Further Evidence of Population Admixture in the Serbian Honey Bee Population
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. DNA Extraction and PCR-RFLP Analysis
2.3. Microsatellite Analysis
2.4. Fragment Analysis
2.5. Statistical Analyses
3. Results
3.1. PCR-RFLP
3.2. Genetic Diversity Analysis
3.2.1. Genetic Diversity Analysis for 14 Microsatellite Loci in the Serbian Sample
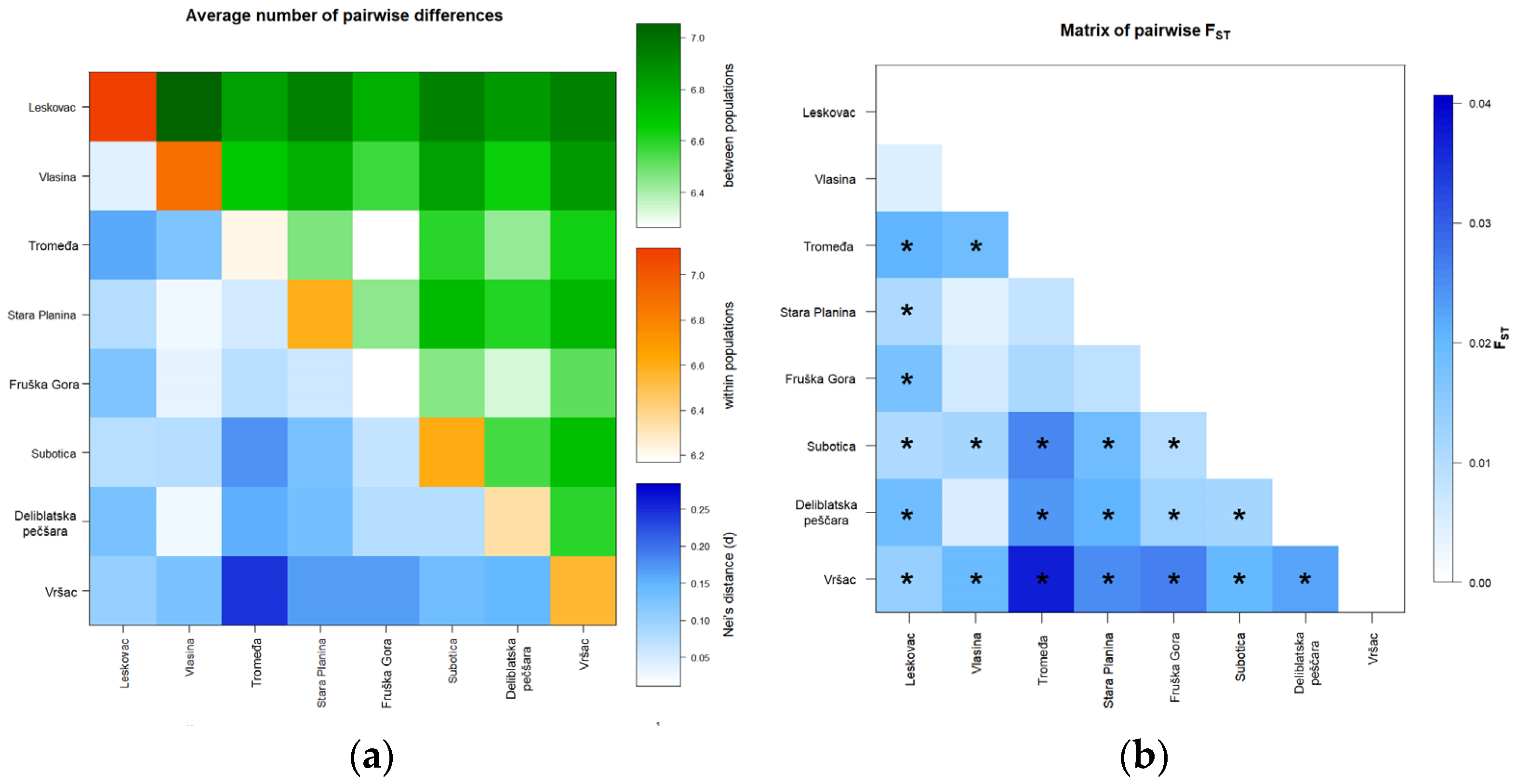
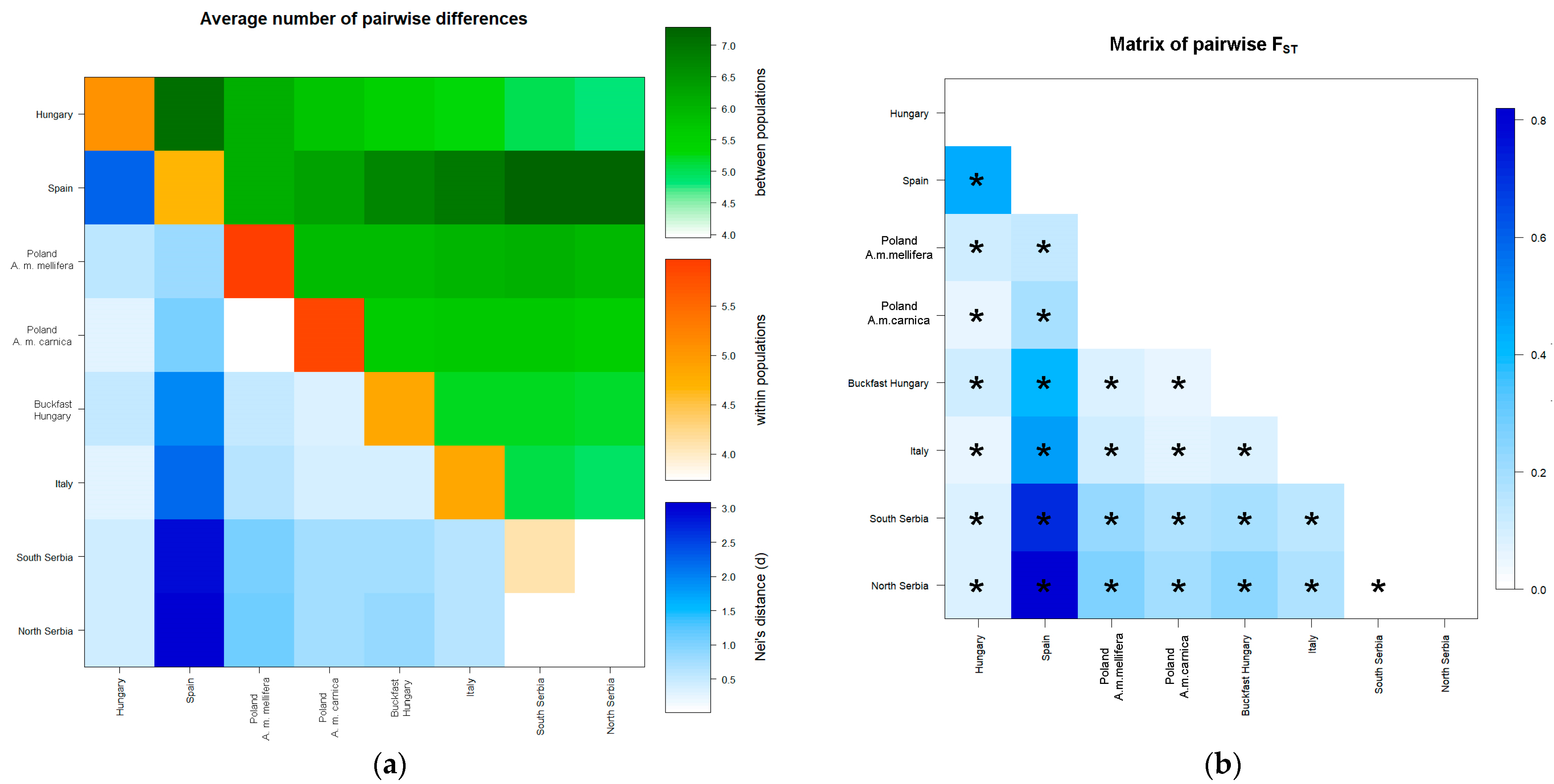
3.2.2. Population Genetic Analysis for Nine Microsatellite Loci in All Sampled Localities
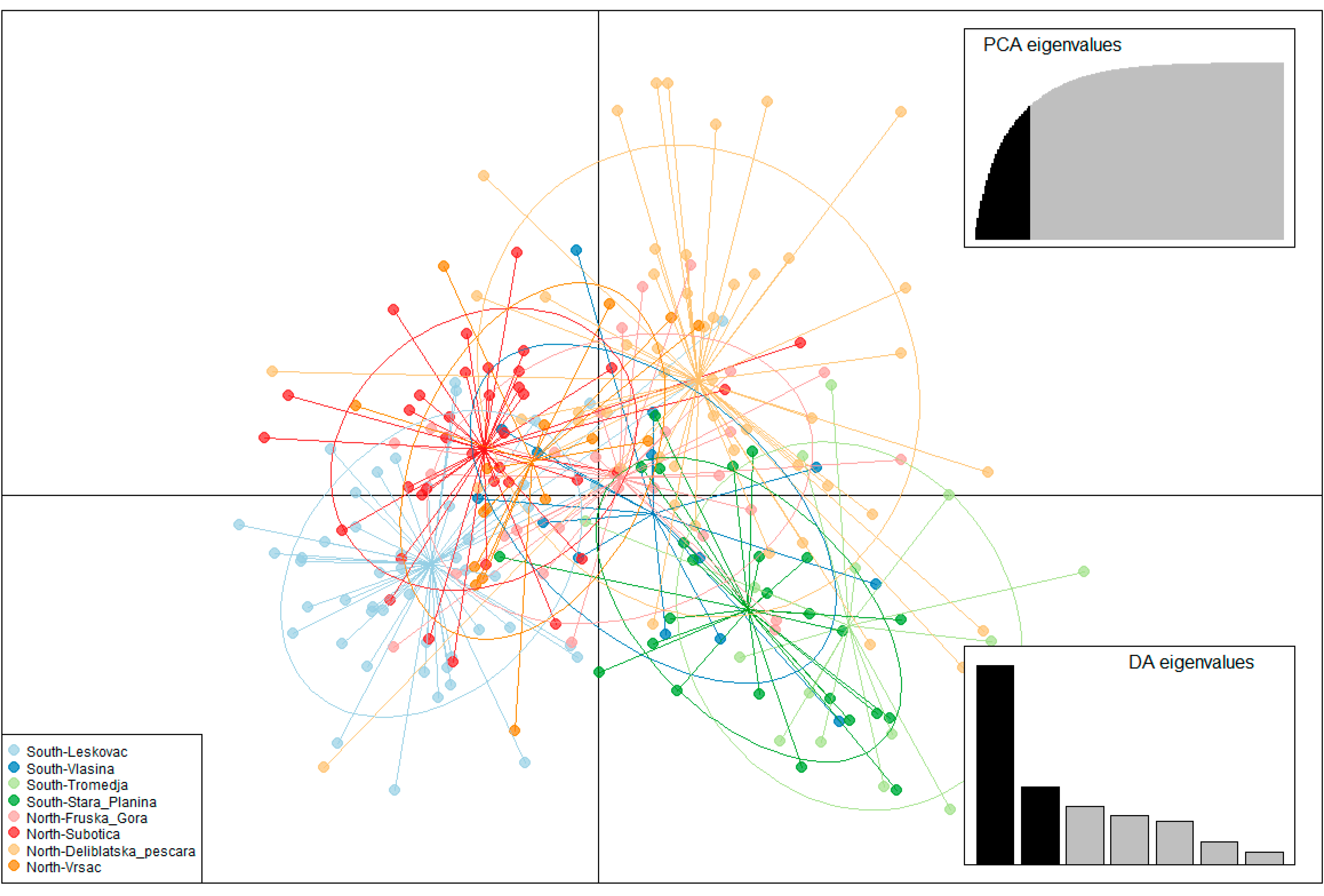
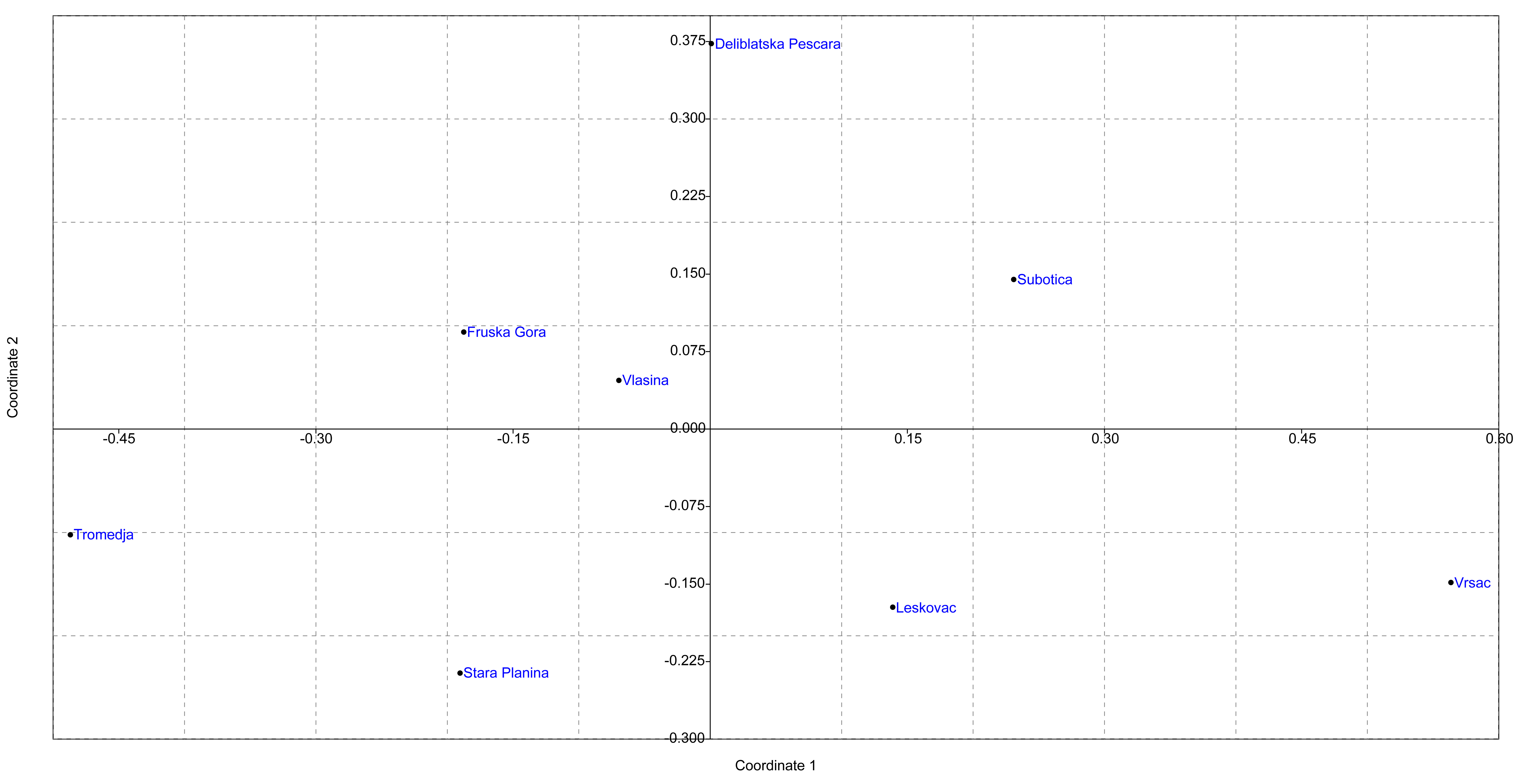
3.3. Population Structure
3.3.1. Population Structure Based on 14 Microsatellite Loci in the Serbian Sample
3.3.2. Population Structure Based on Nine Microsatellite Loci in All Sampled Localities
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Palmer, K.A.; Oldroyd, B.P. Evolution of multiple mating in the genus Apis. Apidologie 2000, 31, 235–248. [Google Scholar] [CrossRef] [Green Version]
- Koeniger, G. Reproduction and mating behavior. In Bee Genetics and Breeding; Elsevier: Amsterdam, The Netherlands, 1986; pp. 255–280. [Google Scholar]
- Kraus, F.; Neumann, P.; Moritz, R. Genetic variance of mating frequency in the honeybee (Apis mellifera L.). Insectes Sociaux 2005, 52, 1–5. [Google Scholar] [CrossRef]
- Leclercq, G.; Gengler, N.; Francis, F. How human reshaped diversity in honey bees (Apis mellifera L.): A review. Entomol. Faun.-Faun. Entomol. 2018, 71, 1–13. [Google Scholar]
- Ruttner, F. Morphometric Analysis and Classification. In Biogeography and Taxonomy of Honeybees; Springer: Berlin/Heidelberg, Germany, 1988; pp. 66–78. [Google Scholar]
- Garnery, L.; Cornuet, J.M.; Solignac, M. Evolutionary history of the honey bee Apis mellifera inferred from mitochondrial DNA analysis. Mol. Ecol. 1992, 1, 145–154. [Google Scholar] [CrossRef]
- Franck, P.; Garnery, L.; Celebrano, G.; Solignac, M.; Cornuet, J.M. Hybrid origins of honeybees from Italy (Apis mellifera ligustica) and Sicily (A. m. sicula). Mol. Ecol. 2000, 9, 907–921. [Google Scholar] [CrossRef]
- Whitfield, C.W.; Behura, S.K.; Berlocher, S.H.; Clark, A.G.; Johnston, J.S.; Sheppard, W.S.; Smith, D.R.; Suarez, A.V.; Weaver, D.; Tsutsui, N.D. Thrice out of Africa: Ancient and recent expansions of the honey bee, Apis mellifera. Science 2006, 314, 642–645. [Google Scholar] [CrossRef] [Green Version]
- Han, F.; Wallberg, A.; Webster, M.T. From where did the W estern honeybee (Apis mellifera) originate? Ecol. Evol. 2012, 2, 1949–1957. [Google Scholar] [CrossRef]
- Tihelka, E.; Cai, C.; Pisani, D.; Donoghue, P.C. Mitochondrial genomes illuminate the evolutionary history of the Western honey bee (Apis mellifera). Sci. Rep. 2020, 10, 14515. [Google Scholar] [CrossRef]
- Cridland, J.M.; Tsutsui, N.D.; Ramírez, S.R. The complex demographic history and evolutionary origin of the western honey bee, Apis mellifera. Genome Biol. Evol. 2017, 9, 457–472. [Google Scholar] [CrossRef] [Green Version]
- Dogantzis, K.A.; Tiwari, T.; Conflitti, I.M.; Dey, A.; Patch, H.M.; Muli, E.M.; Garnery, L.; Whitfield, C.W.; Stolle, E.; Alqarni, A.S. Thrice out of Asia and the adaptive radiation of the western honey bee. Sci. Adv. 2021, 7, eabj2151. [Google Scholar] [CrossRef]
- Ilyasov, R.A.; Lee, M.-l.; Takahashi, J.-I.; Kwon, H.W.; Nikolenko, A.G. A revision of subspecies structure of western honey bee Apis mellifera. Saudi J. Biol. Sci. 2020, 27, 3615–3621. [Google Scholar] [CrossRef] [PubMed]
- Wallberg, A.; Han, F.; Wellhagen, G.; Dahle, B.; Kawata, M.; Haddad, N.; Simões, Z.L.P.; Allsopp, M.H.; Kandemir, I.; De la Rúa, P. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nat. Genet. 2014, 46, 1081–1088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gruber, K.; Schöning, C.; Otte, M.; Kinuthia, W.; Hasselmann, M. Distinct subspecies or phenotypic plasticity? Genetic and morphological differentiation of mountain honey bees in East Africa. Ecol. Evol. 2013, 3, 3204–3218. [Google Scholar] [CrossRef] [PubMed]
- Wallberg, A.; Schoening, C.; Webster, M.T.; Hasselmann, M. Two extended haplotype blocks are associated with adaptation to high altitude habitats in East African honey bees. PLoS Genet. 2017, 13, e1006792. [Google Scholar] [CrossRef]
- Oleksa, A.; Wilde, J.; Tofilski, A.; Chybicki, I.J. Partial reproductive isolation between European subspecies of honey bees. Apidologie 2013, 44, 611–619. [Google Scholar] [CrossRef] [Green Version]
- Oleksa, A.; Chybicki, I.; Tofilski, A.; Burczyk, J. Nuclear and mitochondrial patterns of introgression into native dark bees (Apis mellifera mellifera) in Poland. J. Apic. Res. 2011, 50, 116–129. [Google Scholar] [CrossRef] [Green Version]
- Dall’Olio, R.; Marino, A.; Lodesani, M.; Moritz, R.F. Genetic characterization of Italian honeybees, Apis mellifera ligustica, based on microsatellite DNA polymorphisms. Apidologie 2007, 38, 207–217. [Google Scholar] [CrossRef] [Green Version]
- Jensen, A.B.; Palmer, K.A.; Boomsma, J.J.; Pedersen, B.V. Varying degrees of Apis mellifera ligustica introgression in protected populations of the black honeybee, Apis mellifera mellifera, in northwest Europe. Mol. Ecol. 2005, 14, 93–106. [Google Scholar] [CrossRef]
- Soland-Reckeweg, G.; Heckel, G.; Neumann, P.; Fluri, P.; Excoffier, L. Gene flow in admixed populations and implications for the conservation of the Western honeybee, Apis mellifera. J. Insect Conserv. 2009, 13, 317–328. [Google Scholar] [CrossRef] [Green Version]
- Oleksa, A.; Kusza, S.; Tofilski, A. Mitochondrial DNA Suggests the Introduction of Honeybees of African Ancestry to East-Central Europe. Insects 2021, 12, 410. [Google Scholar] [CrossRef]
- Muñoz, I.; De la Rúa, P. Wide genetic diversity in Old World honey bees threaten by introgression. Apidologie 2021, 52, 200–217. [Google Scholar] [CrossRef]
- Uzunov, A.; Meixner, M.D.; Kiprijanovska, H.; Andonov, S.; Gregorc, A.; Ivanova, E.; Bouga, M.; Dobi, P.; Büchler, R.; Francis, R. Genetic structure of Apis mellifera macedonica in the Balkan Peninsula based on microsatellite DNA polymorphism. J. Apic. Res. 2014, 53, 288–295. [Google Scholar] [CrossRef]
- Muñoz, I.; Dall’Olio, R.; Lodesani, M.; De la Rúa, P. Population genetic structure of coastal Croatian honeybees (Apis mellifera carnica). Apidologie 2009, 40, 617–626. [Google Scholar] [CrossRef] [Green Version]
- Nedić, N.; Francis, R.M.; Stanisavljević, L.; Pihler, I.; Kezić, N.; Bendixen, C.; Kryger, P. Detecting population admixture in honey bees of Serbia. J. Apic. Res. 2014, 53, 303–313. [Google Scholar] [CrossRef]
- Lodesani, M.; Costa, C. Bee breeding and genetics in Europe. Bee World 2003, 84, 69–85. [Google Scholar] [CrossRef]
- Bouga, M.; Alaux, C.; Bienkowska, M.; Büchler, R.; Carreck, N.L.; Cauia, E.; Chlebo, R.; Dahle, B.; Dall’Olio, R.; De la Rúa, P. A review of methods for discrimination of honey bee populations as applied to European beekeeping. J. Apic. Res. 2011, 50, 51–84. [Google Scholar] [CrossRef] [Green Version]
- Oleksa, A.; Gawroński, R.; Tofilski, A. Rural avenues as a refuge for feral honey bee population. J. Insect Conserv. 2013, 17, 465–472. [Google Scholar] [CrossRef] [Green Version]
- Kohl, P.L.; Rutschmann, B. The neglected bee trees: European beech forests as a home for feral honey bee colonies. PeerJ 2018, 6, e4602. [Google Scholar] [CrossRef] [Green Version]
- Requier, F.; Garnery, L.; Kohl, P.L.; Njovu, H.K.; Pirk, C.W.; Crewe, R.M.; Steffan-Dewenter, I. The conservation of native honey bees is crucial. Trends Ecol. Evol. 2019, 34, 789–798. [Google Scholar] [CrossRef]
- Bila Dubaić, J.; Simonović, S.; Plećaš, M.; Stanisavljević, L.; Davidović, S.; Tanasković, M.; Ćetković, A. Unprecedented Density and Persistence of Feral Honey Bees in Urban Environments of a Large SE-European City (Belgrade, Serbia). Insects 2021, 12, 1127. [Google Scholar] [CrossRef]
- Harpur, B.A.; Minaei, S.; Kent, C.F.; Zayed, A. Management increases genetic diversity of honey bees via admixture. Mol. Ecol. 2012, 21, 4414–4421. [Google Scholar] [CrossRef] [PubMed]
- Harpur, B.A.; Minaei, S.; Kent, C.F.; Zayed, A. Admixture increases diversity in managed honey bees: Reply to De la Rúa et al. (2013). Mol. Ecol. 2013, 22, 3211–3215. [Google Scholar] [PubMed]
- Francis, R.M.; Kryger, P.; Meixner, M.; Bouga, M.; Ivanova, E.; Andonov, S.; Berg, S.; Bienkowska, M.; Büchler, R.; Charistos, L. The genetic origin of honey bee colonies used in the COLOSS Genotype-Environment Interactions Experiment: A comparison of methods. J. Apic. Res. 2014, 53, 188–204. [Google Scholar] [CrossRef]
- Bienefeld, K. Breeding success or genetic diversity in honey bees? Bee World 2016, 93, 40–44. [Google Scholar] [CrossRef]
- De la Rúa, P.; Jaffé, R.; Dall’Olio, R.; Muñoz, I.; Serrano, J. Biodiversity, conservation and current threats to European honeybees. Apidologie 2009, 40, 263–284. [Google Scholar] [CrossRef] [Green Version]
- De la Rúa, P.; Jaffé, R.; Muñoz, I.; Serrano, J.; Moritz, R.F.; Kraus, F.B. Conserving genetic diversity in the honeybee: Comments on Harpur et al. (2012). Mol. Ecol. 2013, 22, 3208–3210. [Google Scholar] [PubMed]
- Meixner, M.D.; Costa, C.; Kryger, P.; Hatjina, F.; Bouga, M.; Ivanova, E.; Büchler, R. Conserving diversity and vitality for honey bee breeding. J. Apic. Res. 2010, 49, 85–92. [Google Scholar] [CrossRef]
- Eimanifar, A.; Brooks, S.A.; Bustamante, T.; Ellis, J.D. Population genomics and morphometric assignment of western honey bees (Apis mellifera L.) in the Republic of South Africa. BMC Genom. 2018, 19, 615. [Google Scholar] [CrossRef]
- Eimanifar, A.; Pieplow, J.T.; Asem, A.; Ellis, J.D. Genetic diversity and population structure of two subspecies of western honey bees (Apis mellifera L.) in the Republic of South Africa as revealed by microsatellite genotyping. PeerJ 2020, 8, e8280. [Google Scholar] [CrossRef] [Green Version]
- Bouga, M.; Harizanis, P.C.; Kilias, G.; Alahiotis, S. Genetic divergence and phylogenetic relationships of honey bee Apis mellifera (Hymenoptera: Apidae) populations from Greece and Cyprus using PCR–RFLP analysis of three mtDNA segments. Apidologie 2005, 36, 335–344. [Google Scholar] [CrossRef] [Green Version]
- Sušnik, S.; Kozmus, P.; Poklukar, J.; Meglic, V. Molecular characterisation of indigenous Apis mellifera carnica in Slovenia. Apidologie 2004, 35, 623–636. [Google Scholar] [CrossRef] [Green Version]
- Nedić, N.; Stanisavljević, L.; Mladenović, M.; Stanisavljević, J. Molecular characterization of the honeybee Apis mellifera carnica in Serbia. Arch. Biol. Sci. 2009, 61, 587–598. [Google Scholar] [CrossRef]
- Munoz, I.; Stevanović, J.; Stanimirović, Z.; De la Rua, P. Genetic variation of Apis mellifera from Serbia inferred from mitochondrial analysis. J. Apic. Sci. 2012, 56, 59–69. [Google Scholar]
- Tanasković, M.; Erić, P.; Patenković, A.; Erić, K.; Mihajlović, M.; Tanasić, V.; Stanisavljević, L.; Davidović, S. MtDNA Analysis Indicates Human-Induced Temporal Changes of Serbian Honey Bees Diversity. Insects 2021, 12, 767. [Google Scholar] [CrossRef] [PubMed]
- Stanimirovic, Z.; Stevanovic, J.; Andjelkovic, M. Chromosomal diversity in Apis mellifera carnica from Serbia. Apidologie 2005, 36, 31–42. [Google Scholar] [CrossRef] [Green Version]
- Stevanovic, J. Investigations of Morphometric and Chromosomal Variability in Diversity Preserving of Carniolan Honey Bee (Apis mellifera carnica Pollmann, 1879) in Serbia. Master’s Thesis, Belgrade University, Belgrade, Serbia, 2002. [Google Scholar]
- Stanimirović, Z.Ž.; Pejović, D.; Stevanović, J.B.; Vučinić, M.M.; Mirilović, M. Investigations of hygienic behaviour and disease resistance in organic beekeeping of two honeybee ecogeographic varieties from Serbia. Acta Vet. 2002, 52, 169–179. [Google Scholar]
- Stanimirović, Z.Ž.; Stevanović, J.B.; Ćirković, D. Behavioural defenses of the honey bee ecotype from Sjenica–Pešter against Varroa destructor. Acta Vet. 2005, 55, 69–82. [Google Scholar]
- Kozmus, P.; Stevanović, J.; Stanimirović, Z.; Stojić, V.; Kulišić, Z.; Meglič, V. Analysis of mitochondrial DNA in honey bees (Apis mellifera) from Serbia. Acta Vet. Beogr. 2007, 57, 465–476. [Google Scholar]
- Stanimirović, Z.; Vučinić, M.; Stevanović, J. Biodiversity of the honeybee Apis mellifera, linne (1758), from some Yugoslav regions. II-Ultrastructural chromosomal differences between banat and the Syenichko-Peshterski honeybee ecotypes. Acta Vet. Beogr. 1999, 49, 207–214. [Google Scholar]
- Stevanovic, J.; Stanimirovic, Z.; Radakovic, M.; Kovacevic, S. Biogeographic study of the honey bee (Apis mellifera L.) from Serbia, Bosnia and Herzegovina and Republic of Macedonia based on mitochondrial DNA analyses. Russ. J. Genet. 2010, 46, 603–609. [Google Scholar] [CrossRef]
- Serbia, Statistical Office of the Republic of Serbia. Available online: https://data.stat.gov.rs/Home/Re-sult/130202010207?languageCode=en-US (accessed on 29 December 2021).
- The Official Gazzete of Republic of Serbia, Nos. 41/2009, 93/2012 and 14/2016, Serbia. In Law on Animal Breeding; 2009. Available online: https://www.vet.minpolj.gov.rs/legislativa/zakoni/Zakon%20o%20sto%C4%8Darstvu.pdf (accessed on 14 July 2021). (In Serbian)
- Garnery, L.; Franck, P.; Baudry, E.; Vautrin, D.; Cornuet, J.-M.; Solignac, M. Genetic diversity of the west European honey bee (Apis mellifera mellifera and A. m. iberica) II. Microsatellite loci. Genet. Sel. Evol. 1998, 30, S49–S74. [Google Scholar] [CrossRef]
- Kandemir, I.; Meixner, M.D.; Ozkan, A.; Sheppard, W.S. Genetic characterization of honey bee (Apis mellifera cypria) populations in northern Cyprus. Apidologie 2006, 37, 547–555. [Google Scholar] [CrossRef] [Green Version]
- Péntek-Zakar, E.; Oleksa, A.; Borowik, T.; Kusza, S. Population structure of honey bees in the Carpathian Basin (Hungary) confirms introgression from surrounding subspecies. Ecol. Evol. 2015, 5, 5456–5467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Techer, M.A.; Clémencet, J.; Simiand, C.; Turpin, P.; Garnery, L.; Reynaud, B.; Delatte, H. Genetic diversity and differentiation among insular honey bee populations in the southwest Indian Ocean likely reflect old geographical isolation and modern introductions. PLoS ONE 2017, 12, e0189234. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, D.I.; Ebert, P.R.; Hunt, G.J.; Guzmán-Novoa, E.; Kinnee, S.A.; Page, R.E., Jr. Identification of Africanized honey bees (Hymenoptera: Apidae) incorporating morphometrics and an improved polymerase chain reaction mitotyping procedure. Ann. Entomol. Soc. Am. 1999, 92, 167–174. [Google Scholar] [CrossRef]
- Tanasković, M.; Patenković, A.; Erić, K.; Erić, P.; Stanisavljević, L.; Davidović, S. Microsatellite analysis of Apis mellifera from Northern and Southern parts of Serbia. In Proceedings of the 1st International Electronic Conference on Entomology Session Apiculture and Pollinators, Online, 1–15 July 2021. [Google Scholar]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Kalinowski, S.T. hp-rare 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Notes 2005, 5, 187–189. [Google Scholar] [CrossRef]
- Stoneking, M.; Hedgecock, D.; Higuchi, R.G.; Vigilant, L.; Erlich, H.A. Population variation of human mtDNA control region sequences detected by enzymatic amplification and sequence-specific oligonucleotide probes. Am. J. Hum. Genet. 1991, 48, 370–382. [Google Scholar]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [CrossRef]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Resour. 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [Green Version]
- Ward, J.H., Jr. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- De La Rúa, P.; Galián, J.; Serrano, J.; Moritz, R.F. Genetic structure and distinctness of Apis mellifera L. populations from the Canary Islands. Mol. Ecol. 2001, 10, 1733–1742. [Google Scholar] [CrossRef]
- Coroian, C.O.; Muñoz, I.; Schlüns, E.A.; Paniti-Teleky, O.R.; Erler, S.; Furdui, E.M.; Mărghitaş, L.A.; Dezmirean, D.S.; Schlüns, H.; De La Rua, P. Climate rather than geography separates two E uropean honeybee subspecies. Mol. Ecol. 2014, 23, 2353–2361. [Google Scholar] [CrossRef]


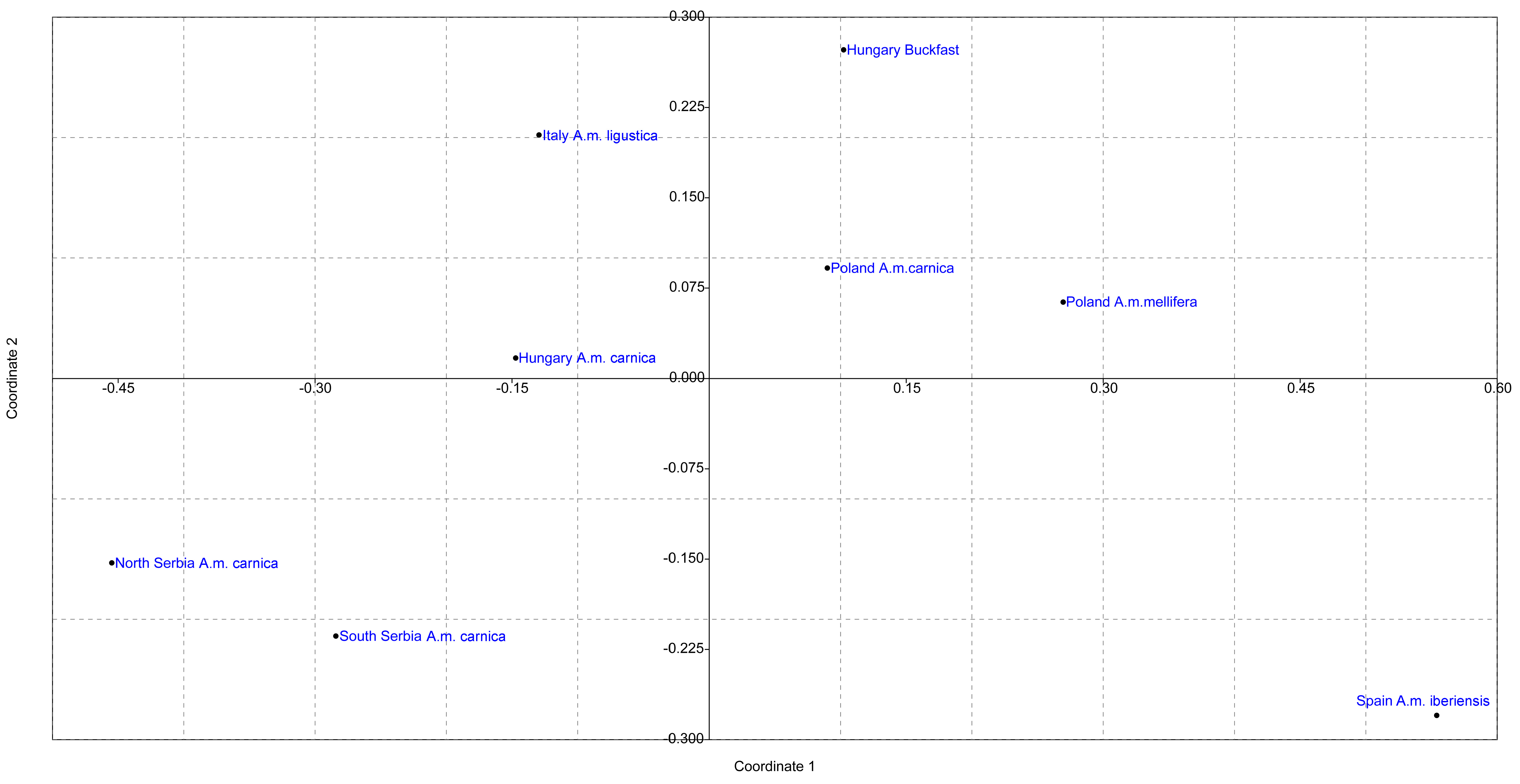

| Locality | N | Na | Agd | Ho | He | Ar | RMP | MPD | G-W | Ar8 | Apr |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Leskovac (L) | 51 | 9 | 13.571 | 0.5984 | 0.5617 | 0.5958 | 0.00980 | 7.7786 | 0.6638 | 3.44 | 0.37 |
| Vlasina (V) | 14 | 5.7140 | 8.429 | 0.5584 | 0.5192 | 0.5888 | 0.03570 | 6.7011 | 0.6350 | 3.41 | 0.33 |
| Tromedja (T) | 15 | 5.1430 | 7.929 | 0.5184 | 0.4698 | 0.5397 | 0.03330 | 6.2207 | 0.6605 | 3.05 | 0.21 |
| Stara Planina (SP) | 25 | 6.7860 | 8.643 | 0.5559 | 0.5291 | 0.5785 | 0.02000 | 5.5592 | 0.6890 | 3.38 | 0.35 |
| Fruska Gora (FG) | 29 | 7.0710 | 10.714 | 0.5063 | 0.4662 | 0.5444 | 0.01780 | 5.5693 | 0.6163 | 3.2 | 0.26 |
| Subotica | 37 | 7.2140 | 10 | 0.5510 | 0.5054 | 0.5552 | 0.01350 | 7.7138 | 0.6667 | 3.19 | 0.22 |
| Deliblatska Pescara (DP) | 50 | 8.2140 | 13.071 | 0.5121 | 0.5098 | 0.5734 | 0.01020 | 4.0968 | 0.6096 | 3.3 | 0.34 |
| Vrsac (Vr) | 16 | 5.7860 | 8.500 | 0.5783 | 0.4311 | 0.5674 | 0.03120 | 6.9395 | 0.6516 | 3.28 | 0.35 |
| Locality | N | Na | Agd | Ho | He | Ar | RMP | MPD | G-W | Ar10 | Apr |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Hungary (A. m. carnica) | 237 | 14 | 0.63694 | 0.89613 | 0.65753 | 35 | 0.0025 | 5.7325 | 0.5158 | 3.85 | 0.27 |
| Spain (A. m. iberiensis) | 10 | 5.3 | 0.73895 | 0.81687 | 0.64477 | 9.375 | 0.0500 | 3.6947 | 0.62704 | 4.18 | 1.22 |
| Poland (A. m. mellifera) | 45 | 9.3 | 0.75006 | 0.86408 | 0.75869 | 12.44 | 0.0111 | 6.7506 | 0.70839 | 4.62 | 0.36 |
| Poland Aug forest | 15 | 6.4 | 0.71239 | 0.88148 | 0.71239 | 7.667 | 0.0333 | 6.4115 | 0.74374 | 4.23 | 0.23 |
| Poland Bialowieza | 15 | 7.4 | 0.74738 | 0.82222 | 0.74738 | 11.667 | 0.0333 | 6.7264 | 0.63242 | 4.66 | 0.21 |
| Poland Siedlice | 15 | 6.1 | 0.75603 | 0.88889 | 0.75648 | 7.556 | 0.0333 | 6.0483 | 0.75483 | 4.35 | 0.06 |
| Poland (A. m. carnica) | 21 | 7.6 | 0.72887 | 0.90476 | 0.74671 | 9.000 | 0.0249 | 6.5598 | 0.78886 | 4.49 | 0.37 |
| Poland Krakow | 15 | 6.9 | 0.74253 | 0.9037 | 0.73498 | 8.667 | 0.0333 | 5.9402 | 0.76265 | 4.47 | 0.19 |
| Poland Wroclaw | 6 | 3.8 | 0.70076 | 0.90741 | 0.70932 | 5.222 | 0.0972 | 5.6061 | 0.65743 | 3.63 | 0.03 |
| Buckfast (Hungary) | 10 | 4.3 | 0.63158 | 0.84321 | 0.64419 | 7.556 | 0.05 | 5.0526 | 0.55482 | 3.57 | 0.13 |
| Italy (A. m. ligustica) | 15 | 5.3 | 0.60977 | 0.91111 | 0.6349 | 6.778 | 0.0333 | 4.8782 | 0.69998 | 3.58 | 0.08 |
| Southern Serbia (A. m. carnica) | 105 | 10.2 | 0.51083 | 0.46459 | 0.52209 | 14.67 | 0.0051 | 4.0866 | 0.69461 | 3.47 | 0.48 |
| Serbia Leskovac | 51 | 8.6 | 0.54021 | 0.50215 | 0.53994 | 12.444 | 0.0106 | 4.3217 | 0.68713 | 3.52 | 0.21 |
| Serbia Vlasina | 14 | 5.3 | 0.50970 | 0.44445 | 0.50970 | 8.222 | 0.0357 | 4.5873 | 0.64462 | 3.38 | 0.13 |
| Serbia Tromedja | 15 | 4.8 | 0.42644 | 0.41235 | 0.44805 | 7.889 | 0.0333 | 3.4115 | 0.63851 | 2.98 | 0.08 |
| Serbia Stara Planina | 25 | 6.7 | 0.46898 | 0.42468 | 0.51138 | 9.222 | 0.0208 | 2.8139 | 0.62132 | 3.44 | 0.20 |
| Northern Serbia (A. m. carnica) | 131 | 10.4 | 0.46634 | 0.40063 | 0.47838 | 13.333 | 0.0044 | 3.7307 | 0.72894 | 3.23 | 0.36 |
| Serbia Fruska Gora | 29 | 6 | 0.38355 | 0.38641 | 0.438 | 9.111 | 0.0196 | 2.6848 | 0.5726 | 3.02 | 0.08 |
| Serbia Subotica | 36 | 6.7 | 0.47344 | 0.44136 | 0.47344 | 9.556 | 0.0143 | 4.261 | 0.6193 | 3.11 | 0.06 |
| Serbia Deliblatska Pescara | 50 | 7.9 | 0.3918 | 0.40292 | 0.49063 | 12.889 | 0.0104 | 1.959 | 0.6065 | 3.28 | 0.13 |
| Serbia Vrsac | 16 | 5.2 | 0.49568 | 0.33029 | 0.49716 | 8.667 | 0.0312 | 3.4698 | 0.59755 | 3.23 | 0.26 |
| Source of Variation | d.f. | SS | Variance Components | Percentage of Variation |
|---|---|---|---|---|
| Among localities | 7 | 43.424 | 0.047 | 1.42 (p = 0.005) |
| Among individuals within localities | 229 | 797.685 | 0.184 | 5.49 (p = 0.0001) |
| Within individuals | 237 | 738.685 | 3.116 | 93.1 (p = 0.000) |
| Total | 473 | 1579.61 | 3.347 |
| Source of Variation | d.f. | SS | Variance Components | Percentage of Variation |
|---|---|---|---|---|
| Among localities | 16 | 286.781 | 0.2801 | 10.79 (p = 0.00) |
| Among individuals within localities | 557 | 1034.257 | −0.4601 | −17.72 |
| Within individuals | 574 | 1594.000 | 2.7770 | 106.93 |
| Total | 1147 | 2915.037 | 2.5970 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanasković, M.; Erić, P.; Patenković, A.; Erić, K.; Mihajlović, M.; Tanasić, V.; Kusza, S.; Oleksa, A.; Stanisavljević, L.; Davidović, S. Further Evidence of Population Admixture in the Serbian Honey Bee Population. Insects 2022, 13, 180. https://doi.org/10.3390/insects13020180
Tanasković M, Erić P, Patenković A, Erić K, Mihajlović M, Tanasić V, Kusza S, Oleksa A, Stanisavljević L, Davidović S. Further Evidence of Population Admixture in the Serbian Honey Bee Population. Insects. 2022; 13(2):180. https://doi.org/10.3390/insects13020180
Chicago/Turabian StyleTanasković, Marija, Pavle Erić, Aleksandra Patenković, Katarina Erić, Milica Mihajlović, Vanja Tanasić, Szilvia Kusza, Andrzej Oleksa, Ljubiša Stanisavljević, and Slobodan Davidović. 2022. "Further Evidence of Population Admixture in the Serbian Honey Bee Population" Insects 13, no. 2: 180. https://doi.org/10.3390/insects13020180










