Cell Line Platforms Support Research into Arthropod Immunity
Simple Summary
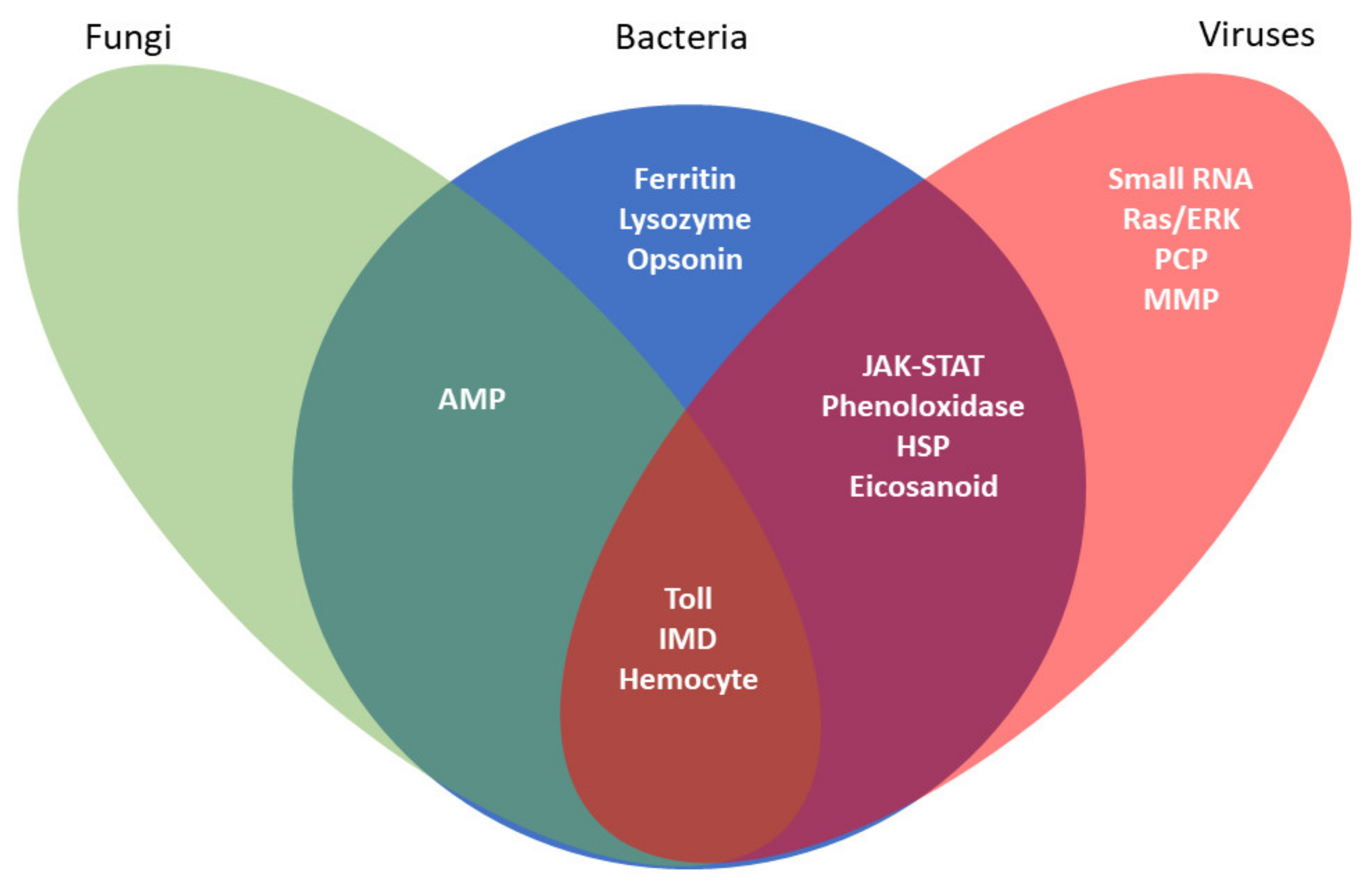
Abstract
1. Introduction
2. Pathogen-Associated Molecular Patterns (PAMPs), Pattern Recognition Receptors (PRRs), and Opsonins
| Order | Species of Origin | Stage/Tissue of Origin | Cell Line Designation | Research Focus | References |
|---|---|---|---|---|---|
| Coleoptera | Anthonomus grandis | Embryo | BRL-AG-1 | Humoral Responses | [37] |
| A. grandis | Embryo | BRL-AG-3A | Humoral Responses | [37] | |
| A. grandis | Embryo | BRL-AG-3C | Humoral Responses | [37] | |
| Tribolium castaneum | Pupa/Adult | BCIRL-TcA-CLG1 | Signaling Pathways | [38] | |
| Diptera | Aedes aegypti | Neonate larva | Aag-2 | Signaling Pathways, Cellular Responses, Humoral Responses | [37,39,40,41,42,43,44,45,46,47,48] |
| Ae. aegypti | Neonate larva | AF5 and subline AF319 | Signaling Pathways | [43,45] | |
| Ae. aegypti | Neonate larva | ATC-10 (CCL-125) | Signaling Pathways, Humoral Responses | [42,45,49,50] | |
| Aedes albopictus | Neonate larva | C6/36 | PAMPs, Signaling Pathways, Humoral Responses | [35,42,44,45,46,51,52,53,54,55,56,57] | |
| Ae. albopictus | Neonate larva | C6/36 HT | Signaling Pathways | [39] | |
| Ae. albopictus | Neonate larva | C7-10 | Signaling Pathways, Cellular Responses, Humoral Responses | [37,42,45,58,59,60] | |
| Ae. albopictus | Neonate larva | U4.4 | Signaling Pathways | [42,44,45,61] | |
| Anophelesgambiae | Neonate larva | 4a-2 | Humoral Responses | [37] | |
| A. gambiae | Neonate larva | 4a-3A | Humoral Responses | [37,62] | |
| A. gambiae | Neonate larva | 4a-3B | Humoral Responses | [37] | |
| A. gambiae | Neonate larva | Sua1B | Signaling Pathways, Cellular Responses, Humoral Responses | [37,62,63] | |
| A. gambiae | Neonate larva | Sua5.1* | Opsonins, Signaling Pathways | [64] | |
| Anopheles stephensi | 1st Instar larva | 4a-3A | Signaling Pathways | [62] | |
| A. stephensi | 1st Instar larva | 4a-3B | Signaling Pathways | [65] | |
| A. stephensi | 1st Instar larva | LSTM-AS-43 (MSQ43) | Signaling Pathways, Humoral Responses | [62] | |
| Culex quinquefasciatus | Ovary (adult) | Hsu | Signaling Pathways | [56] | |
| Culex tarsalis | Embryo | CT | Signaling Pathways | [56] | |
| Lutzomyia longipalpis | Embryo | LL5 | Signaling Pathways | [66,67] | |
| Sarcophaga peregrina | Embryo | NIH-Sape-4 | Humoral Responses | [37,68] | |
| Hemiptera | Anasa tristis | Embryo | BCIRL-AtE-CLG15A | Signaling Pathways | [38] |
| Lepidoptera | Opodiphthera (Antheraea) eucalypti | Pupal ovaries | Ae | Cellular Responses | [69] |
| Bombyx mori | Ovary (larval) | Bm5 | Signaling Pathways | [55,70,71] | |
| B. mori | Ovary | BmN | Signaling Pathways | [72] | |
| B. mori | Ovary | BmN4 | Signaling Pathways | [55] | |
| B. mori | Ovary (larval) | BmN-SWU1 | Signaling Pathways | [73] | |
| B. mori | Ovary | NISES-BoMo-Cam1 | PAMPs, Signaling Pathways | [34] | |
| Choristoneurafumiferana | Midgut (larval) | IPRI-CF-1 | Signaling Pathways | [74] | |
| C. fumiferana | Midgut | FPMI-CF-203 | Signaling Pathways | [55] | |
| Chrysodeixis (Pseudoplusia) includens | Embryo | UGA-CiE1 | Cellular Responses, Humoral Responses | [75] | |
| Estigmene acraea | Hemocyte (larval) | BTI-EA-1174-A | Cellular Responses, Humoral Responses | [37,76] | |
| Helicoverpa zea | Ovary (pupal) | BCIRL-HzAM1 | Signaling Pathways, Humoral Responses | [77,78,79,80] | |
| H. zea | Midgut (larval) | RP-HzGUT-AW1 | Signaling Pathways | [55] | |
| Helithis virescens | Ovary (pupal) | BCIRL-HvAM1 | Signaling Pathways | [38] | |
| Lymantria dispar | Ovary (pupal) | IPLB-Ld-652Y | Cellular Responses | [81] | |
| Malacosoma disstria | Hemocyte (larval) | IPRI-Md-66 | Signaling Pathways, Cellular Responses, Humoral Responses | [82] | |
| M. disstria | Hemocyte (larval) | IPRI-Md-108 | Signaling Pathways, Cellular Responses, Humoral Responses | [82] | |
| Manduca sexta | Embryo | MRRL-CHE-20 | Signaling Pathways | [74] | |
| Perina nuda | Ovary (pupal) | NTU-Pn-HH | Cellular Responses | [81] | |
| Plodia interpunctella | Unspecified | KSU-P15.3 | Signaling Pathways | [74] | |
| Spodoptera exigua | Embryo/neonate larva | Se301 | Cellular Responses | [69] | |
| S. exigua | Hemocyte (larval) | SeHe920-1a | Cellular Responses | [69] | |
| S. exigua | Neonate larva | UCR-Se-1 | Cellular Responses | [81] | |
| Spodoptera frugiperda | Ovary (pupal) | IPLB-Sf-5-5C | Cellular Responses | [81] | |
| S. frugiperda | Ovary (pupal) | Sf21 | Signaling Pathways | [55] | |
| S. frugiperda | Ovary (pupal) | Sf9 | Signaling Pathways, Humoral Responses | [83,84,85,86,87] | |
| S. frugiperda | Ovary (pupal) | Sf9-SF (serum-free) | Signaling Pathways | [55] | |
| Spodoptera littoralis | Ovary | Sl2 | Signaling Pathways | [55] | |
| Spodoptera litura | Ovary (pupal) | IBL-Sl-1A | Cellular Responses | [81] | |
| Trichoplusia ni | Embryo | High Five (BTI-TN-5B1-4) | Signaling Pathways, Cellular Responses | [55,69,71,87,88] | |
| T. ni | Embryo | High Five-SF (serum-free) | Signaling Pathways, Humoral Responses | [55] |
3. Signaling Pathways and Signaling Molecules Involved in Antimicrobial and Antiviral Responses
3.1. Phagocytosis Related Signaling Pathways
3.2. Signaling Pathways Associated with Antimicrobial Humoral Responses
3.3. Eicosanoid-Related Signaling
4. Antiviral Signaling Pathways
4.1. sRNA-Related Pathways
4.2. Non-RNA-Related Antiviral Signaling Pathways
4.3. Other Uses of Cell Lines in Viral Pathway Studies
5. Antimicrobial Cellular Responses
Phagocytosis
6. Antimicrobial and Antiviral Humoral Responses
6.1. Antimicrobial Peptides (AMPS)
6.2. Lysozymes
6.3. Other Immune-Related Proteins
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Lemaitre, B.; Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef]
- Kanost, M.R.; Jiang, H. Clip-domain serine proteases as immune factors in insect hemolymph. Curr. Opin. Insect Sci. 2015, 11, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.; Kim, Y. Prostaglandins and other eicosanoids in insects: Biosynthesis and biological actions. Front. Physiol. 2019, 9, 1927. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Stanley, D. Eicosanoid signaling in insect immunology: New genes and unresolved issues. Genes 2021, 12, 211. [Google Scholar] [CrossRef] [PubMed]
- Rheins, L.A.; Karp, R.D.; Butz, A. Induction of specific humoral immunity to soluble proteins in the American cockroach (Periplaneta americana). I. Nature of the primary response. Dev. Comp. Immunol. 1980, 4, 447–458. [Google Scholar] [CrossRef]
- Cooper, D.; Eleftherianos, I. Memory and specificity in the insect immune system: Current perspectives and future challenges. Front. Immunol. 2017, 8, 539. [Google Scholar] [CrossRef] [PubMed]
- Tassetto, M.; Kunitomi, M.; Andino, R. Circulating immune cells mediate a systemic RNAi-based adaptive antiviral response in Drosophila. Cell 2017, 169, 314–325.e313. [Google Scholar] [CrossRef]
- Sheehan, G.; Farrell, G.; Kavanagh, K. Immune priming: The secret weapon of the insect world. Virulence 2020, 11, 238–246. [Google Scholar] [CrossRef]
- Bonning, B.C.; Saleh, M.-C. The interplay between viruses and RNAi pathways in insects. Annu. Rev. Entomol. 2021, 66, 61–79. [Google Scholar] [CrossRef]
- Dunn, P.E.; Drake, D.R. Fate of bacteria injected into naive and immunized larvae of the tobacco hornworm Manduca sexta. J. Invertebr. Pathol. 1983, 41, 77–85. [Google Scholar] [CrossRef]
- Hillyer, J.F. Insect immunology and hematopoiesis. Dev. Comp. Immunol. 2016, 58, 102–118. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, Y.; Zhang, R.; Zhang, J. The diversity of pattern recognition receptors (PRRs) involved with insect defense against pathogens. Curr. Opin. Insect Sci. 2019, 33, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Su, F.; Li, Q.; Zhang, J.; Li, Y.; Tang, T.; Hu, Q.; Yu, X.Q. Pattern recognition receptors in Drosophila immune responses. Dev. Comp. Immunol. 2020, 102, 103468. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ren, M.; Liu, X.; Xia, H.; Chen, K. Peptidoglycan recognition proteins in insect immunity. Mol. Immunol. 2019, 106, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Stokes, B.A.; Yadav, S.; Shokal, U.; Smith, L.C.; Eleftherianos, I. Bacterial and fungal pattern recognition receptors in homologous innate signaling pathways of insects and mammals. Front. Microbiol. 2015, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Strand, M.R. The insect cellular immune response. Insect Sci. 2008, 15, 1–14. [Google Scholar] [CrossRef]
- Merchant, D.; Ertl, R.L.; Rennard, S.I.; Stanley, D.W.; Miller, J.S. Eicosanoids mediate insect hemocyte migration. J. Insect Physiol. 2008, 54, 215–221. [Google Scholar] [CrossRef]
- Wu, Q.; Patocka, J.; Kuca, K. Insect antimicrobial peptides, a mini review. Toxins 2018, 10, 461. [Google Scholar] [CrossRef]
- Yakovlev, A.Y.; Nesin, A.P.; Simonenko, N.P.; Gordya, N.A.; Tulin, D.V.; Kruglikova, A.A.; Chernysh, S.I. Fat body and hemocyte contribution to the antimicrobial peptide synthesis in Calliphora vicina R.-D. (Diptera: Calliphoridae) larvae. Vitr. Cell. Dev. Biol. Anim. 2017, 53, 33–42. [Google Scholar] [CrossRef]
- Fauvarque, M.O.; Williams, M.J. Drosophila cellular immunity: A story of migration and adhesion. J. Cell Sci. 2011, 124, 1373–1382. [Google Scholar] [CrossRef]
- Trammell, C.E.; Goodman, A.G. Emerging mechanisms of insulin-mediated antiviral immunity in Drosophila melanogaster. Front. Immunol. 2019, 10, 2973. [Google Scholar] [CrossRef] [PubMed]
- Smagghe, G.; Goodman, C.L.; Stanley, D. Cell-based screening systems for insecticides. Insect cell culture and applications to research and pest management. Vitr. Cell. Dev. Biol. Anim. 2009, 45, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Smagghe, G.; Swevers, L. Advanced Technologies for Managing Insect Pests; Ishaaya, I., Palli, S.R., Horowitz, A.R., Eds.; Springer Science and Business Media: Dordrecht, The Netherlands; Heidelberg, Germany; New York, NY, USA; London, UK, 2012; pp. 107–134. [Google Scholar] [CrossRef]
- Arunkarthick, S.; Asokan, R.; Aravintharaj, R.; Niveditha, M.; Kumar, N.K.K. A review of insect cell culture: Establishment, maintenance and applications in entomological research. J. Entomol. Sci. 2017, 52, 261–273. [Google Scholar] [CrossRef]
- Yee, C.M.; Zak, A.J.; Hill, B.D.; Wen, F. The coming age of insect cells for manufacturing and development of protein therapeutics. Ind. Eng. Chem. Res. 2018, 57, 10061–10070. [Google Scholar] [CrossRef]
- Rubio, N.R.; Fish, K.D.; Trimmer, B.A.; Kaplan, D.L. In vitro insect muscle for tissue engineering applications. ACS Biomater. Sci. Eng. 2019, 5, 1071–1082. [Google Scholar] [CrossRef]
- Stanley, D.; Kim, Y. Eicosanoid signaling in insects: From discovery to plant protection. Crit. Rev. Plant. Sci. 2014, 33, 20–63. [Google Scholar] [CrossRef]
- Hwang, S.; Bang, K.; Lee, J.; Cho, S. Circulating hemocytes from larvae of the Japanese rhinoceros beetle Allomyrina dichotoma (Linnaeus) (Coleoptera: Scarabaeidae) and the cellular immune response to microorganisms. PLoS ONE 2015, 10, e0128519. [Google Scholar] [CrossRef]
- Duressa, T.F.; Huybrechts, R. Development of primary cell cultures using hemocytes and phagocytic tissue cells of Locusta migratoria: An application for locust immunity studies. Vitr. Cell. Dev. Biol. Anim. 2016, 52, 100–106. [Google Scholar] [CrossRef]
- Richardson, R.T.; Ballinger, M.N.; Qian, F.; Christman, J.W.; Johnson, R.M. Morphological and functional characterization of honey bee, Apis mellifera, hemocyte cell communities. Apidologie 2018, 49, 397–410. [Google Scholar] [CrossRef]
- Guo, Y.; Goodman, C.L.; Stanley, D.W.; Bonning, B.C. Cell lines for honey bee virus research. Viruses 2020, 12, 236. [Google Scholar] [CrossRef]
- Luhur, A.; Klueg, K.M.; Roberts, J.; Zelhof, A.C. Thawing, culturing, and cryopreserving Drosophila cell lines. J. Vis. Exp. 2019. [Google Scholar] [CrossRef] [PubMed]
- Valanne, S.; Hultmark, D.; Vesala, L. Recent Advances in Drosophila Cellular and Humoral Innate Immunity; Frontiers Media SA: Lausanne, Switzerland, 2020; pp. 1–219. [Google Scholar] [CrossRef]
- Ha Lee, J.; Hee Lee, I.; Noda, H.; Mita, K.; Taniai, K. Verification of elicitor efficacy of lipopolysaccharides and peptidoglycans on antibacterial peptide gene expression in Bombyx mori. Insect Biochem. Mol. Biol. 2007, 37, 1338–1347. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, T.; Kobayashi, M.; Eshita, Y.; Shirato, K.; Kimura, T.; Ako, Y.; Miyoshi, H.; Takasaki, T.; Kurane, I.; Kariwa, H.; et al. Involvement of the JNK-like protein of the Aedes albopictus mosquito cell line, C6/36, in phagocytosis, endocytosis and infection of West Nile virus. Insect Mol. Biol. 2003, 12, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Shaw, D.K.; Wang, X.; Brown, L.J.; Chávez, A.S.O.; Reif, K.E.; Smith, A.A.; Scott, A.J.; McClure, E.E.; Boradia, V.M.; Hammond, H.L.; et al. Infection-derived lipids elicit an immune deficiency circuit in arthropods. Nat. Commun. 2017, 8, 14401. [Google Scholar] [CrossRef]
- Fallon, A.M.; Sun, D. Exploration of mosquito immunity using cells in culture. Insect Biochem. Mol. Biol. 2001, 31, 263–278. [Google Scholar] [CrossRef]
- Li, Y.F.; Zhang, H.; Ringbauer, J.A., Jr.; Goodman, C.L.; Lincoln, T.R.; Zhou, K.; Stanley, D. Prostaglandin actions in established insect cell lines. Vitr. Cell. Dev. Biol. Anim. 2017, 53, 421–429. [Google Scholar] [CrossRef]
- Trujillo-Ocampo, A.; Cazares-Raga, F.E.; Del Angel, R.M.; Medina-Ramirez, F.; Santos-Argumedo, L.; Rodriguez, M.H.; Hernandez-Hernandez, F.C. Participation of 14-3-3ε and 14-3-3ζ proteins in the phagocytosis, component of cellular immune response, in Aedes mosquito cell lines. Parasites Vectors 2017, 10, 362. [Google Scholar] [CrossRef]
- Barletta, A.B.; Silva, M.C.; Sorgine, M.H. Validation of Aedes aegypti Aag-2 cells as a model for insect immune studies. Parasites Vectors 2012, 5, 148. [Google Scholar] [CrossRef]
- Zhang, R.; Zhu, Y.; Pang, X.; Xiao, X.; Zhang, R.; Cheng, G. Regulation of antimicrobial peptides in Aedes aegypti Aag2 cells. Front. Cell. Infect. Microbiol. 2017, 7, 22. [Google Scholar] [CrossRef]
- Morazzani, E.M.; Wiley, M.R.; Murreddu, M.G.; Adelman, Z.N.; Myles, K.M. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog. 2012, 8, e1002470. [Google Scholar] [CrossRef]
- Varjak, M.; Donald, C.L.; Mottram, T.J.; Sreenu, V.B.; Merits, A.; Maringer, K.; Schnettler, E.; Kohl, A. Characterization of the Zika virus induced small RNA response in Aedes aegypti cells. PLoS Negl. Trop. Dis. 2017, 11, e0006010. [Google Scholar] [CrossRef] [PubMed]
- Vodovar, N.; Bronkhorst, A.W.; van Cleef, K.W.R.; Miesen, P.; Blanc, H.; van Rij, R.P.; Saleh, M.-C. Arbovirus-derived piRNAs exhibit a ping-pong signature in mosquito cells. PLoS ONE 2012, 7, e30861. [Google Scholar] [CrossRef] [PubMed]
- Sigle, L.T.; McGraw, E.A. Expanding the canon: Non-classical mosquito genes at the interface of arboviral infection. Insect Biochem. Mol. Biol. 2019, 109, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.Q.; Chen, S.Q.; Bai, H.Q.; Wei, Q.M.; Zhang, S.N.; Chen, C.; Zhu, Y.H.; Yi, T.W.; Guo, X.P.; Chen, S.Y.; et al. The Ras/ERK signaling pathway couples antimicrobial peptides to mediate resistance to dengue virus in Aedes mosquitoes. PLoS Negl. Trop. Dis. 2020, 14, e0008660. [Google Scholar] [CrossRef] [PubMed]
- Russell, T.A.; Ayaz, A.; Davidson, A.D.; Fernandez-Sesma, A.; Maringer, K. Imd pathway-specific immune assays reveal NF-κB stimulation by viral RNA PAMPs in Aedes aegypti Aag2 cells. PLoS Negl. Trop. Dis. 2021, 15, e0008524. [Google Scholar] [CrossRef]
- Gao, Y.; Fallon, A.M. Immune activation upregulates lysozyme gene expression in Aedes aegypti mosquito cell culture. Insect Mol. Biol. 2000, 9, 553–558. [Google Scholar] [CrossRef]
- Geiser, D.L.; Zhou, G.; Mayo, J.J.; Winzerling, J.J. The effect of bacterial challenge on ferritin regulation in the yellow fever mosquito, Aedes aegypti. Insect Sci. 2013, 20, 601–619. [Google Scholar] [CrossRef]
- Colpitts, T.M.; Cox, J.; Vanlandingham, D.L.; Feitosa, F.M.; Cheng, G.; Kurscheid, S.; Wang, P.H.; Krishnan, M.N.; Higgs, S.; Fikrig, E. Alterations in the Aedes aegypti transcriptome during infection with West Nile, dengue and yellow fever viruses. PLoS Pathog. 2011, 7, e1002189. [Google Scholar] [CrossRef]
- Barbosa, J.A.; Rebello, M.A. Effect of prostaglandin A1 in the induction of stress proteins in Aedes albopictus cells. Braz. J. Med. Biol. Res. 1998, 31, 499–503. [Google Scholar] [CrossRef]
- de Meneses, M.D.; Rebello, M.A. Effect of prostaglandin A1, arsenite and aspirin on stress proteins response in mosquito cells. Z. Nat. C J. Biosci. 2001, 56, 298–302. [Google Scholar] [CrossRef]
- Burlandy, F.M.; Ferreira, D.F.; Rebello, M.A. Inhibition of vesicular stomatitis virus replication by prostaglandin A1 in Aedes albopictus cells. Z. Nat. C J. Biosci. 2004, 59, 127–131. [Google Scholar] [CrossRef]
- Souza-Neto, J.A.; Sim, S.; Dimopoulos, G. An evolutionary conserved function of the JAK-STAT pathway in anti-dengue defense. Proc. Natl. Acad. Sci. USA 2009, 106, 17841–17846. [Google Scholar] [CrossRef] [PubMed]
- Swevers, L.; Ioannidis, K.; Kolovou, M.; Zografidis, A.; Labropoulou, V.; Santos, D.; Wynant, N.; Broeck, J.V.; Wang, L.; Cappelle, K.; et al. Persistent RNA virus infection of lepidopteran cell lines: Interactions with the RNAi machinery. J. Insect Physiol 2016, 93–94, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Ruckert, C.; Prasad, A.N.; Garcia-Luna, S.M.; Robison, A.; Grubaugh, N.D.; Weger-Lucarelli, J.; Ebel, G.D. Small RNA responses of Culex mosquitoes and cell lines during acute and persistent virus infection. Insect Biochem. Mol. Biol. 2019, 109, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, A.; Gupta, K.; Shah, R.; van Hoek, M.L. Antimicrobial activity of mosquito cecropin peptides against Francisella. Dev. Comp. Immunol. 2016, 63, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Fallon, A.M. Characterization of genomic DNA encoding cecropins from an Aedes albopictus mosquito cell line. Insect Mol. Biol. 2002, 11, 21–30. [Google Scholar] [CrossRef]
- Hernandez, V.P.; Higgins, L.; Fallon, A.M. Characterization and cDNA cloning of an immune-induced lysozyme from cultured Aedes albopictus mosquito cells. Dev. Comp. Immunol. 2003, 27, 11–20. [Google Scholar] [CrossRef]
- Nasr, N.M.; Fallon, A.M. Detection of lysozyme-like enzymatic activity secreted by an immune-responsive mosquito cell line. J. Invertebr. Pathol. 2003, 82, 162–166. [Google Scholar] [CrossRef]
- Rodriguez-Andres, J.; Rani, S.; Varjak, M.; Chase-Topping, M.E.; Beck, M.H.; Ferguson, M.C.; Schnettler, E.; Fragkoudis, R.; Barry, G.; Merits, A.; et al. Phenoloxidase activity acts as a mosquito innate immune response against infection with Semliki Forest virus. PLoS Pathog. 2012, 8, e1002977. [Google Scholar] [CrossRef]
- Luna, C.; Hoa, N.T.; Lin, H.; Zhang, L.; Nguyen, H.L.; Kanzok, S.M.; Zheng, L. Expression of immune responsive genes in cell lines from two different Anopheline species. Insect Mol. Biol. 2006, 15, 721–729. [Google Scholar] [CrossRef]
- Hoa, N.T.; Keene, K.M.; Olson, K.E.; Zheng, L. Characterization of RNA interference in an Anopheles gambiae cell line. Insect Biochem. Mol. Biol. 2003, 33, 949–957. [Google Scholar] [CrossRef]
- Moita, L.F.; Vriend, G.; Mahairaki, V.; Louis, C.; Kafatos, F.C. Integrins of Anopheles gambiae and a putative role of a new beta integrin, BINT2, in phagocytosis of E. coli. Insect Biochem. Mol. Biol. 2006, 36, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.M.; Dimopoulos, G.; Blass, C.; Kafatos, F.C. A hemocyte-like cell line established from the malaria vector Anopheles gambiae expresses six prophenoloxidase genes. J. Biol. Chem. 1999, 274, 11727–11735. [Google Scholar] [CrossRef]
- Tinoco-Nunes, B.; Telleria, E.L.; da Silva-Neves, M.; Marques, C.; Azevedo-Brito, D.A.; Pitaluga, A.N.; Traub-Csekö, Y.M. The sandfly Lutzomyia longipalpis LL5 embryonic cell line has active Toll and Imd pathways and shows immune responses to bacteria, yeast and Leishmania. Parasites Vectors 2016, 9, 222. [Google Scholar] [CrossRef] [PubMed]
- Pitaluga, A.N.; Mason, P.W.; Traub-Cseko, Y.M. Non-specific antiviral response detected in RNA-treated cultured cells of the sandfly, Lutzomyia longipalpis. Dev. Comp. Immunol. 2008, 32, 191–197. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, K.; Natori, S. Purification of three antibacterial proteins from the culture medium of NIH-Sape-4, an embryonic cell line of Sarcophaga peregrina. J. Biol. Chem. 1988, 263, 17112–17116. [Google Scholar] [CrossRef]
- Chisa, Y.A.; Imanishi, S.; Iiyama, K.; Kawarabata, T. Establishment of phagocytic cell lines from larval hemocytes of the beet armyworm, Spodoptera exigua. Vitr. Cell. Dev. Biol. Anim. 2004, 40, 183–186. [Google Scholar] [CrossRef]
- Liu, J.; Kolliopoulou, A.; Smagghe, G.; Swevers, L. Modulation of the transcriptional response of innate immune and RNAi genes upon exposure to dsRNA and LPS in silkmoth-derived Bm5 cells overexpressing BmToll9-1 receptor. J. Insect Physiol. 2014, 66, 10–19. [Google Scholar] [CrossRef]
- Santos, D.; Wynant, N.; Van den Brande, S.; Verdonckt, T.W.; Mingels, L.; Peeters, P.; Kolliopoulou, A.; Swevers, L.; Vanden Broeck, J. Insights into RNAi-based antiviral immunity in Lepidoptera: Acute and persistent infections in Bombyx mori and Trichoplusia ni cell lines. Sci. Rep. 2018, 8, 2423. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhu, F.; Xiao, R.; Ge, Q.; Tang, H.; Kong, M.; Taha, R.H.; Chen, K. Increased expression of Suppressor of cytokine signaling 2 (BmSOCS2) is correlated with suppression of Bombyx mori nucleopolyhedrovirus replication in silkworm larval tissues and cells. J. Invertebr. Pathol. 2020, 174, 107419. [Google Scholar] [CrossRef]
- Yu, B.; Sang, Q.; Pan, G.; Li, C.; Zhou, Z. A Toll-Spätzle pathway in the immune response of Bombyx mori. Insects 2020, 11, 586. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.E.; Howard, R.W. Inhibitors of eicosanoid biosynthesis and their effect upon Bacillus thuringiensis δ-endotoxin response in cultured insect cells and developing larvae. Curr. Microbiol. 1996, 32, 1–6. [Google Scholar] [CrossRef]
- Johnson, J.A.; Bitra, K.; Zhang, S.; Wang, L.; Lynn, D.E.; Strand, M.R. The UGA-CiE1 cell line from Chrysodeixis includens exhibits characteristics of granulocytes and is permissive to infection by two viruses. Insect Biochem. Mol. Biol. 2010, 40, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Wittwer, D.; Weise, C.; Gotz, P.; Wiesner, A. LPS (lipopolysaccharide)-activated immune responses in a hemocyte cell line from Estigmene acraea (Lepidoptera). Dev. Comp. Immunol. 1997, 21, 323–336. [Google Scholar] [CrossRef]
- Stanley, D.W.; Goodman, C.; An, S.; McIntosh, A.; Song, Q. Prostaglandins A1 and E1 influence gene expression in an established insect cell line (BCIRL-HzAM1 cells). Insect Biochem. Mol. Biol. 2008, 38, 275–284. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.W.; Goodman, C.; An, S.; Song, Q. Prostaglandin A2 influences gene expression in an established insect cell line (BCIRL-HzAM1) cells. J. Insect Physiol. 2012, 58, 837–849. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.; Goodman, C.L.; Ringbauer, J.A.; Song, Q. Prostaglandins influence protein phosphorylation in established insect cell line. Arch. Insect Biochem. Physiol. 2020, 105, e21725. [Google Scholar] [CrossRef]
- Breitenbach, J.E.; Popham, H.J. Baculovirus replication induces the expression of heat shock proteins in vivo and in vitro. Arch. Virol. 2013, 158, 1517–1522. [Google Scholar] [CrossRef]
- Yang, H.-N.; Lo, C.-F.; Lin, C.-Y.; Tsae, P.-F.; Wang, C.-H. In vitro phagocytosis of occlusion bodies of nucleopolyhedroviruses by insect cell lines. Appl. Entomol. Zool. 2001, 36, 59–69. [Google Scholar] [CrossRef][Green Version]
- Lapointe, J.F.; Dunphy, G.B.; Giannoulis, P.; Mandato, C.A.; Nardi, J.B.; Gharib, O.H.; Niven, D.F. Cell lines, Md108 and Md66, from the hemocytes of Malacosoma disstria (Lepidoptera) display aspects of plasma-free innate non-self activities. J. Invertebr. Pathol. 2011, 108, 180–193. [Google Scholar] [CrossRef]
- Rao, X.J.; Xu, X.X.; Yu, X.Q. Manduca sexta moricin promoter elements can increase promoter activities of Drosophila melanogaster antimicrobial peptide genes. Insect Biochem. Mol. Biol. 2011, 41, 982–992. [Google Scholar] [CrossRef] [PubMed]
- Mehrabadi, M.; Hussain, M.; Matindoost, L.; Asgari, S.; Sandri-Goldin, R.M. The baculovirus antiapoptotic p35 protein functions as an inhibitor of the host RNA interference antiviral response. J. Virol. 2015, 89, 8182–8192. [Google Scholar] [CrossRef] [PubMed]
- Mehrabadi, M.; Hussain, M.; Asgari, S. MicroRNAome of Spodoptera frugiperda cells (Sf9) and its alteration following baculovirus infection. J. Gen. Virol. 2013, 94, 1385–1397. [Google Scholar] [CrossRef] [PubMed]
- Karamipour, N.; Fathipour, Y.; Talebi, A.A.; Asgari, S.; Mehrabadi, M. Small interfering RNA pathway contributes to antiviral immunity in Spodoptera frugiperda (Sf9) cells following Autographa californica multiple nucleopolyhedrovirus infection. Insect Biochem. Mol. Biol. 2018, 101, 24–31. [Google Scholar] [CrossRef]
- Svensson, I.; Calles, K.; Lindskog, E.; Henriksson, H.; Eriksson, U.; Haggstrom, L. Antimicrobial activity of conditioned medium fractions from Spodoptera frugiperda Sf9 and Trichoplusia ni Hi5 insect cells. Appl. Microbiol. Biotechnol. 2005, 69, 92–98. [Google Scholar] [CrossRef]
- Beck, M.H.; Strand, M.R. A novel polydnavirus protein inhibits the insect prophenoloxidase activation pathway. Proc. Natl. Acad. Sci. USA 2007, 104, 19267–19272. [Google Scholar] [CrossRef]
- Kurtti, T.J.; Keyhani, N.O. Intracellular infection of tick cell lines by the entomopathogenic fungus Metarhizium anisopliae. Microbiology 2008, 154, 1700–1709. [Google Scholar] [CrossRef][Green Version]
- Teixeira, R.C.; Baeta, B.A.; Ferreira, J.S.; Medeiros, R.C.; Maya-Monteiro, C.M.; Lara, F.A.; Bell-Sakyi, L.; Fonseca, A.H. Fluorescent membrane markers elucidate the association of Borrelia burgdorferi with tick cell lines. Braz. J. Med. Biol. Res. 2016, 49. [Google Scholar] [CrossRef]
- Mattila, J.T.; Munderloh, U.G.; Kurtti, T.J. Phagocytosis of the Lyme disease spirochete, Borrelia burgdorferi, by cells from the ticks, Ixodes scapularis and Dermacentor andersoni, infected with an endosymbiont, Rickettsia peacockii. J. Insect Sci. 2007, 7, 58. [Google Scholar] [CrossRef]
- Simser, J.A.; Macaluso, K.R.; Mulenga, A.; Azad, A.F. Immune-responsive lysozymes from hemocytes of the American dog tick, Dermacentor variabilis and an embryonic cell line of the Rocky Mountain wood tick, D. andersoni. Insect Biochem. Mol. Biol. 2004, 34, 1235–1246. [Google Scholar] [CrossRef]
- Tonk, M.; Cabezas-Cruz, A.; Valdes, J.J.; Rego, R.O.; Rudenko, N.; Golovchenko, M.; Bell-Sakyi, L.; de la Fuente, J.; Grubhoffer, L. Identification and partial characterisation of new members of the Ixodes ricinus defensin family. Gene 2014, 540, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Weisheit, S.; Villar, M.; Tykalova, H.; Popara, M.; Loecherbach, J.; Watson, M.; Ruzek, D.; Grubhoffer, L.; de la Fuente, J.; Fazakerley, J.K.; et al. Ixodes scapularis and Ixodes ricinus tick cell lines respond to infection with tick-borne encephalitis virus: Transcriptomic and proteomic analysis. Parasites Vectors 2015, 8, 599. [Google Scholar] [CrossRef]
- Garcia, S.; Billecocq, A.; Crance, J.M.; Prins, M.; Garin, D.; Bouloy, M. Viral suppressors of RNA interference impair RNA silencing induced by a Semliki Forest virus replicon in tick cells. J. Gen. Virol. 2006, 87, 1985–1989. [Google Scholar] [CrossRef] [PubMed]
- Esteves, E.; Lara, F.A.; Lorenzini, D.M.; Costa, G.H.; Fukuzawa, A.H.; Pressinotti, L.N.; Silva, J.R.; Ferro, J.A.; Kurtti, T.J.; Munderloh, U.G.; et al. Cellular and molecular characterization of an embryonic cell line (BME26) from the tick Rhipicephalus (Boophilus) microplus. Insect Biochem. Mol. Biol. 2008, 38, 568–580. [Google Scholar] [CrossRef] [PubMed]
- Tootle, T.L.; Spradling, A.C. Drosophila Pxt: A cyclooxygenase-like facilitator of follicle maturation. Development 2008, 135, 839–847. [Google Scholar] [CrossRef]
- Park, J.; Stanley, D.; Kim, Y. Roles of Peroxinectin in PGE2-Mediated Cellular Immunity in Spodoptera exigua. PLoS ONE 2014, 9, e105717. [Google Scholar] [CrossRef]
- Calderwood, S.K.; Gong, J.; Murshid, A. Extracellular HSPs: The complicated roles of extracellular HSPs in immunity. Front. Immunol. 2016, 7, 159. [Google Scholar] [CrossRef]
- Wronska, A.K.; Bogus, M.I. Heat shock proteins (HSP 90, 70, 60, and 27) in Galleria mellonella (Lepidoptera) hemolymph are affected by infection with Conidiobolus coronatus (Entomophthorales). PLoS ONE 2020, 15, e0228556. [Google Scholar] [CrossRef] [PubMed]
- Sandamalika, W.M.G.; Priyathilaka, T.T.; Lee, S.; Yang, H.; Lee, J. Immune and xenobiotic responses of glutathione S-Transferase theta (GST-θ) from marine invertebrate disk abalone (Haliotis discus discus): With molecular characterization and functional analysis. Fish. Shellfish Immunol. 2019, 91, 159–171. [Google Scholar] [CrossRef]
- Walker, T.; Jeffries, C.L.; Mansfield, K.L.; Johnson, N. Mosquito cell lines: History, isolation, availability and application to assess the threat of arboviral transmission in the United Kingdom. Parasites Vectors 2014, 7, 382. [Google Scholar] [CrossRef]
- Gammon, D.B.; Mello, C.C. RNA interference-mediated antiviral defense in insects. Curr. Opin. Insect Sci. 2015, 8, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Leggewie, M.; Schnettler, E. RNAi-mediated antiviral immunity in insects and their possible application. Curr. Opin. Virol. 2018, 32, 108–114. [Google Scholar] [CrossRef]
- Xu, J.; Hopkins, K.; Sabin, L.; Yasunaga, A.; Subramanian, H.; Lamborn, I.; Gordesky-Gold, B.; Cherry, S. ERK signaling couples nutrient status to antiviral defense in the insect gut. Proc. Natl. Acad. Sci. USA 2013, 110, 15025–15030. [Google Scholar] [CrossRef] [PubMed]
- Ahlers, L.R.H.; Trammell, C.E.; Carrell, G.F.; Mackinnon, S.; Torrevillas, B.K.; Chow, C.Y.; Luckhart, S.; Goodman, A.G. Insulin potentiates JAK/STAT signaling to broadly inhibit flavivirus replication in insect vectors. Cell Rep. 2019, 29, 1946–1960.e1945. [Google Scholar] [CrossRef]
- Hillyer, J.F.; Strand, M.R. Mosquito hemocyte-mediated immune responses. Curr. Opin. Insect Sci. 2014, 3, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Wittwer, C.T.; Reed, G.H.; Gundry, C.N.; Vandersteen, J.G.; Pryor, R.J. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin. Chem. 2003, 49, 853–860. [Google Scholar] [CrossRef] [PubMed]
- Nairz, M.; Weiss, G. Iron in infection and immunity. Mol. Asp. Med. 2020, 75, 100864. [Google Scholar] [CrossRef] [PubMed]
- Patel, S. A critical review on serine protease: Key immune manipulator and pathology mediator. Allergol Immunopathol. 2017, 45, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Stuart, T.; Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 2019, 20, 257–272. [Google Scholar] [CrossRef]
- Merkling, S.H.; Lambrechts, L. Taking insect immunity to the single-cell level. Trends Immunol. 2020, 41, 190–199. [Google Scholar] [CrossRef]
- Tachibana, C. Beyond CRISPR: What’s current and upcoming in genome editing. Science 2019, 365, 1481–1483. [Google Scholar] [CrossRef][Green Version]
- Li, H.; Yang, Y.; Hong, W.; Huang, M.; Wu, M.; Zhao, X. Applications of genome editing technology in the targeted therapy of human diseases: Mechanisms, advances and prospects. Signal. Transduct. Target. Ther. 2020, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Reardon, S. Step aside CRISPR, RNA editing is taking off. Nature 2020, 578, 24–27. [Google Scholar] [CrossRef] [PubMed]
| Term | Abbreviation |
|---|---|
| anti-microbial peptides | AMPs |
| Argonaute | Ago |
| Dicer2 | Dcr2 |
| Double-stranded RNA | dsRNA |
| Extracellular receptor kinase | ERK |
| Heat shock protein | HSP |
| Immune deficiency | IMD |
| Janus Kinase and Signal Transducer and Activator of Transcription | JAK/STAT |
| Jun N-terminal Kinase | JNK |
| Lipopolysaccharides | LPS |
| Mitogen-Activated Protein Kinase | MAPK |
| Pathogen-associated molecular pattern | PAMP |
| Pattern recognition receptor | PRRs |
| Peptidoglycan | PGN |
| Phenoloxidase or prophenoloxidase | PO or PPO |
| Piwi RNA | piRNA |
| Prostaglandin | PG |
| Post-infection | PI |
| Relish | REL |
| small interfering RNA | siRNA |
| Single-stranded RNA | ssRNA |
| RNA Interference | RNAi |
| Small RNA | sRNA |
| thiol-ester motif-containing protein | TEP |
| viral-derived piwi-associated RNA | vpiRNA |
| viral small interfering RNAs | vsiRNA |
| Order | Species of Origin | Stage/Tissue of Origin | Cell Line Designation | Research Focus | References |
|---|---|---|---|---|---|
| Parasitiformes | Amblyomma americanum | Embryo | AAE2 | Cellular Responses | [89] |
| Amblyomma variegatum | Molting larva | AVL/CTVM17 | Cellular Responses | [90] | |
| Dermacentor andersoni | Embryo | DAE15 | Cellular Responses | [91,92] | |
| D. andersoni | Embryo | DAE100 | Humoral Responses | [92] | |
| Hyalomma anatolicum | Embryo | HAE/CTVM8 | Cellular Responses | [90] | |
| Ixodes ricinus | Embryo | IRE/CTVM19 | Cellular Responses | [90,93,94] | |
| Ixodes scapularis | Embryo | IDE8 | Cellular Responses, Humoral Responses | [90,94] | |
| I. scapularis | Embryo | IDE12 | Cellular Responses | [89,91] | |
| I. scapularis | Embryo | ISE6 | PAMPs, Signaling Pathways, Cellular Responses | [36,90,91,95] | |
| Ixodes ricinus | Embryo | IRE/CTVM19 | Humoral Responses | [93,94] | |
| Rhipicephalus appendiculatus | Molting nymph | RA243 | Cellular Responses | [90] | |
| R. appendiculatus | Embryo | RAE/CTVM1 | Cellular Responses | [90] | |
| Rhipicephalus (Boophilus) microplus | Embryo | BME/CTVM2 | Cellular Responses | [90] | |
| R. microplus | Embryo | BME26 | Cellular Responses, Humoral Responses | [96] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goodman, C.L.; Kang, D.S.; Stanley, D. Cell Line Platforms Support Research into Arthropod Immunity. Insects 2021, 12, 738. https://doi.org/10.3390/insects12080738
Goodman CL, Kang DS, Stanley D. Cell Line Platforms Support Research into Arthropod Immunity. Insects. 2021; 12(8):738. https://doi.org/10.3390/insects12080738
Chicago/Turabian StyleGoodman, Cynthia L., David S. Kang, and David Stanley. 2021. "Cell Line Platforms Support Research into Arthropod Immunity" Insects 12, no. 8: 738. https://doi.org/10.3390/insects12080738
APA StyleGoodman, C. L., Kang, D. S., & Stanley, D. (2021). Cell Line Platforms Support Research into Arthropod Immunity. Insects, 12(8), 738. https://doi.org/10.3390/insects12080738






