.; Lo, N.; Simpson, S.J.; Sword, G.A. Evidence for Widespread Genomic Methylation in the Migratory Locust, Locusta migratoria (Orthoptera: Acrididae). PLoS ONE 2011, 6, e28167. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Fang, X.; Yang, P.; Jiang, X.; Jiang, F.; Zhao, D.; Li, B.; Cui, F.; Wei, J.; Ma, C.; et al. The locust genome provides insight into swarm formation and long-distance flight. Nat. Commun. 2014, 5, 2957. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Chen, S.; Yang, P.; Ma, Z.; Kang, L. Characterization and comparative profiling of the small RNA transcriptomes in two phases of locust. Genome Biol. 2009, 10, R6. [Google Scholar] [CrossRef] [Green Version]
- Müller, C.B.; Williams, I.S.; Hardie, J. The role of nutrition, crowding and interspecific interactions in the development of winged aphids. Ecol. Entomol. 2001, 26, 330–340. [Google Scholar] [CrossRef]
- Ishikawa, A.; Miura, T. Transduction of high-density signals across generations in aphid wing polyphenism. Physiol. Èntomol. 2013, 38, 150–156. [Google Scholar] [CrossRef]
- Vellichirammal, N.N.; Madayiputhiya, N.; Brisson, J.A. The genomewide transcriptional response underlying the pea aphid wing polyphenism. Mol. Ecol. 2016, 25, 4146–4160. [Google Scholar] [CrossRef] [Green Version]
- Vellichirammal, N.N.; Gupta, P.; Hall, T.A.; Brisson, J.A. Ecdysone signaling underlies the pea aphid transgenerational wing polyphenism. Proc. Natl. Acad. Sci. USA 2017, 114, 1419–1423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grantham, M.E.; Brisson, J. Extensive Differential Splicing Underlies Phenotypically Plastic Aphid Morphs. Mol. Biol. Evol. 2018, 35, 1934–1946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavanchy, G.; Schwander, T. Hybridogenesis. Curr. Biol. 2019, 29, 539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, H.; Simola, D.F.; Bonasio, R.; Liebig, J.; Berger, S.; Reinberg, D. Eusocial insects as emerging models for behavioural epigenetics. Nat. Rev. Genet. 2014, 15, 677–688. [Google Scholar] [CrossRef] [PubMed]
- Liang, Z.; Nguyen, T.; Mattila, H.R.; Rodriguez-Zas, S.L.; Seeley, T.D.; Robinson, G.E. Molecular Determinants of Scouting Behavior in Honey Bees. Science 2012, 335, 1225–1228. [Google Scholar] [CrossRef]
- Brian, M.V. Studies of caste differentiation in Myrmica rubra L. 4. Controlled larval nutrition. Insectes Sociaux 1956, 3, 369–394. [Google Scholar] [CrossRef]
- Brian, M.V. Caste differentiation and division of labour. In Social Insects; Elsevier Inc.: Amsterdam, The Netherlands, 1979; Volume 1. [Google Scholar]
- Chittka, A.; Wurm, Y.; Chittka, L. Epigenetics: The making of ant castes. Curr. Biol. 2012, 22, R835–R838. [Google Scholar] [CrossRef] [Green Version]
- Beshers, S.N.; Fewell, J.H. Models of division of labor in social insects. Annu. Rev. Entomol. 2001, 46, 413–440. [Google Scholar] [CrossRef] [Green Version]
- Gospocic, J.; Shields, E.J.; Glastad, K.; Lin, Y.; Penick, C.; Yan, H.; Mikheyev, A.; Linksvayer, T.; Garcia, B.A.; Berger, S.; et al. The Neuropeptide Corazonin Controls Social Behavior and Caste Identity in Ants. Cell 2017, 170, 748–759. [Google Scholar] [CrossRef]
- Kamakura, M. Royalactin induces queen differentiation in honeybees. Nature 2011, 473, 478–483. [Google Scholar] [CrossRef]
- Mutti, N.S.; Dolezal, A.G.; Wolschin, F.; Mutti, J.S.; Gill, K.S.; Amdam, G.V. IRS and TOR nutrient-signaling pathways act via juvenile hormone to influence honey bee caste fate. J. Exp. Biol. 2011, 214, 3977–3984. [Google Scholar] [CrossRef] [Green Version]
- Kucharski, R.; Maleszka, R.; Foret, S. Nutritional Control of Reproductive Status in Honeybees via DNA Methylation. Science 2008, 319, 1827–1830. [Google Scholar] [CrossRef] [Green Version]
- Glastad, K.; Hunt, B.G.; Yi, S.V.; Goodisman, M.A.D. DNA methylation in insects: On the brink of the epigenomic era. Insect Mol. Biol. 2011, 20, 553–565. [Google Scholar] [CrossRef]
- Elango, N.; Hunt, B.; Goodisman, M.A.D.; Yi, S.V. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc. Natl. Acad. Sci. USA 2009, 106, 11206–11211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyko, F.; Foret, S.; Kucharski, R.; Wolf, S.; Falckenhayn, C.; Maleszka, R. The honey bee epigenomes: Differential methylation of brain DNA in queens and workers. PLoS Biol. 2010, 8, e1000506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foret, S.; Kucharski, R.; Pellegrini, M.; Feng, S.; Jacobsen, S.E.; Robinson, G.E.; Maleszka, R. DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. Proc. Natl. Acad. Sci. USA 2012, 109, 4968–4973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li-Byarlay, H.; Li, Y.; Stroud, H.; Feng, S.; Newman, T.C.; Kaneda, M.; Hou, K.K.; Worley, K.C.; Elsik, C.; Wickline, S.A.; et al. RNA interference knockdown of DNA methyl-transferase 3 affects gene alternative splicing in the honey bee. Proc. Natl. Acad. Sci. USA 2013, 110, 12750–12755. [Google Scholar] [CrossRef] [Green Version]
- Bonasio, R.; Li, Q.; Lian, J.; Mutti, N.S.; Jin, L.; Zhao, H.; Zhang, P.; Wen, P.; Xiang, H.; Ding, Y.; et al. Genome-wide and Caste-Specific DNA Methylomes of the Ants Camponotus floridanus and Harpegnathos saltator. Curr. Biol. 2012, 22, 1755–1764. [Google Scholar] [CrossRef] [Green Version]
- Glastad, K.; Gokhale, K.; Liebig, J.; Goodisman, M.A.D. The caste- and sex-specific DNA methylome of the termite Zootermopsis nevadensis. Sci. Rep. 2016, 6, 37110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickman, M.J.; Kucharski, R.; Maleszka, R.; Hurd, P.J. Extensive histone post-translational modification in honey bees. Insect Biochem. Mol. Biol. 2013, 43, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Spannhoff, A.; Kim, Y.K.; Raynal, N.J.M.; Gharibyan, V.; Su, M.; Zhou, Y.; Li, J.; Castellano, S.; Sbardella, G.; Issa, J.-P.; et al. Histone deacetylase inhibitor activity in royal jelly might facilitate caste switching in bees. EMBO Rep. 2011, 12, 238–243. [Google Scholar] [CrossRef] [Green Version]
- Wojciechowski, M.; Lowe, R.; Maleszka, J.; Conn, D.; Maleszka, R.; Hurd, P.J. Phenotypically distinct female castes in honey bees are defined by alternative chromatin states during larval development. Genome Res. 2018, 28, 1532–1542. [Google Scholar] [CrossRef] [Green Version]
- Bonasio, R. The role of chromatin and epigenetics in the polyphenisms of ant castes. Brief. Funct. Genom. 2014, 13, 235–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simola, D.F.; Wissler, L.; Donahue, G.; Waterhouse, R.; Helmkampf, M.; Roux, J.; Nygaard, S.; Glastad, K.; Hagen, D.E.; Viljakainen, L.; et al. Social insect genomes exhibit dramatic evolution in gene composition and regulation while preserving regulatory features linked to sociality. Genome Res. 2013, 23, 1235–1247. [Google Scholar] [CrossRef] [Green Version]
- Simola, D.F.; Ye, C.; Mutti, N.S.; Dolezal, K.; Bonasio, R.; Liebig, J.; Reinberg, D.; Berger, S.L. A chromatin link to caste identity in the carpenter ant Camponotus floridanus. Genome Res. 2013, 23, 486–496. [Google Scholar] [CrossRef] [Green Version]
- Gräff, J.; Tsai, L.-H. Histone acetylation: Molecular mnemonics on the chromatin. Nat. Rev. Neurosci. 2013, 14, 97–111. [Google Scholar] [CrossRef]
- Bonasio, R.; Zhang, G.; Ye, C.; Mutti, N.S.; Fang, X.; Qin, N.; Donahue, G.; Yang, P.; Li, Q.; Li, C.; et al. Genomic Comparison of the Ants Camponotus floridanus and Harpegnathos saltator. Science 2010, 329, 1068–1071. [Google Scholar] [CrossRef] [Green Version]
- Behura, S.K.; Whitfield, C.W. Correlated expression patterns of microRNA genes with age-dependent behavioural changes in honeybee. Insect Mol. Biol. 2010, 19, 431–439. [Google Scholar] [CrossRef]
- Greenberg, J.K.; Xia, J.; Zhou, X.; Thatcher, S.R.; Gu, X.; Ament, S.A.; Newman, T.C.; Green, P.J.; Zhang, W.; Robinson, G.E.; et al. Behavioral plasticity in honey bees is associated with differences in brain microRNA transcriptome. Genes Brain Behav. 2012, 11, 660–670. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Peng, W.; Li, Z.; Li, W.; Li, L.; Pan, J.; Zhang, S.; Miao, Y.; Chen, S.; Su, S. Next-generation small RNA sequencing for microRNAs profiling in Apis mellifera: Comparison between nurses and foragers. Insect Mol. Biol. 2012, 21, 297–303. [Google Scholar] [CrossRef]
- Guo, X.; Su, S.; Skogerboe, G.; Dai, S.; Li, W.; Li, Z.; Liu, F.; Ni, R.; Guo, Y.; Chen, S.; et al. Recipe for a Busy Bee: MicroRNAs in Honey Bee Caste Determination. PLoS ONE 2013, 8, e81661. [Google Scholar] [CrossRef] [PubMed]
- Collins, D.H.; Mohorianu, I.; Beckers, M.; Moulton, V.; Dalmay, T.; Bourke, A.F.G. MicroRNAs associated with caste determination and differentiation in a primitively eusocial insect. Sci. Rep. 2017, 7, 45674. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Humann, F.C.; Tiberio, G.J.; Hartfelder, K. Sequence and Expression Characteristics of Long Noncoding RNAs in Honey Bee Caste Development—Potential Novel Regulators for Transgressive Ovary Size. PLoS ONE 2013, 8, e78915. [Google Scholar] [CrossRef] [Green Version]
| Insect Species | Phenotypes | Environmental Signal(s) | Neuro-Endocrine System | Epigenetic Mechanism Studied | Key References | ||||
|---|---|---|---|---|---|---|---|---|---|
| Chromatin Regulation | DNA Methylation | sRNAs | lncRNAs | ||||||
| Seasonal polyphenism | Bicyclus anynana (Nymphalida) | Wings spots patterns | Temperature | Ecdysteroids | - | - | - | - | - |
| Acyrthosiphon pisum Myzus persicae (Aphidida) | Clonal Sexual | Photoperiod | Dopamine, Insulin, Juvenile hormone | Morph-specific open chromatin profile of the X chromosome | Morph-specific DNA methylation profile of the X chromosome | - | - | [44,45] | |
| Dispersal polyphenism | Schistocerca gregaria Locusta migratoria (Acrididae) | Solitary Gregarious | Visual, olfactory and physical contacts | Corazonin, Dopamine, Juvenile hormone, Serotonin | H3 phosphorylation in gregarious locusts brains | Differentially methylated genes between phases | Phase-specific sRNAs profiles | - | [47,58,59,60,61] |
| Acyrthosiphon pisum (Aphididae) | Winged Wingless | Crowding, Predators, Food quality | Ecdysone, Juvenile hormone | - | - | - | - | [65] | |
| Caste polyphenism | Apis mellifera (Apidae) | Queen Worker | Larval diet, Genetic factors | Insulin, Juvenile hormone | HDACi in royal jelly Caste-specific patterns of H3K4me3, H3K27ac, H3K36me3 | Differentially methylated genes between castes. Correlation between methylation and alternative splicing | Caste-specific miRNAs profiles | Caste specific lncRNAs (lncov1, lncov2) | [75,79,80,81,82,85,86,87,93,94,95,96,99] |
| Camponotus floridanus Harpegnathos saltator (Formicidae) | Queen Male Worker (Gamergate) | Larval diet Pheromones Genetic factors | Corazonin | H3K27ac strongly associated with caste identity Differential binding of CBP | Differentially methylated genes between castes. Correlation between methylation and alternative splicing | Caste-specific miRNAs profiles | - | [83,89,90,92] | |
| Zootermopsis nevadensis (Archotermopsidae) | Queen Worker | Larval diet | Juvenile hormone | - | Differentially methylated genes between castes. Correlation between methylation and alternative splicing | - | - | [84] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Richard, G.; Jaquiéry, J.; Le Trionnaire, G. Contribution of Epigenetic Mechanisms in the Regulation of Environmentally-Induced Polyphenism in Insects. Insects 2021, 12, 649. https://doi.org/10.3390/insects12070649
Richard G, Jaquiéry J, Le Trionnaire G. Contribution of Epigenetic Mechanisms in the Regulation of Environmentally-Induced Polyphenism in Insects. Insects. 2021; 12(7):649. https://doi.org/10.3390/insects12070649
Chicago/Turabian StyleRichard, Gautier, Julie Jaquiéry, and Gaël Le Trionnaire. 2021. "Contribution of Epigenetic Mechanisms in the Regulation of Environmentally-Induced Polyphenism in Insects" Insects 12, no. 7: 649. https://doi.org/10.3390/insects12070649
APA StyleRichard, G., Jaquiéry, J., & Le Trionnaire, G. (2021). Contribution of Epigenetic Mechanisms in the Regulation of Environmentally-Induced Polyphenism in Insects. Insects, 12(7), 649. https://doi.org/10.3390/insects12070649






