Meta-Analysis of Immune Induced Gene Expression Changes in Diverse Drosophila melanogaster Innate Immune Responses
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Drosophila Melanogaster Genome Data
2.2. Meta-Analysis of Gene Expression Studies
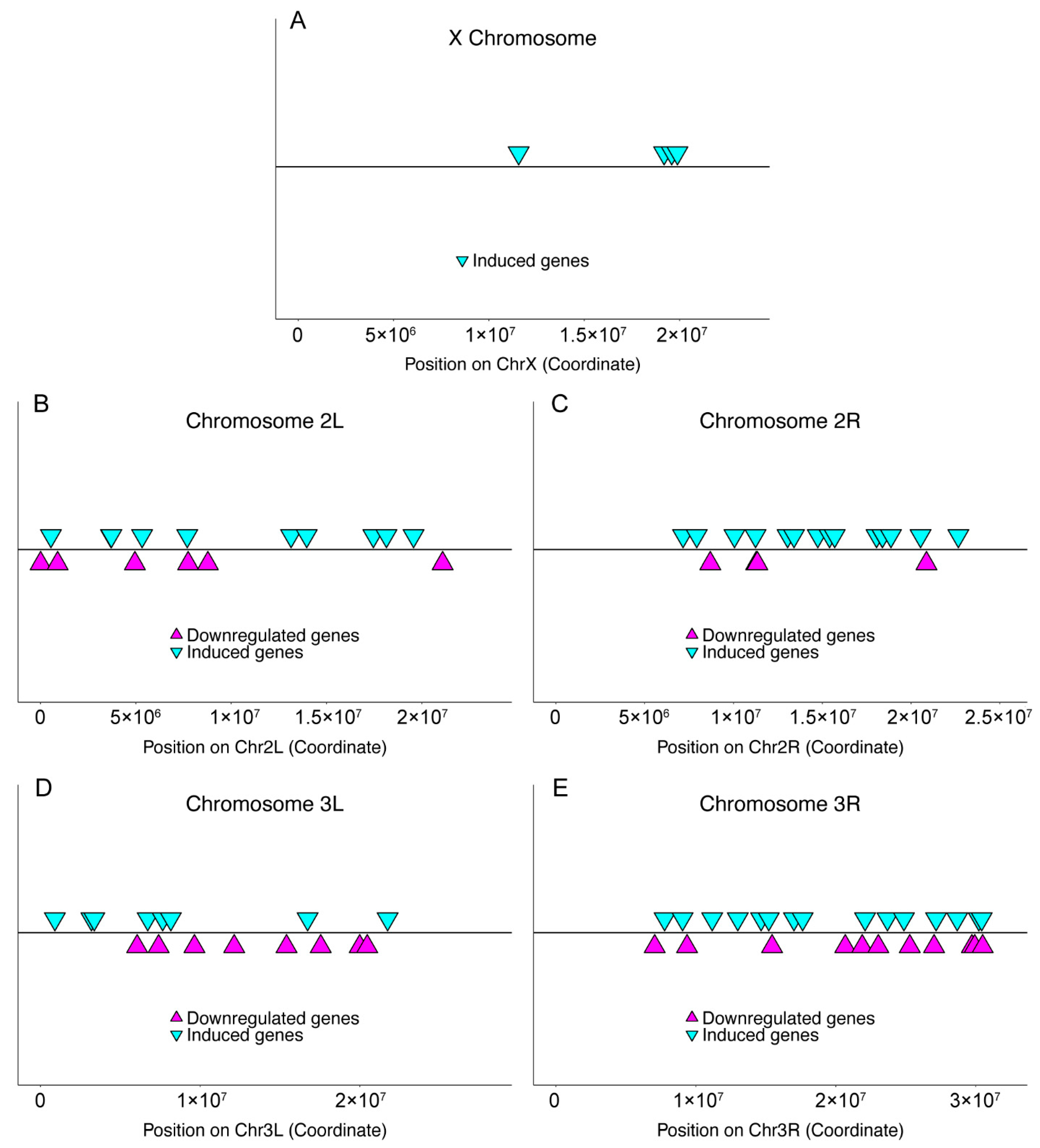
2.3. Chromosomal Distribution
2.4. Gene Locus Uniformity and Clustering
2.5. Motif Finding
2.6. Statistics
3. Results
3.1. Meta-Analysis of Genome-Wide Transcript Levels following Pathogen Infection
3.2. Infection-Induced Genes in Host Immunity
3.3. Predicted Functions of Infection Induced Genes
3.4. Motif Finding Analysis of Infection Induced Genes
3.5. Analysis of Downregulated Transcripts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Danilova, N. The evolution of immune mechanisms. J. Exp. Zoöl. Part B Mol. Dev. Evol. 2006, 306, 496–520. [Google Scholar] [CrossRef] [PubMed]
- Chaplin, D.D. Overview of the immune response. J. Allergy Clin. Immunol. 2010, 125, S3–S23. [Google Scholar] [CrossRef] [PubMed]
- Fairfax, B.P.; Knight, J.C. Genetics of gene expression in immunity to infection. Curr. Opin. Immunol. 2014, 30, 63–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Cao, X. Epigenetic regulation of the innate immune response to infection. Nat. Rev. Immunol. 2019, 19, 417–432. [Google Scholar] [CrossRef]
- Groitl, B.; Dahl, J.-U.; Schroeder, J.W.; Jakob, U. Pseudomonas aeruginosa defense systems against microbicidal oxidants. Mol. Microbiol. 2017, 106, 335–350. [Google Scholar] [CrossRef] [Green Version]
- Kar, S.; Mai, H.-J.; Khalouf, H.; Ben Abdallah, H.; Flachbart, S.; Fink-Straube, C.; Bräutigam, A.; Xiong, G.; Shang, L.; Panda, S.K.; et al. Comparative Transcriptomics of Lowland Rice Varieties Uncovers Novel Candidate Genes for Adaptive Iron Excess Tolerance. Plant Cell Physiol. 2021, 62, 624–640. [Google Scholar] [CrossRef]
- Nica, A.C.; Dermitzakis, E.T. Using gene expression to investigate the genetic basis of complex disorders. Hum. Mol. Genet. 2008, 17, R129–R134. [Google Scholar] [CrossRef] [Green Version]
- Petruccelli, E.; Brown, T.; Waterman, A.; Ledru, N.; Kaun, K.R. Alcohol Causes Lasting Differential Transcription in Drosophila Mushroom Body Neurons. Genetics 2020, 215, 103–116. [Google Scholar] [CrossRef] [Green Version]
- Scheid, A.D.; Van Keulen, V.P.; Felts, S.J.; Neier, S.C.; Middha, S.; Nair, A.A.; Techentin, R.W.; Gilbert, B.K.; Jen, J.; Neuhauser, C.; et al. Gene Expression Signatures Characterized by Longitudinal Stability and Interindividual Variability Delineate Baseline Phenotypic Groups with Distinct Responses to Immune Stimulation. J. Immunol. 2018, 200, 1917–1928. [Google Scholar] [CrossRef]
- Mola, S.; Foisy, S.; Boucher, G.; Major, F.; Beauchamp, C.; Karaky, M.; Goyette, P.; Lesage, S.; Rioux, J.D. A transcriptome-based approach to identify functional modules within and across primary human immune cells. PLoS ONE 2020, 15, e0233543. [Google Scholar] [CrossRef]
- Ding, P.; Ngou, B.P.M.; Furzer, O.J.; Sakai, T.; Shrestha, R.K.; MacLean, D.; Jones, J.D.G. High-resolution expression profiling of selected gene sets during plant immune activation. Plant Biotechnol. J. 2020, 18, 1610–1619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgerton, E.B.; McCrea, A.R.; Berry, C.T.; Kwok, J.Y.; Thompson, L.K.; Watson, B.; Fuller, E.M.; Nolan, T.J.; Lok, J.B.; Povelones, M. Activation of mosquito immunity blocks the development of transmission-stage filarial nematodes. Proc. Natl. Acad. Sci. USA 2020, 117, 3711–3717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brutscher, L.M.; Daughenbaugh, K.F.; Flenniken, M.L. Virus and dsRNA-triggered transcriptional responses reveal key components of honey bee antiviral defense. Sci. Rep. 2017, 7, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Fors, L.; Slotte, T.; Theopold, U.; Binzer-Panchal, M.; Wheat, C.W.; Hambäck, P.A. Differential Expression of Immune Genes between Two Closely Related Beetle Species with Different Immunocompetence following Attack by Asecodes parviclava. Genome Biol. Evol. 2020, 12, 522–534. [Google Scholar] [CrossRef] [PubMed]
- Roesel, C.L.; Vollmer, S.V. Differential gene expression analysis of symbiotic and aposymbiotic Exaiptasia anemones under immune challenge with Vibrio coralliilyticus. Ecol. Evol. 2019, 9, 8279–8293. [Google Scholar] [CrossRef] [Green Version]
- Byadgi, O.; Chen, Y.-C.; Maekawa, S.; Wang, P.-C.; Chen, S.-C. Immune-Related Functional Differential Gene Expression in Koi Carp (Cyprinus carpio) after Challenge with Aeromonas sobria. Int. J. Mol. Sci. 2018, 19, 2107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robledo, D.; Taggart, J.B.; Ireland, J.H.; McAndrew, B.J.; Starkey, W.G.; Haley, C.S.; Hamilton, A.; Guy, D.R.; Mota-Velasco, J.C.; Gheyas, A.A.; et al. Gene expression comparison of resistant and susceptible Atlantic salmon fry challenged with Infectious Pancreatic Necrosis virus reveals a marked contrast in immune response. BMC Genom. 2016, 17, 279. [Google Scholar] [CrossRef] [Green Version]
- De Gregorio, E.; Spellman, P.T.; Tzou, P.; Rubin, G.; Lemaitre, B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002, 21, 2568–2579. [Google Scholar] [CrossRef] [Green Version]
- Schlenke, T.A.; Morales, J.; Govind, S.; Clark, A. Contrasting Infection Strategies in Generalist and Specialist Wasp Parasitoids of Drosophila melanogaster. PLoS Pathog. 2007, 3, e158. [Google Scholar] [CrossRef]
- Brennan, C.A.; Anderson, K.V. Drosophila: The Genetics of Innate Immune Recognition and Response. Annu. Rev. Immunol. 2004, 22, 457–483. [Google Scholar] [CrossRef]
- Crum-Cianflone, N.F. Bacterial, Fungal, Parasitic, and Viral Myositis. Clin. Microbiol. Rev. 2008, 21, 473–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemaitre, B.; Hoffmann, J. The Host Defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, S.; Plüddemann, A.; Estrada, F.M. Macrophage heterogeneity in tissues: Phenotypic diversity and functions. Immunol. Rev. 2014, 262, 36–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemaitre, B.; Reichhart, J.-M.; Hoffmann, J.A. Drosophila host defense: Differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl. Acad. Sci. USA 1997, 94, 14614–14619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koonin, E.V. Evolution of RNA- and DNA-guided antivirus defense systems in prokaryotes and eukaryotes: Common ancestry vs. convergence. Biol. Direct 2017, 12, 5. [Google Scholar] [CrossRef] [Green Version]
- Weber, F. Antiviral Innate Immunity: Introduction. Encycl. Virol. 2020, 577–583. [Google Scholar] [CrossRef]
- Beutler, B.; Jiang, Z.; Georgel, P.; Crozat, K.; Croker, B.; Rutschmann, S.; Du, X.; Hoebe, K. GENETIC ANALYSIS OF HOST RESISTANCE: Toll-Like Receptor Signaling and Immunity at Large. Annu. Rev. Immunol. 2006, 24, 353–389. [Google Scholar] [CrossRef] [Green Version]
- Lemaitre, B.; Nicolas, E.; Michaut, L.; Reichhart, J.-M.; Hoffmann, J.A. The Dorsoventral Regulatory Gene Cassette spätzle/Toll/cactus Controls the Potent Antifungal Response in Drosophila Adults. Cell 1996, 86, 973–983. [Google Scholar] [CrossRef] [Green Version]
- Medzhitov, R. Toll-like receptors and innate immunity. Nat. Rev. Immunol. 2001, 1, 135–145. [Google Scholar] [CrossRef]
- O’Shea, J.J.; Plenge, R. JAK and STAT Signaling Molecules in Immunoregulation and Immune-Mediated Disease. Immunity 2012, 36, 542–550. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, J.A.; Kafatos, F.C.; Janeway, C.A.; Ezekowitz, R.A.B. Phylogenetic Perspectives in Innate Immunity. Science 1999, 284, 1313–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howell, L.; Sampson, C.J.; Xavier, M.J.; Bolukbasi, E.; Heck, M.M.S.; Williams, M.J. A directed miniscreen for genes involved in the Drosophila anti-parasitoid immune response. Immunogenetics 2011, 64, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Stroschein-Stevenson, S.L.; Foley, E.; O’Farrell, P.H.; Johnson, A.D. Identification of Drosophila Gene Products Required for Phagocytosis of Candida albicans. PLoS Biol. 2005, 4, e4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Gregorio, E.; Spellman, P.T.; Rubin, G.M.; Lemaitre, B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. USA 2001, 98, 12590–12595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Felix, T.M.; Hughes, K.A.; Stone, E.A.; Drnevich, J.M.; Leips, J. Age-Specific Variation in Immune Response in Drosophila melanogaster Has a Genetic Basis. Genetics 2012, 191, 989–1002. [Google Scholar] [CrossRef] [Green Version]
- Pal, S.; Wu, J.; Wu, L.P. Microarray analyses reveal distinct roles for Rel proteins in the Drosophila immune response. Dev. Comp. Immunol. 2008, 32, 50–60. [Google Scholar] [CrossRef] [Green Version]
- Dostert, C.; Jouanguy, E.; Irving, P.; Troxler, L.; Galiana-Arnoux, D.; Hetru, C.; Hoffmann, J.A.; Imler, J.-L. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of drosophila. Nat. Immunol. 2005, 6, 946–953. [Google Scholar] [CrossRef]
- Huang, Z.; Kingsolver, M.B.; Avadhanula, V.; Hardy, R.W. An Antiviral Role for Antimicrobial Peptides during the Arthropod Response to Alphavirus Replication. J. Virol. 2013, 87, 4272–4280. [Google Scholar] [CrossRef] [Green Version]
- Kemp, C.; Mueller, S.; Goto, A.; Barbier, V.; Paro, S.; Bonnay, F.; Dostert, C.; Troxler, L.; Hetru, C.; Meignin, C.; et al. Broad RNA Interference–Mediated Antiviral Immunity and Virus-Specific Inducible Responses in Drosophila. J. Immunol. 2012, 190, 650–658. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.J.; Mondal, A.; Small, C.; Paddibhatla, I.; Kawaguchi, A.; Govind, S. A database for the analysis of immunity genes in Drosophila. Fly 2011, 5, 155–161. [Google Scholar] [CrossRef] [Green Version]
- Toro-Domínguez, D.; García, J.A.V.; Martorell-Marugán, J.; Román-Montoya, Y.; Alarcón-Riquelme, M.E.; Carmona-Sáez, P. A survey of gene expression meta-analysis: Methods and applications. Brief. Bioinform. 2020, 22, 1694–1705. [Google Scholar] [CrossRef] [PubMed]
- Larkin, A.; Marygold, S.J.; Antonazzo, G.; Attrill, H.; dos Santos, G.; Garapati, P.V.; Goodman, J.L.; Gramates, L.S.; Millburn, G.; Strelets, V.B.; et al. FlyBase: Updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 2020, 49, D899–D907. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Del Carratore, F.; Jankevics, A.; Eisinga, R.; Heskes, T.; Hong, F.; Breitling, R. RankProd 2.0: A refactored bioconductor package for detecting differentially expressed features in molecular profiling datasets. Bioinformatics 2017, 33, 2774–2775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, F.; Breitling, R.; McEntee, C.W.; Wittner, B.S.; Nemhauser, J.L.; Chory, J. RankProd: A bioconductor package for detecting differentially expressed genes in meta-analysis. Bioinformatics 2006, 22, 2825–2827. [Google Scholar] [CrossRef] [Green Version]
- Obregón, F.P.; Soto, P.; Lavín, J.L.; Cortazar, A.R.; Barrio, R.; Aransay, A.M.; Cantera, R. Cluster Locator, online analysis and visualization of gene clustering. Bioinformatics 2018, 34, 3377–3379. [Google Scholar] [CrossRef]
- Piipari, M.; Down, T.A.; Saini, H.; Enright, A.; Hubbard, T.J. iMotifs: An integrated sequence motif visualization and analysis environment. Bioinformatics 2010, 26, 843–844. [Google Scholar] [CrossRef]
- Ryan, S.M.; Wildman, K.; Oceguera-Perez, B.; Barbee, S.; Mortimer, N.T.; Vrailas-Mortimer, A.D. Evolutionarily conserved transcription factors drive the oxidative stress response in Drosophila. J. Exp. Biol. 2020, 223, jeb221622. [Google Scholar] [CrossRef]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Stamatoyannopoulos, J.A.; Bailey, T.L.; Noble, W.S. Quantifying similarity between motifs. Genome Biol. 2007, 8, R24. [Google Scholar] [CrossRef] [Green Version]
- Shazman, S.; Lee, H.; Socol, Y.; Mann, R.S.; Honig, B. OnTheFly: A database of Drosophila melanogaster transcription factors and their binding sites. Nucleic Acids Res. 2013, 42, D167–D171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009; ISBN 978-0-387-98141-3. [Google Scholar]
- Breitling, R.; Armengaud, P.; Amtmann, A.; Herzyk, P. Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004, 573, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Riddle, N.C.; Elgin, S.C.R. The Drosophila Dot Chromosome: Where Genes Flourish Amidst Repeats. Genetics 2018, 210, 757–772. [Google Scholar] [CrossRef] [Green Version]
- Kaufman, T.C. A Short History and Description of Drosophila melanogaster Classical Genetics: Chromosome Aberrations, Forward Genetic Screens, and the Nature of Mutations. Genetics 2017, 206, 665–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill-Burns, E.M.; Clark, A.G. X-Linked Variation in Immune Response in Drosophila melanogaster. Genetics 2009, 183, 1477–1491. [Google Scholar] [CrossRef] [Green Version]
- Hedengren, M.; Borge, K.; Hultmark, D. Expression and Evolution of the Drosophila Attacin/Diptericin Gene Family. Biochem. Biophys. Res. Commun. 2000, 279, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-N.; Yu, B.; Han, G.-Q.; Chen, D.-W. Molecular cloning, expression in Escherichia coli of Attacin A gene from Drosophila and detection of biological activity. Mol. Biol. Rep. 2009, 37, 2463–2469. [Google Scholar] [CrossRef]
- Lin, S.J.H.; Fulzele, A.; Cohen, L.B.; Bennett, E.J.; Wasserman, S.A. Bombardier Enables Delivery of Short-Form Bomanins in the Drosophila Toll Response. Front. Immunol. 2020, 10, 3040. [Google Scholar] [CrossRef]
- Clemmons, A.W.; Lindsay, S.A.; Wasserman, S.A. An Effector Peptide Family Required for Drosophila Toll-Mediated Immunity. PLoS Pathog. 2015, 11, e1004876. [Google Scholar] [CrossRef] [Green Version]
- Hanson, M.A.; Dostálová, A.; Ceroni, C.; Poidevin, M.; Kondo, S.; Lemaitre, B. Synergy and remarkable specificity of antimicrobial peptides in vivo using a systematic knockout approach. eLife 2019, 8, e44341. [Google Scholar] [CrossRef]
- Lindsay, S.A.; Lin, S.J.; Wasserman, S.A. Short-Form Bomanins Mediate Humoral Immunity in Drosophila. J. Innate Immun. 2018, 10, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, B.; Kromer-Metzger, E.; Michaut, L.; Nicolas, E.; Meister, M.; Georgel, P.; Reichhart, J.M.; Hoffmann, J.A. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc. Natl. Acad. Sci. USA 1995, 92, 9465–9469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samakovlis, C.; Kimbrell, D.A.; Kylsten, P.; Engström, A.; Hultmark, D. The immune response in Drosophila: Pattern of cecropin expression and biological activity. EMBO J. 1990, 9, 2969–2976. [Google Scholar] [CrossRef] [PubMed]
- Tryselius, Y.; Samakovlis, C.; Kimbrell, D.A.; Hultmark, D. CecC, a cecropin gene expressed during metamorphosis in Drosophila pupae. JBIC J. Biol. Inorg. Chem. 1992, 204, 395–399. [Google Scholar] [CrossRef]
- Tzou, P.; Reichhart, J.-M.; Lemaitre, B. Constitutive expression of a single antimicrobial peptide can restore wild-type resistance to infection in immunodeficient Drosophila mutants. Proc. Natl. Acad. Sci. USA 2002, 99, 2152–2157. [Google Scholar] [CrossRef] [Green Version]
- Wicker, C.; Reichhart, J.M.; Hoffmann, D.; Hultmark, D.; Samakovlis, C.; Hoffmann, J.A. Insect immunity. Characterization of a Drosophila cDNA encoding a novel member of the diptericin family of immune peptides. J. Biol. Chem. 1990, 265, 22493–22498. [Google Scholar] [CrossRef]
- Lee, J.H.; Cho, K.S.; Lee, J.; Yoo, J.; Lee, J.; Chung, J. Diptericin-like protein: An immune response gene regulated by the anti-bacterial gene induction pathway in Drosophila. Gene 2001, 271, 233–238. [Google Scholar] [CrossRef]
- Chiu, H.; Ring, B.C.; Sorrentino, R.P.; Kalamarz, M.; Garza, D.; Govind, S. dUbc9 negatively regulates the Toll-NF-κB pathways in larval hematopoiesis and drosomycin activation in Drosophila. Dev. Biol. 2005, 288, 60–72. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Zhu, S. Functional role of charged residues in drosomycin, a Drosophila antifungal peptide. Dev. Comp. Immunol. 2010, 34, 953–958. [Google Scholar] [CrossRef]
- Ayres, J.S.; Freitag, N.; Schneider, D.S. Identification of Drosophila Mutants Altering Defense of and Endurance to Listeria monocytogenes Infection. Genetics 2008, 178, 1807–1815. [Google Scholar] [CrossRef] [Green Version]
- Hanson, M.A.; Cohen, L.B.; Marra, A.; Iatsenko, I.; Wasserman, S.A.; Lemaitre, B. The Drosophila Baramicin polypeptide gene protects against fungal infection. PLoS Pathog. 2021, 17, e1009846. [Google Scholar] [CrossRef] [PubMed]
- Ha, E.-M.; Oh, C.-T.; Ryu, J.-H.; Bae, Y.-S.; Kang, S.-W.; Jang, I.-H.; Brey, P.T.; Lee, W.-J. An Antioxidant System Required for Host Protection against Gut Infection in Drosophila. Dev. Cell 2005, 8, 125–132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keebaugh, E.S.; Schlenke, T.A. Adaptive Evolution of a Novel Drosophila Lectin Induced by Parasitic Wasp Attack. Mol. Biol. Evol. 2011, 29, 565–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goto, A.; Yano, T.; Terashima, J.; Iwashita, S.; Oshima, Y.; Kurata, S. Cooperative Regulation of the Induction of the Novel Antibacterial Listericin by Peptidoglycan Recognition Protein LE and the JAK-STAT Pathway. J. Biol. Chem. 2010, 285, 15731–15738. [Google Scholar] [CrossRef] [Green Version]
- Pal, S.; Leger, R.J.S.; Wu, L. Fungal Peptide Destruxin A Plays a Specific Role in Suppressing the Innate Immune Response in Drosophila melanogaster. J. Biol. Chem. 2007, 282, 8969–8977. [Google Scholar] [CrossRef] [Green Version]
- Brun, S.; Vidal, S.; Spellman, P.; Takahashi, K.; Tricoire, H.; Lemaitre, B. The MAPKKK Mekk1 regulates the expression of Turandot stress genes in response to septic injury in Drosophila. Genes Cells 2006, 11, 397–407. [Google Scholar] [CrossRef]
- Levashina, E.A.; Ohresser, S.; Bulet, P.; Reichhart, J.-M.; Hetru, C.; Hoffmann, J.A. Metchnikowin, a Novel Immune-Inducible Proline-Rich Peptide from Drosophila with Antibacterial and Antifungal Properties. Eur. J. Biochem. 1995, 233, 694–700. [Google Scholar] [CrossRef]
- Levashina, E.; Ohresser, S.; Lemaitre, B.; Imler, J.-L. Two distinct pathways can control expression of the gene encoding the Drosophila antimicrobial peptide metchnikowin. J. Mol. Biol. 1998, 278, 515–527. [Google Scholar] [CrossRef] [Green Version]
- Levashina, E.A.; Langley, E.; Green, C.; Gubb, D.; Ashburner, M.; Hoffmann, J.A.; Reichhart, J.-M. Constitutive Activation of Toll-Mediated Antifungal Defense in Serpin-Deficient Drosophila. Science 1999, 285, 1917–1919. [Google Scholar] [CrossRef]
- Kurucz, É.; Markus, R.; Zsámboki, J.; Folkl-Medzihradszky, K.; Darula, Z.; Vilmos, P.; Udvardy, A.; Krausz, I.; Lukacsovich, T.; Gateff, E.; et al. Nimrod, a Putative Phagocytosis Receptor with EGF Repeats in Drosophila Plasmatocytes. Curr. Biol. 2007, 17, 649–654. [Google Scholar] [CrossRef] [Green Version]
- Somogyi, K.; Sipos, B.; Pénzes, Z.; Andó, I. A conserved gene cluster as a putative functional unit in insect innate immunity. FEBS Lett. 2010, 584, 4375–4378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchon, N.; Poidevin, M.; Kwon, H.-M.; Guillou, A.; Sottas, V.; Lee, B.-L.; Lemaitre, B. A single modular serine protease integrates signals from pattern-recognition receptors upstream of the Drosophila Toll pathway. Proc. Natl. Acad. Sci. USA 2009, 106, 12442–12447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gottar, M.; Gobert, V.; Michel, T.; Belvin, M.; Duyk, G.; Hoffmann, J.A.; Ferrandon, D.; Royet, J. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nat. Cell Biol. 2002, 416, 640–644. [Google Scholar] [CrossRef] [PubMed]
- Mellroth, P.; Steiner, H. PGRP-SB1: An N-acetylmuramoyl l-alanine amidase with antibacterial activity. Biochem. Biophys. Res. Commun. 2006, 350, 994–999. [Google Scholar] [CrossRef]
- Zaidman-Rémy, A.; Poidevin, M.; Hervé, M.; Welchman, D.P.; Paredes, J.C.; Fahlander, C.; Steiner, H.; Mengin-Lecreulx, D.; Lemaitre, B. Drosophila Immunity: Analysis of PGRP-SB1 Expression, Enzymatic Activity and Function. PLoS ONE 2011, 6, e17231. [Google Scholar] [CrossRef]
- Bischoff, V.; Vignal, C.; Boneca, I.G.; Michel, T.; Hoffmann, J.A.; Royet, J. Function of the drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat. Immunol. 2004, 5, 1175–1180. [Google Scholar] [CrossRef]
- Dushay, M.S.; Asling, B.; Hultmark, D. Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc. Natl. Acad. Sci. USA 1996, 93, 10343–10347. [Google Scholar] [CrossRef] [Green Version]
- Hedengren, M.; Bengtåsling, B.; Dushay, M.S.; Ando, I.; Ekengren, S.; Wihlborg, M.; Hultmark, D. Relish, a Central Factor in the Control of Humoral but Not Cellular Immunity in Drosophila. Mol. Cell 1999, 4, 827–837. [Google Scholar] [CrossRef]
- Seong, C.-S.; Varela-Ramirez, A.; Tang, X.; Anchondo, B.; Magallanes, D.; Aguilera, R.J. Cloning and Characterization of a Novel Drosophila Stress Induced DNase. PLoS ONE 2014, 9, e103564. [Google Scholar] [CrossRef] [Green Version]
- Castillejo-López, C.; Häcker, U. The serine protease Sp7 is expressed in blood cells and regulates the melanization reaction in Drosophila. Biochem. Biophys. Res. Commun. 2005, 338, 1075–1082. [Google Scholar] [CrossRef]
- Dudzic, J.P.; Hanson, M.A.; Iatsenko, I.; Kondo, S.; Lemaitre, B. More Than Black or White: Melanization and Toll Share Regulatory Serine Proteases in Drosophila. Cell Rep. 2019, 27, 1050–1061.e3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kambris, Z.; Brun, S.; Jang, I.-H.; Nam, H.-J.; Romeo, Y.; Takahashi, K.; Lee, W.-J.; Ueda, R.; Lemaitre, B. Drosophila Immunity: A Large-Scale In Vivo RNAi Screen Identifies Five Serine Proteases Required for Toll Activation. Curr. Biol. 2006, 16, 808–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, I.-H.; Chosa, N.; Kim, S.-H.; Nam, H.-J.; Lemaitre, B.; Ochiai, M.; Kambris, Z.; Brun, S.; Hashimoto, C.; Ashida, M.; et al. A Spätzle-Processing Enzyme Required for Toll Signaling Activation in Drosophila Innate Immunity. Dev. Cell 2006, 10, 45–55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dostálová, A.; Rommelaere, S.; Poidevin, M.; Lemaitre, B. Thioester-containing proteins regulate the Toll pathway and play a role in Drosophila defence against microbial pathogens and parasitoid wasps. BMC Biol. 2017, 15, 79. [Google Scholar] [CrossRef] [PubMed]
- Boutros, M.; Agaisse, H.; Perrimon, N. Sequential Activation of Signaling Pathways during Innate Immune Responses in Drosophila. Dev. Cell 2002, 3, 711–722. [Google Scholar] [CrossRef] [Green Version]
- Zhong, W.; McClure, C.D.; Evans, C.R.; Mlynski, D.T.; Immonen, E.; Ritchie, M.G.; Priest, N.K. Immune anticipation of mating in Drosophila: Turandot M promotes immunity against sexually transmitted fungal infections. Proc. R. Soc. B Boil. Sci. 2013, 280, 20132018. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Berkey, C.D.; Watnick, P.I. The Drosophila Protein Mustard Tailors the Innate Immune Response Activated by the Immune Deficiency Pathway. J. Immunol. 2012, 188, 3993–4000. [Google Scholar] [CrossRef] [Green Version]
- Irving, P.; Troxler, L.; Heuer, T.S.; Belvin, M.; Kopczynski, C.; Reichhart, J.-M.; Hoffmann, J.A.; Hetru, C. A genome-wide analysis of immune responses in Drosophila. Proc. Natl. Acad. Sci. USA 2001, 98, 15119–15124. [Google Scholar] [CrossRef] [Green Version]
- Hanson, M.A.; Lemaitre, B.; Unckless, R.L. Dynamic Evolution of Antimicrobial Peptides Underscores Trade-Offs Between Immunity and Ecological Fitness. Front. Immunol. 2019, 10, 2620. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.; Jiang, H. Building a platform for predicting functions of serine protease-related proteins in Drosophila melanogaster and other insects. Insect Biochem. Mol. Biol. 2018, 103, 53–69. [Google Scholar] [CrossRef]
- Ligoxygakis, P.; Pelte, N.; Hoffmann, J.A.; Reichhart, J.-M. Activation of Drosophila Toll During Fungal Infection by a Blood Serine Protease. Science 2002, 297, 114–116. [Google Scholar] [CrossRef]
- Patrnogic, J.; Leclerc, V. The serine protease homolog spheroide is involved in sensing of pathogenic Gram-positive bacteria. PLoS ONE 2017, 12, e0188339. [Google Scholar] [CrossRef] [Green Version]
- Kr, P.; Lee, J.; Mortimer, N.T. The S1A Protease Family Members CG10764 and CG4793 Regulate Cellular Immunity in Dro-sophila. Micropublication Biol. 2021, 2021. [Google Scholar] [CrossRef]
- Hayden, M.S.; Ghosh, S. NF-κB in immunobiology. Cell Res. 2011, 21, 223–244. [Google Scholar] [CrossRef] [Green Version]
- Hogan, P.G. Calcium–NFAT transcriptional signalling in T cell activation and T cell exhaustion. Cell Calcium 2017, 63, 66–69. [Google Scholar] [CrossRef] [Green Version]
- Villarino, A.; Kanno, Y.; Ferdinand, J.R.; O’Shea, J.J. Mechanisms of Jak/STAT Signaling in Immunity and Disease. J. Immunol. 2014, 194, 21–27. [Google Scholar] [CrossRef] [Green Version]
- Gallant, S.; Gilkeson, G. ETS transcription factors and regulation of immunity. Arch. Immunol. Ther. Exp. 2006, 54, 149–163. [Google Scholar] [CrossRef]
- Copley, R.R.; Totrov, M.; Linnell, J.; Field, S.; Ragoussis, J.; Udalova, I.A. Functional conservation of Rel binding sites in drosophilid genomes. Genome Res. 2007, 17, 1327–1335. [Google Scholar] [CrossRef] [Green Version]
- Immarigeon, C.; Bernat-Fabre, S.; Guillou, E.; Verger, A.; Prince, E.; Benmedjahed, M.A.; Payet, A.; Couralet, M.; Monte, D.; Villeret, V.; et al. Mediator complex subunit Med19 binds directly GATA transcription factors and is required with Med1 for GATA-driven gene regulation in vivo. J. Biol. Chem. 2020, 295, 13617–13629. [Google Scholar] [CrossRef]
- Juven-Gershon, T.; Hsu, J.-Y.; Theisen, J.; Kadonaga, J.T. The RNA polymerase II core promoter—The gateway to transcription. Curr. Opin. Cell Biol. 2008, 20, 253–259. [Google Scholar] [CrossRef] [Green Version]
- Ngoc, L.V.; Kassavetis, G.A.; Kadonaga, J.T. The RNA Polymerase II Core Promoter in Drosophila. Genetics 2019, 212, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Reichhart, J.M.; Georgel, P.; Meister, M.; Lemaitre, B.; Kappler, C.; Hoffmann, J.A. Expression and nuclear translocation of the rel/NF-kappa B-related morphogen dorsal during the immune response of Drosophila. C R Acad. Sci. III 1993, 316, 1218–1224. [Google Scholar] [PubMed]
- Petersen, U.; Björklund, G.; Ip, Y.; Engström, Y. The dorsal-related immunity factor, Dif, is a sequence-specific trans-activator of Drosophila Cecropin gene expression. EMBO J. 1995, 14, 3146–3158. [Google Scholar] [CrossRef] [PubMed]
- Petersen, U.-M.; Kadalayil, L.; Rehorn, K.-P.; Hoshizaki, D.K.; Reuter, R.; Engström, Y. Serpent regulates Drosophila immunity genes in the larval fat body through an essential GATA motif. EMBO J. 1999, 18, 4013–4022. [Google Scholar] [CrossRef] [Green Version]
- Clark, R.I.; Tan, S.W.; Péan, C.B.; Roostalu, U.; Vivancos, V.; Bronda, K.; Pilátová, M.; Fu, J.; Walker, D.W.; Berdeaux, R.; et al. MEF2 Is an In Vivo Immune-Metabolic Switch. Cell 2013, 155, 435–447. [Google Scholar] [CrossRef] [Green Version]
- Bajgar, A.; Kucerova, K.; Jonatova, L.; Tomcala, A.; Schneedorferova, I.; Okrouhlik, J.; Dolezal, T. Extracellular Adenosine Mediates a Systemic Metabolic Switch during Immune Response. PLoS Biol. 2015, 13, e1002135. [Google Scholar] [CrossRef] [Green Version]
- Mihajlovic, Z.; Tanasic, D.; Bajgar, A.; Perez-Gomez, R.; Steffal, P.; Krejci, A. Lime is a new protein linking immunity and metabolism in Drosophila. Dev. Biol. 2019, 452, 83–94. [Google Scholar] [CrossRef]
- DiAngelo, J.R.; Bland, M.L.; Bambina, S.; Cherry, S.; Birnbaum, M.J. The immune response attenuates growth and nutrient storage in Drosophila by reducing insulin signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 20853–20858. [Google Scholar] [CrossRef] [Green Version]
- Schwenke, R.A.; Lazzaro, B.P.; Wolfner, M.F. Reproduction–Immunity Trade-Offs in Insects. Annu. Rev. Èntomol. 2016, 61, 239–256. [Google Scholar] [CrossRef] [Green Version]
- Martínez, B.A.; Hoyle, R.G.; Yeudall, S.; Granade, M.E.; Harris, T.E.; Castle, J.D.; Leitinger, N.; Bland, M.L. Innate immune signaling in Drosophila shifts anabolic lipid metabolism from triglyceride storage to phospholipid synthesis to support immune function. PLoS Genet. 2020, 16, e1009192. [Google Scholar] [CrossRef]
- Hart, B. Biological basis of the behavior of sick animals. Neurosci. Biobehav. Rev. 1988, 12, 123–137. [Google Scholar] [CrossRef]
- Murray, M.J.; Murray, A.B. Anorexia of infection as a mechanism of host defense. Am. J. Clin. Nutr. 1979, 32, 593–596. [Google Scholar] [CrossRef] [PubMed]
- Adamo, S.A. Parasitic suppression of feeding in the tobacco hornworm, Manduca sexta: Parallels with feeding depression after an immune challenge. Arch. Insect Biochem. Physiol. 2005, 60, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Surendran, S.; Hückesfeld, S.; Wäschle, B.; Pankratz, M.J. Pathogen induced food evasion behavior in Drosophila larvae. J. Exp. Biol. 2017, 220, 1774–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayres, J.S.; Schneider, D.S. The Role of Anorexia in Resistance and Tolerance to Infections in Drosophila. PLoS Biol. 2009, 7, e1000150. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.; Liu, C.; Bai, X.; Li, X.; Li, J.; Zhang, Z.; Zhang, Y.; Guo, J.; Li, Y. Drosophila FIT is a protein-specific satiety hormone essential for feeding control. Nat. Commun. 2017, 8, 14161. [Google Scholar] [CrossRef] [Green Version]
- Erkosar, B.; Kolly, S.; van der Meer, J.R.; Kawecki, T.J. Adaptation to Chronic Nutritional Stress Leads to Reduced Dependence on Microbiota in Drosophila melanogaster. mBio 2017, 8, e01496-17. [Google Scholar] [CrossRef] [Green Version]
- Lemaitre, B.; Miguel-Aliaga, I. The Digestive Tract of Drosophila melanogaster. Annu. Rev. Genet. 2013, 47, 377–404. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.; Huen, S.C.; Luan, H.H.; Yu, S.; Zhang, C.; Gallezot, J.-D.; Booth, C.J.; Medzhitov, R. Opposing Effects of Fasting Metabolism on Tissue Tolerance in Bacterial and Viral Inflammation. Cell 2016, 166, 1512–1525.e12. [Google Scholar] [CrossRef] [Green Version]
- Howick, V.M.; Lazzaro, B.P. Genotype and diet shape resistance and tolerance across distinct phases of bacterial infection. BMC Evol. Biol. 2014, 14, 56. [Google Scholar] [CrossRef] [Green Version]
- Coustau, C.; Carton, Y.; Nappl, A.; Shotkoski, F.; Ffrench-Constant, R. Differential induction of antibacterial transcripts in Drosophila susceptible and resistant to parasitism by Leptopilina boulardi. Insect Mol. Biol. 1996, 5, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, E.; Nappi, A.J.; Lemaitre, B. Expression of antimicrobial peptide genes after infection by parasitoid wasps in Drosophila. Dev. Comp. Immunol. 1996, 20, 175–181. [Google Scholar] [CrossRef] [Green Version]
- Leulier, F.; Parquet, C.; Pili-Floury, S.; Ryu, J.-H.; Caroff, M.; Lee, W.-J.; Mengin-Lecreulx, D.; Lemaitre, B. The Drosophila immune system detects bacteria through specific peptidoglycan recognition. Nat. Immunol. 2003, 4, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Kraaijeveld, A.R.; Layen, S.J.; Futerman, P.H.; Godfray, H.C.J. Lack of Phenotypic and Evolutionary Cross-Resistance against Parasitoids and Pathogens in Drosophila melanogaster. PLoS ONE 2012, 7, e53002. [Google Scholar] [CrossRef] [Green Version]
- Bentz, M.L.; Humphrey, E.A.; Harshman, L.G.; Wayne, M.L. Sigma Virus (DMelSV) Incidence in Lines of Drosophila melanogaster Selected for Survival following Infection with Bacillus cereus. Psyche A J. Èntomol. 2017, 2017, e3593509. [Google Scholar] [CrossRef] [Green Version]
- Cattadori, I.; Boag, B.; Hudson, P. Parasite co-infection and interaction as drivers of host heterogeneity. Int. J. Parasitol. 2008, 38, 371–380. [Google Scholar] [CrossRef]
- Salam, N.; Mustafa, S.; Hafiz, A.; Chaudhary, A.A.; Deeba, F.; Parveen, S. Global prevalence and distribution of coinfection of malaria, dengue and chikungunya: A systematic review. BMC Public 2018, 18, 710. [Google Scholar] [CrossRef]
- Dieme, C.; Zmarlak, N.M.; Brito-Fravallo, E.; Travaillé, C.; Pain, A.; Cherrier, F.; Genève, C.; Alvarez, E.C.; Riehle, M.M.; Vernick, K.D.; et al. Exposure of Anopheles mosquitoes to trypanosomes reduces reproductive fitness and enhances susceptibility to Plasmodium. PLoS Negl. Trop. Dis. 2020, 14, e0008059. [Google Scholar] [CrossRef] [Green Version]
- Pokutnaya, D.; Molaei, G.; Weinberger, D.M.; Vossbrinck, C.R.; Diaz, A.J. Prevalence of Infection and Co-Infection and Presence of Rickettsial Endosymbionts in Ixodes scapularis (Acari: Ixodidae) in Connecticut, USA. J. Parasitol. 2020, 106, 30–37. [Google Scholar] [CrossRef]
- Sheehan, G.; Tully, L.; Kavanagh, K.A. Candida albicans increases the pathogenicity of Staphylococcus aureus during polymicrobial infection of Galleria mellonella larvae. Microbiology 2020, 166, 375–385. [Google Scholar] [CrossRef]
- Diaz, J.H. Tickborne Coinfections in the United States. J. La. State Med. Soc. 2016, 168, 44–53. [Google Scholar] [PubMed]
- Markow, T.A. The secret lives of Drosophila flies. eLife 2015, 4, e06793. [Google Scholar] [CrossRef] [PubMed]
- Small, C.; Paddibhatla, I.; Rajwani, R.; Govind, S. An Introduction to Parasitic Wasps of Drosophila and the Antiparasite Immune Response. J. Vis. Exp. 2012, 63, e3347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galko, M.J.; Krasnow, M.A. Cellular and Genetic Analysis of Wound Healing in Drosophila Larvae. PLoS Biol. 2004, 2, e239. [Google Scholar] [CrossRef] [Green Version]
- Schlamp, F.; Delbare, S.Y.N.; Early, A.; Wells, M.T.; Basu, S.; Clark, A.G. Dense time-course gene expression profiling of the Drosophila melanogaster innate immune response. BMC Genom. 2021, 22, 304. [Google Scholar] [CrossRef]
- Senger, K.; Armstrong, G.W.; Rowell, W.; Kwan, J.M.; Markstein, M.; Levine, M. Immunity Regulatory DNAs Share Common Organizational Features in Drosophila. Mol. Cell 2004, 13, 19–32. [Google Scholar] [CrossRef] [Green Version]
- Senger, K.; Harris, K.; Levine, M. GATA factors participate in tissue-specific immune responses in Drosophila larvae. Proc. Natl. Acad. Sci. USA 2006, 103, 15957–15962. [Google Scholar] [CrossRef] [Green Version]
- Tanji, T.; Hu, X.; Weber, A.N.R.; Ip, Y.T. Toll and IMD Pathways Synergistically Activate an Innate Immune Response in Drosophila melanogaster. Mol. Cell. Biol. 2007, 27, 4578–4588. [Google Scholar] [CrossRef] [Green Version]
- Unckless, R.L.; Rottschaefer, S.M.; Lazzaro, B.P. The Complex Contributions of Genetics and Nutrition to Immunity in Drosophila melanogaster. PLoS Genet. 2015, 11, e1005030. [Google Scholar] [CrossRef] [Green Version]




| GEO Accession | Pathogen Type | Pathogen | Host Stage | Reference |
|---|---|---|---|---|
| - 1 | Bacteria | Escherichia coli + Micrococcus luteus | Adult | [34] |
| - 1 | Bacteria | E. coli, M. luteus + Enterococcus faecalis | Adult | [18] |
| GSE37708 | Bacteria | E. coli | Adult | [35] |
| GSE5489 | Bacteria | E. coli | Larva | [36] |
| - 1 | Fungus | Beauvaria bassiana | Adult | [34] |
| - 1 | Fungus | Aspergillus fumigatus | Adult | [18] |
| GSE2828 | Virus | Drosophila C Virus | Adult | [37] |
| GSE42726 | Virus | Sindbis virus (transgenic Drosophila model) | Adult | [38] |
| GSE31542 | Virus | Flock House Virus | Adult | [39] |
| GSE31542 | Virus | Sindbis virus | Adult | [39] |
| GSE25522 | Parasite | Ganaspis xanothopoda | Larva | [40] |
| GSE8938 | Parasite | Leptopilina boulardi | Larva | [19] |
| Sample | X | 2L | 2R | 3L | 3R | 4 | U | Total |
|---|---|---|---|---|---|---|---|---|
| Dataset | 1793 | 1951 | 2129 | 2103 | 2734 | 65 | 43 | 10,818 |
| Induced genes | 4 b | 11 | 22 a | 8 | 17 | 0 | 0 | 62 |
| Downregulated genes | 0 b | 6 | 5 b | 9 | 11 | 0 | 0 | 31 |
| Chromosome Arm | D Up | p Value Up | D Down | p Value Down |
|---|---|---|---|---|
| X | 0.62 | 0.05 | - | - |
| 2L | 0.17 | 0.87 | 0.41 | 0.20 |
| 2R | 0.24 | 0.15 | 0.51 | 0.10 |
| 3L | 0.41 | 0.10 | 0.25 | 0.52 |
| 3R | 0.17 | 0.66 | 0.31 | 0.19 |
| Gene Name | Function | Immune Pathway | References |
|---|---|---|---|
| AttA | Antimicrobial peptide | IMD | [58,59] |
| AttD | Antimicrobial peptide | IMD | [58] |
| Bbd | Production of AMP-like peptides | Toll | [60] |
| BomBc1 | AMP-like | Toll | [61,62] |
| BomBc2 | AMP-like | Toll | [61,62] |
| BomBc3 | AMP-like | Toll | [61,62] |
| BomS1 | AMP-like | Toll | [61,62,63] |
| BomS2 | AMP-like | Toll | [61,62,63] |
| BomS3 | AMP-like | Toll | [61,62,63] |
| BomS5 | AMP-like | Toll | [61,62,63] |
| BomS6 | AMP-like | Toll | [61,62,63] |
| BomT2 | AMP-like | Toll | [61,62] |
| BomT3 | AMP-like | Toll | [61,62] |
| CecB | Antimicrobial peptide | IMD | [64,65] |
| CecC | Antimicrobial peptide | IMD | [64,66] |
| Def | Antimicrobial peptide | IMD, Toll | [18,67] |
| DptA | Antimicrobial peptide | IMD | [64,68] |
| DptB | Antimicrobial peptide | IMD | [64,69] |
| Drs | Antimicrobial peptide | Toll | [70,71] |
| Ets21C | Transcription factor | IMD | [72] |
| BaraA | Antimicrobial peptide | Toll | [73] |
| Irc | Oxidant detoxification | - | [74] |
| lectin-24A | Carbohydrate binding | - | [75] |
| Listericin | Antimicrobial peptide | JAK-STAT | [76] |
| mat | - | JAK-STAT | [77,78] |
| Mtk | Antimicrobial peptide | IMD, Toll | [79,80] |
| nec | Serpin | Toll | [81] |
| NimB1 | Pathogen recognition (predicted) | - | [82,83] |
| PGRP-SA | Pathogen recognition | Toll | [84,85] |
| PGRP-SB1 | Antimicrobial effector | IMD | [86,87] |
| PGRP-SD | Pathogen recognition | Toll | [88] |
| Rel | Transcription factor | IMD | [89,90] |
| Sid | DNA endonuclease | Toll | [18,91] |
| Sp7 | S1A Serine Protease | Toll | [92,93] |
| SPE | S1A Serine Protease | Toll | [94,95] |
| Tep2 | Thioester-containing Protein | Toll | [96] |
| TotM | - | JAK-STAT | [97,98] |
| CG13675 | Chitin Binding | IMD | [99] |
| CG14957 | Chitin Binding | JNK | [78] |
| CG3505 | S1A Serine Protease | Toll/IMD | [100] |
| CG5909 | S1A Serine Protease | Toll/IMD | [18] |
| Gene Set | Motif 1 | Motif 2 | Both | Neither | Total |
|---|---|---|---|---|---|
| Induced | 36 * | 39 * | 22 * | 9 * | 62 |
| Unchanged | 14 | 15 | 2 | 35 | 62 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waring, A.L.; Hill, J.; Allen, B.M.; Bretz, N.M.; Le, N.; Kr, P.; Fuss, D.; Mortimer, N.T. Meta-Analysis of Immune Induced Gene Expression Changes in Diverse Drosophila melanogaster Innate Immune Responses. Insects 2022, 13, 490. https://doi.org/10.3390/insects13050490
Waring AL, Hill J, Allen BM, Bretz NM, Le N, Kr P, Fuss D, Mortimer NT. Meta-Analysis of Immune Induced Gene Expression Changes in Diverse Drosophila melanogaster Innate Immune Responses. Insects. 2022; 13(5):490. https://doi.org/10.3390/insects13050490
Chicago/Turabian StyleWaring, Ashley L., Joshua Hill, Brooke M. Allen, Nicholas M. Bretz, Nguyen Le, Pooja Kr, Dakota Fuss, and Nathan T. Mortimer. 2022. "Meta-Analysis of Immune Induced Gene Expression Changes in Diverse Drosophila melanogaster Innate Immune Responses" Insects 13, no. 5: 490. https://doi.org/10.3390/insects13050490
APA StyleWaring, A. L., Hill, J., Allen, B. M., Bretz, N. M., Le, N., Kr, P., Fuss, D., & Mortimer, N. T. (2022). Meta-Analysis of Immune Induced Gene Expression Changes in Diverse Drosophila melanogaster Innate Immune Responses. Insects, 13(5), 490. https://doi.org/10.3390/insects13050490






