Bombyx mori Nucleopolyhedrovirus p26 Is Associated with Viral Late Stage Replication
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Computer-Assisted Sequence Analysis
2.3. Expression of p26 in E. coli and Generation of Anti-p26 Serum
2.4. RT-PCR
2.5. Immunodetection of p26
2.6. Immunofluorescence Microscopy
2.7. EGFP-p26 Over-Expression in BmN Cells
2.8. RNAi-Based Knockdown
2.9. Verification of p26 Knockdown
2.10. Infectious Virus Titration
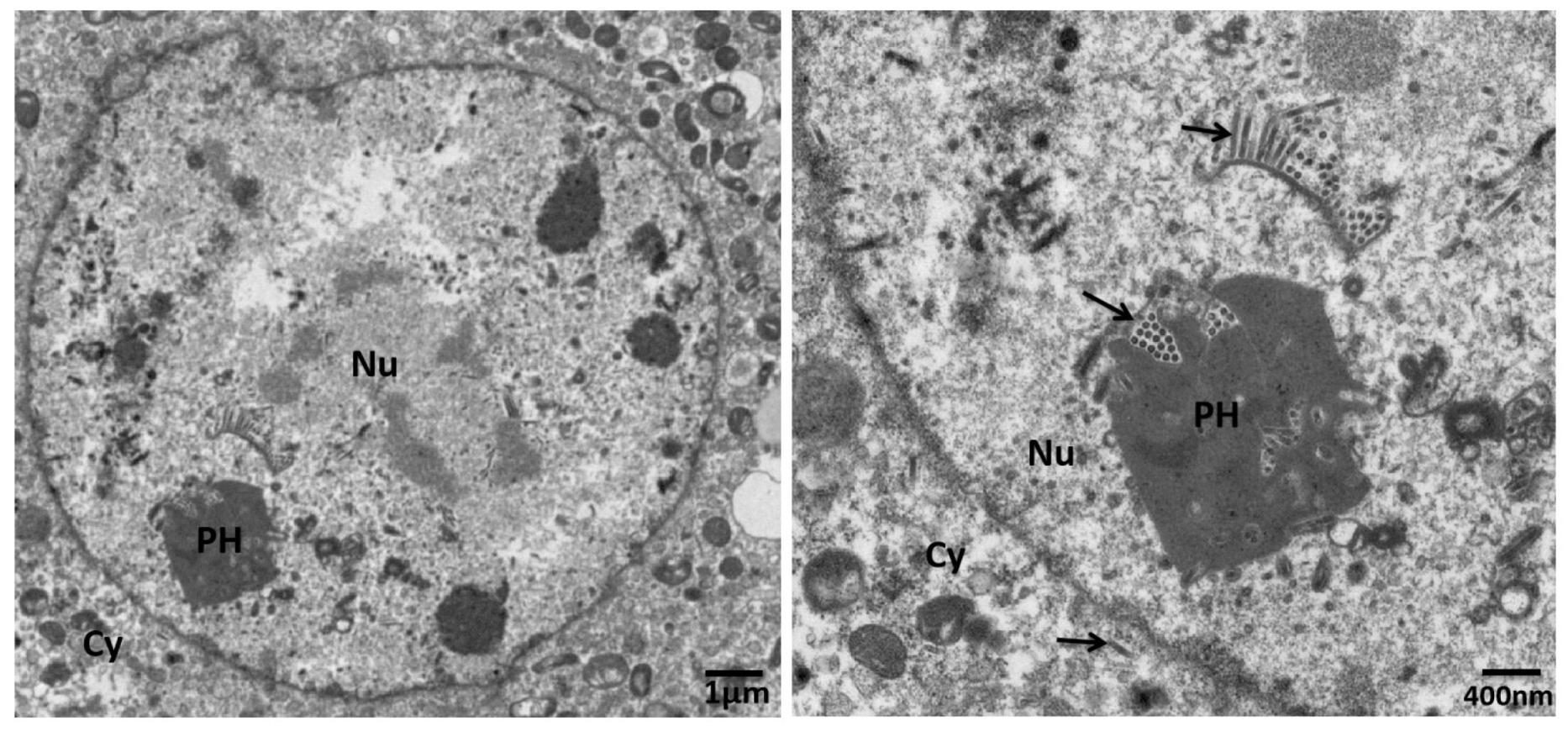
2.11. Electron Microscopy
2.12. Transcription of gp64 and p74
3. Results
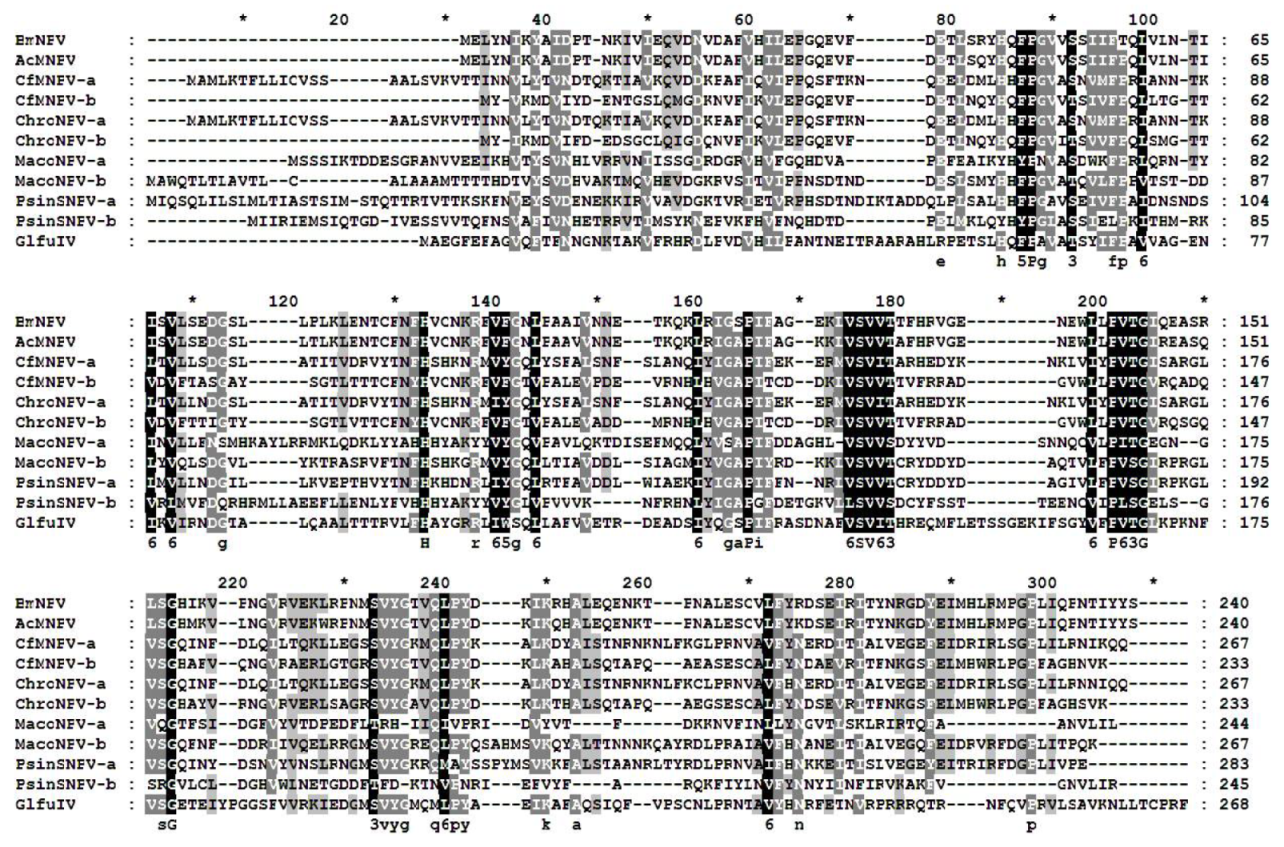
3.1. Sequence Analysis of p26
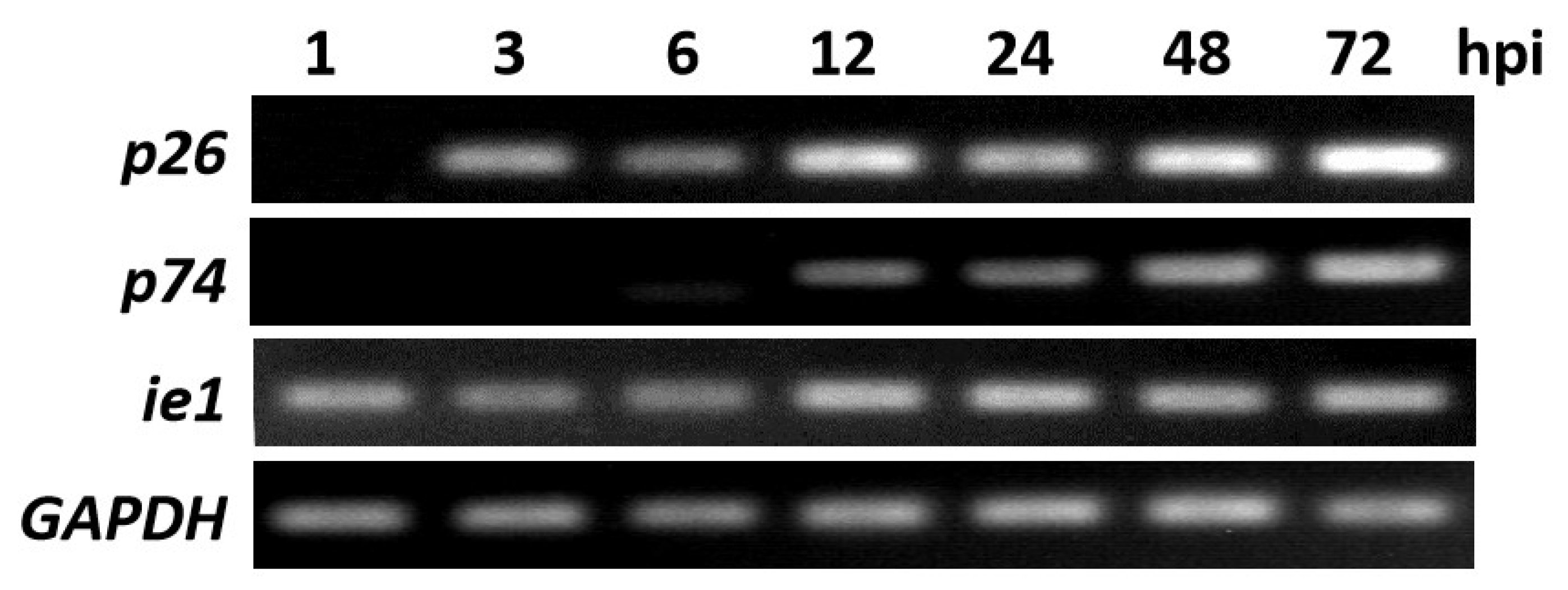
3.2. Transcription of p26
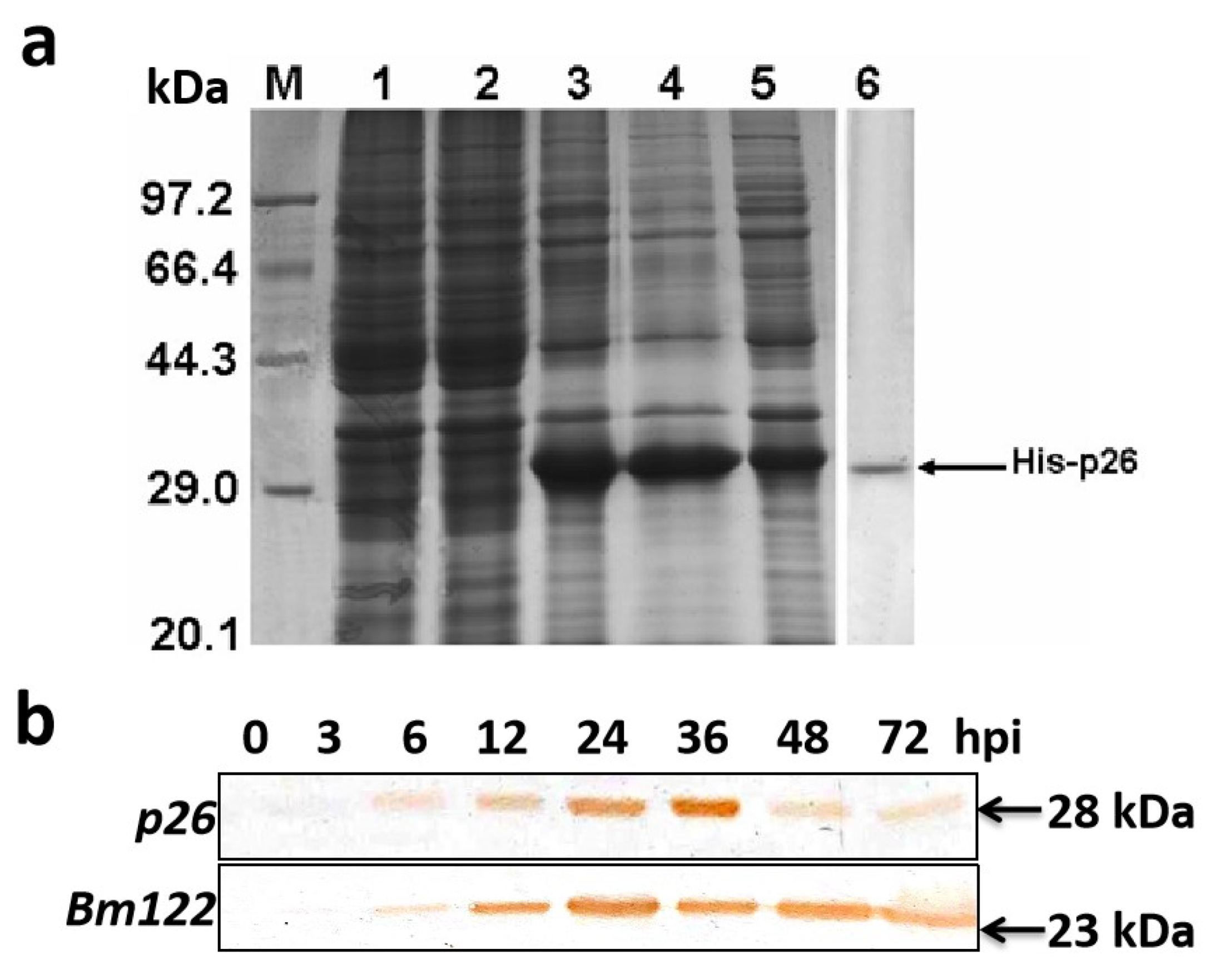
3.3. Immunodetection of p26
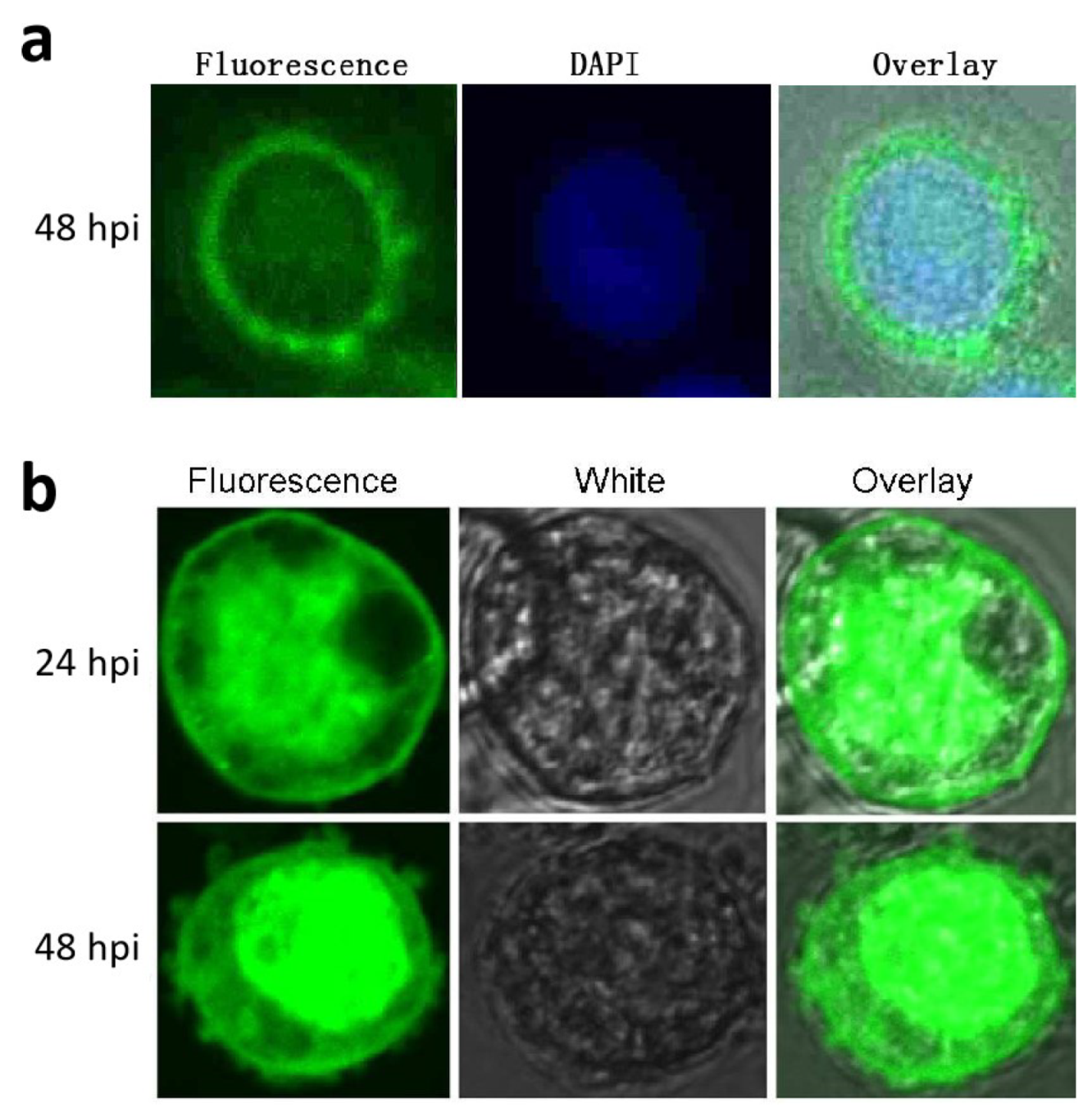
3.4. Subcellular Localization of p26
3.5. Knockdown of p26
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
Appendix A
References
- Blissard, G.W.; Rohrmann, G.F. Baculovirus diversity and molecular biology. Annu. Rev. Entomol. 1990, 35, 127–155. [Google Scholar] [CrossRef]
- Hoopes, R.R.; Rohrmann, G.F. In vitro transcription of baculovirus immediate early genes: Accurate mRNA initiation by nuclear extracts from both insect and human cells. Proc. Natl. Acad. Sci. USA 1991, 88, 4513–4517. [Google Scholar] [CrossRef] [PubMed]
- Huijskens, I.; Li, L.; Willis, L.G. Theilmann DA. Role of AcMNPV IE0 in baculovirus very late gene activation. Virology 2014, 323, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Yamada, Y.; Matsuyama, T.; Quan, G.X.; Kanda, T.; Tamura, T.; Sahara, K.; Asano, S.; Bando, H. Use of an N-terminal half truncated IE1 as an antagonist of IE1, an essential regulatory protein in baculovirus. Virus Res. 2002, 90, 253–261. [Google Scholar] [CrossRef]
- Guarino, L.A.; Xu, B.; Jin, J.; Dong, W.A. virus-encoded RNA polymerase purified from baculovirus-infected cells. J. Virol. 1998, 72, 7985–7991. [Google Scholar] [CrossRef]
- Lu, A.; Miller, L.K. The roles of eighteen baculovirus late expression factor genes in transcription DNA, replication. J. Virol. 1995, 69, 975–982. [Google Scholar] [CrossRef]
- Gomi, S.; Majima, K.; Maeda, S. Sequence analysis of the genome of Bombyx mori nucleopolyhedrovirus. J. Gen. Virol. 1999, 80 Pt 5, 1323–1337. [Google Scholar] [CrossRef]
- Liu, A.; Qin, J.C.; Rankin, C.; Hardin, S.E.; Weaver, R.F. Nucleotide sequence of a portion of the Autographa californica nuclear polyhedrosis virus genome containing the EcoRI site-rich region (hr5) and an open reading frame just 5′ of the p10 gene. J. Gen. Virol. 1986, 67 Pt 11, 2565–2570. [Google Scholar] [CrossRef]
- Huht, N.E.; Weaver, R.F. Categorizing some early and late transcripts directed by the Autographa californica nuclear polyhedrosis virus. J. Gen. Virol. 1990, 71, 2195–2200. [Google Scholar]
- Chen, Y.R.; Zhong, S.; Fei, Z.; Hashimoto, Y.; Xiang, J.Z.; Zhang, S.; Blissard, G.W. The transcriptome of the baculovirus Autographa californica multiple nucleopolyhedrovirus in Trichoplusia ni cells. J. Virol. 2013, 87, 6391–6405. [Google Scholar] [CrossRef] [PubMed]
- Glocker, B.; Hoopes, R.R., Jr.; Rohrmann, G.F. In vitro transactivation of baculovirus early genes by nuclear extracts from Autographa californica nuclear polyhedrosis virus-infected Spodoptera frugiperda cells. J. Virol. 1992, 66, 3476–3484. [Google Scholar] [CrossRef] [PubMed]
- Rodems, S.M.; Friesen, P.D. The hr5 transcriptional enhancer stimulates early expression from the Autographa californica nuclear polyhedrosis virus genome but is not required for virus replication. J. Virol. 1993, 67, 5776–5785. [Google Scholar] [CrossRef] [PubMed]
- Simon, O.; Williams, T.; Caballero, P.; Possee, R.D. Effects of Acp26 on in vitro and in vivo productivity, pathogenesis and virulence of Autographa californica multiple nucleopolyhedrovirus. Virus Res. 2008, 136, 202–205. [Google Scholar] [CrossRef]
- Wang, L.; Salem, T.Z.; Campbell, D.J.; Turney, C.M.; Kumar, C.M.; Cheng, X.W. Characterization of a virion occlusion-defective Autographa californica multiple nucleopolyhedrovirus mutant lacking the p26, p10 and p74 genes. J. Gen. Virol. 2009, 90 Pt 7, 1641–1648. [Google Scholar] [CrossRef]
- Ono, C.; Kamagata, T.; Taka, H.; Sahara, K.; Asano, S.; Bando, H. Phenotypic grouping of 141 BmNPVs lacking viral gene sequences. Virus Res. 2012, 165, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.Q.; Zhao, J.F.; Shao, Y.M.; Tian, C.H.; Zhang, C.X. Characterization of an early gene orf122 from Bombyx mori nucleopolyhedrovirus. Mol. Biol. Rep. 2009, 36, 543–548. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, C.X. HearSNPV orf83 encodes a late nonstructural protein with an active chitin-binding domain. Virus Res. 2006, 117, 237–243. [Google Scholar] [CrossRef]
- Ge, J.Q.; Yang, Z.N.; Tang, X.D.; Xu, H.J.; Hong, J.; Chen, J.G.; Zhang, C.X. Characterization of a nucleopolyhedrovirus with a deletion of the baculovirus core gene Bm67. J. Gen. Virol. 2008, 89 Pt 3, 766–774. [Google Scholar] [CrossRef]
- Monsma, S.A.; Oomens, A.G.; Blissard, G.W. The GP64 envelope fusion protein is an essential baculovirus protein required for cell-to-cell transmission of infection. J. Virol. 1996, 70, 4607–4616. [Google Scholar] [CrossRef]
- Faulkner, P.; Kuzio, J.; Williams, G.V.; Wilson, J.A. Analysis of p74, a PDV envelope protein of Autographa californica nucleopolyhedrovirus required for occlusion body infectivity in vivo. J. Gen. Virol. 1997, 78 Pt 12, 3091–3100. [Google Scholar] [CrossRef]
- Guo, H.; Jiang, L.; Xia, Q. Selection of reference genes for analysis of stress-responsive genes after challenge with viruses and temperature changes in the silkworm Bombyx mori. Mol. Genet. Genomics 2016, 291, 999–1004. [Google Scholar] [CrossRef]
- De Jong, J.G.; Lauzon, H.A.; Dominy, C.; Poloumienko, A.; Carstens, E.B.; Arif, B.M.; Krell, P.J. Analysis of the Choristoneura fumiferana nucleopolyhedrovirus genome. J. Gen. Virol. 2005, 86 Pt 4, 929–943. [Google Scholar] [CrossRef]
- Thumbi, D.K.; Beliveau, C.; Cusson, M.; Lapointe, R.; Lucarotti, C.J. Comparative genome sequence analysis of Choristoneura occidentalis Freeman and C. rosaceana Harris (Lepidoptera: Tortricidae) alphabaculoviruses. PLoS ONE 2013, 8, e68968. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Li, Q.; Donly, C.; Li, L.; Willis, L.G.; Theilmann, D.A.; Erlandson, M. Sequence and organization of the Mamestra configurata nucleopolyhedrovirus genome. Virology 2002, 294, 106–121. [Google Scholar] [CrossRef]
- Craveiro, S.R.; Inglis, P.W.; Togawa, R.C.; Grynberg, P.; Melo, F.L.; Ribeiro, Z.M.; Ribeiro, B.M.; Bao, S.N.; Castro, M.E. The genome sequence of Pseudoplusia includens single nucleopolyhedrovirus and an analysis of p26 gene evolution in the baculoviruses. BMC Genom. 2015, 16, 127. [Google Scholar] [CrossRef] [PubMed]
- Van Mierlo, J.T.; van Cleef, K.W.; van Rij, R.P. Defense and counterdefense in the RNAi-based antiviral immune system in insects. Methods Mol. Biol. 2011, 721, 3–22. [Google Scholar] [PubMed]
- Jayachandran, B.; Hussain, M.; Asgari, S. RNA interference as a cellular defense mechanism against the DNA virus baculovirus. J. Virol. 2012, 86, 13729–13734. [Google Scholar] [CrossRef] [PubMed]
- Huh, N.E.; Weaver, R.F. Identifying the RNA polymerases that synthesize specific transcripts of the Autographa californica nuclear polyhedrosis virus. J. Gen. Virol. 1990, 71 Pt 1, 195–201. [Google Scholar] [CrossRef]
- Braunagel, S.C.; Russell, W.K.; Rosas-Acosta, G.; Russell, D.H.; Summers, M.D. Determination of the protein composition of the occlusion-derived virus of Autographa californica nucleopolyhedrovirus. Proc. Natl. Acad. Sci. USA 2003, 100, 9797–9802. [Google Scholar] [CrossRef]
- Goenka, S.; Weaver, R.F. The p26 gene of the Autographa californica nucleopolyhedrovirus: Timing of transcription, and cellular localization and dimerization of product. Virus Res. 2008, 131, 136–144. [Google Scholar] [CrossRef]
- Beliveau, C.; Cohen, A.; Stewart, D.; Periquet, G.; Djoumad, A.; Kuhn, L.; Stoltz, D.; Boyle, B.; Volkoff, A.N.; Herniou, E.A.; et al. Genomic and Proteomic Analyses Indicate that Banchine and Campoplegine Polydnaviruses Have Similar, if Not Identical, Viral Ancestors. J. Virol. 2015, 89, 8909–8921. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.W.; Peiffer, M.; Hoover, K.; Rosa, C.; Acevedo, F.E.; Felton, G.W. Symbiotic polydnavirus of a parasite manipulates caterpillar and plant immunity. Proc. Natl. Acad. Sci. USA 2018, 115, 5199–5204. [Google Scholar] [CrossRef] [PubMed]

| Name | Sequence | Target |
|---|---|---|
| p26FW | 5’-CGGATCCATGGAATTGTATAATATTAAAT-3’ | p26 |
| p26RW | 5’-CAAGCTTTTAGCTGTAATATATTGTGTTG-3’ | |
| T7p26iFW | 5’-TAATACGACTCACTATAGGGTTTCCTGGCGTCGTTAGTTC-3’ | p26 |
| T7p26iRW | 5’-TAATACGACTCACTATAGGGTTGCACAGTCCCGTAAACAG-3’ | |
| T7GFPiFW | 5’-TAATACGACTCACTATAGGGTGGTAAAAGGACAGGGCCATC-3’ | gfp |
| T7GFPiRW | 5’-TAATACGACTCACTATAGGGCCATGGCCAACACTTGTCAC-3’ | |
| Qp26FW | 5’-TGTAATAGAGCAAGTCGACAATGTG-3’ | p26 |
| Qp26RW | 5’-TGGTACCGGCTTAGCGTTTC-3’ | |
| Qie1FW | 5’-AACATTTGCACGGTCGCTTC-3’ | ie1 |
| Qie1RW | 5’- GGTCGGAGAACCTGTTGGAA-3’ | |
| Qgp64FW | 5’-ACGGCATCAGCAAAAACGTG-3’ | gp64 |
| Qgp64RW | 5’-AAGGTGGACGAGCGTTTGAT-3’ | |
| Qp74FW | 5’-TCTGTAGTGGTATCGCGCAC-3’ | p74 |
| Qp74RW | 5’-AGCGCCTTCCAGCATACTAC-3’ | |
| QBmGapdhFW | 5’-AGGGCAGTGTTGAGGTTCAG-3’ | gapdh |
| QBmGapdhRW | 5’-GGCCTTAGGGTCCCTTTCTG-3’ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ge, J.-Q.; Wang, Z.-H.; Chen, X.; Chen, H.; Huang, J. Bombyx mori Nucleopolyhedrovirus p26 Is Associated with Viral Late Stage Replication. Insects 2021, 12, 707. https://doi.org/10.3390/insects12080707
Ge J-Q, Wang Z-H, Chen X, Chen H, Huang J. Bombyx mori Nucleopolyhedrovirus p26 Is Associated with Viral Late Stage Replication. Insects. 2021; 12(8):707. https://doi.org/10.3390/insects12080707
Chicago/Turabian StyleGe, Jun-Qing, Zhu-Hong Wang, Xi Chen, Hua Chen, and Jian Huang. 2021. "Bombyx mori Nucleopolyhedrovirus p26 Is Associated with Viral Late Stage Replication" Insects 12, no. 8: 707. https://doi.org/10.3390/insects12080707
APA StyleGe, J.-Q., Wang, Z.-H., Chen, X., Chen, H., & Huang, J. (2021). Bombyx mori Nucleopolyhedrovirus p26 Is Associated with Viral Late Stage Replication. Insects, 12(8), 707. https://doi.org/10.3390/insects12080707






