In Silico Characterization and Gene Expression Analysis of Toll Signaling Pathway-Related Genes in Diaphorina citri
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. D. citri Colonies
2.2. Protein–Protein BLAST (BLASTp) Analysis
2.3. Sequence Alignment and Phylogenetic Tree
2.4. Evolutionary Analysis of Protein AA Sequences by Maximum Likelihood Method
2.5. Generation of Three-Dimensional (3D) Structure Model
2.6. Gene Expression Analysis Using Quantitative Real-Time PCR (RT-PCR)
2.7. Statistical Analysis
3. Results
3.1. In Silico Identification and Toll Pathway-Related Genes in D. citri
3.2. Characterization of Diaphorina citri Pelle Protein (DcPelle)
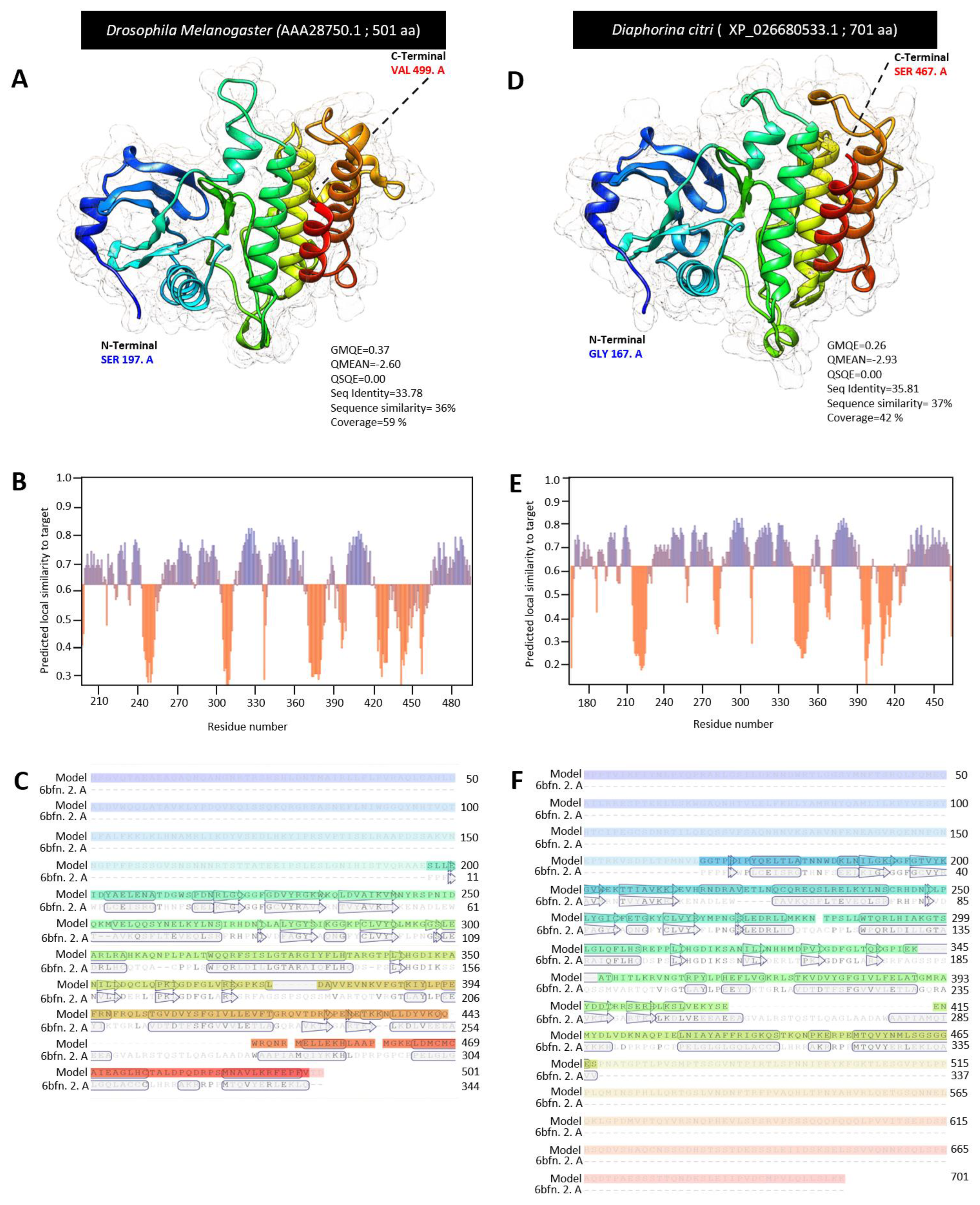
3.3. Characterization of Diaphorina citri Tube Protein (DcTube)
3.4. Characterization of Diaphorina citri Dorsal Protein (DcDorsal)
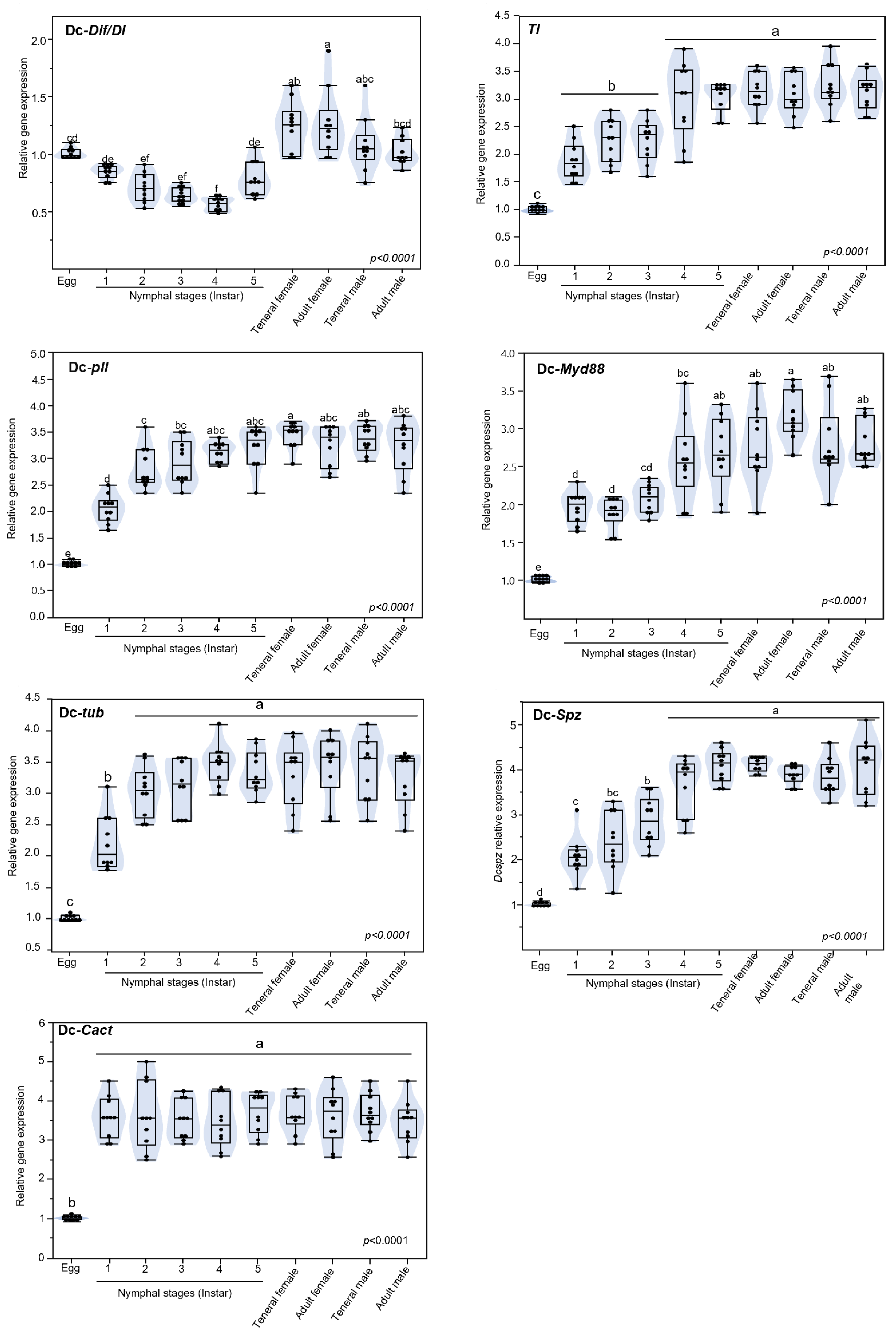
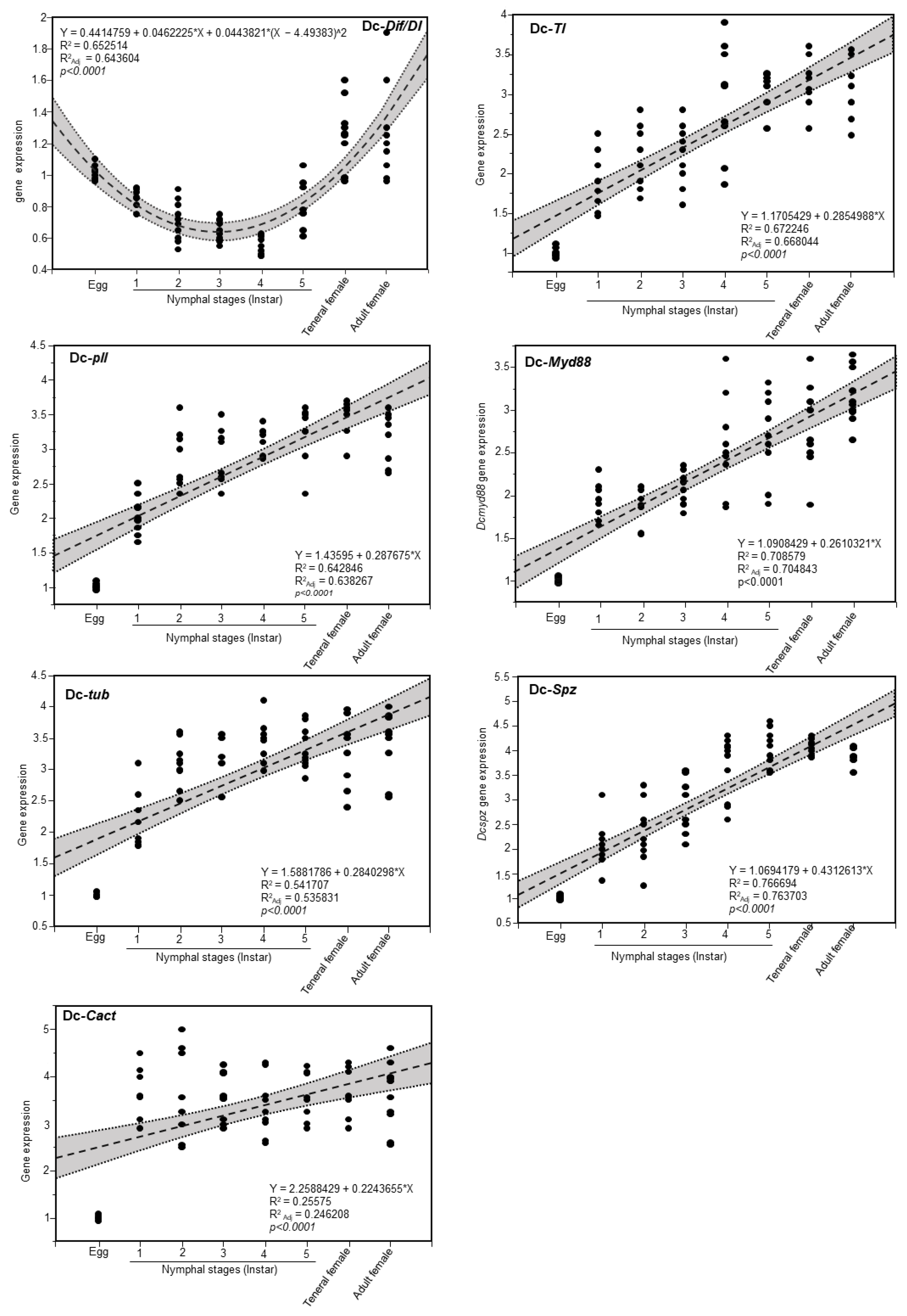
3.5. Expression of Toll Pathway-Related Genes at Different Developmental Stage of D. citri
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bové, J.M. Huanglongbing: A destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 2006, 88, 7–37. [Google Scholar]
- Da Grw, V. Citrus Greening Disease. Annu. Rev. Phytopathol. 1991, 29, 109–145. [Google Scholar]
- Halbert, S.E.; Manjunath, K.L. Asian citrus psyllids (Sternorrhyncha: Psyllidae) and greening disease of citrus: A literature review and assessment of risk in Florida. Fla. Entomol. 2004, 87, 330–353. [Google Scholar] [CrossRef]
- Coletta-Filho, H.D.; Targon, M.L.P.N.; Takita, M.A.; De Negri, J.D.; Pompeu, J.; Machado, M.A.; do Amaral, A.M.; Muller, G.W. First Report of the Causal Agent of Huanglongbing (“Candidatus Liberibacter asiaticus”) in Brazil. Plant Dis. 2004, 88, 1382. [Google Scholar] [CrossRef]
- Teixeira, D.D.C.; Danet, J.L.; Eveillard, S.; Martins, E.C.; Waldir, C.D.J., Jr.; Yamamoto, P.T.; Lopes, S.A.; Bassanezi, R.B.; Ayres, A.J.; Saillard, C.; et al. Citrus huanglongbing in São Paulo State, Brazil: PCR detection of the ‘Candidatus’ Liberibacter species associated with the disease. Mol. Cell. Probes 2005, 19, 173–179. [Google Scholar] [CrossRef]
- Halbert, S. Second International Citrus Canker and Huanglongbing Research Workshop 50. Proc. Second Int. Citrus Canker Huanglongbing Res. Work. H-3 2005. [Google Scholar]
- Wang, N.; Trivedi, P. Citrus Huanglongbing: A Newly Relevant Disease Presents Unprecedented Challenges. Phytopathology 2013, 103, 652–665. [Google Scholar] [CrossRef]
- Li, S.; Wu, F.; Duan, Y.; Singerman, A.; Guan, Z. Citrus greening: Management strategies and their economic impact. HortScience 2020, 55, 604–612. [Google Scholar] [CrossRef]
- Jagoueix, S.; Bove, J.M.; Garnier, M. The phloem-limited bacterium of greening disease of citrus is a member of the α subdivision of the Proteobacteria. Int. J. Syst. Bacteriol. 1994, 44, 379–386. [Google Scholar] [CrossRef]
- Inoue, H.; Ohnishi, J.; Ito, T.; Tomimura, K.; Miyata, S.; Iwanami, T.; Ashihara, W. Enhanced proliferation and efficient transmission of Candidatus Liberibacter asiaticus by adult Diaphorina citri after acquisition feeding in the nymphal stage. Ann. Appl. Biol. 2009, 155, 29–36. [Google Scholar] [CrossRef]
- Ammar, E.-D.; George, J.; Sturgeon, K.; Stelinski, L.L.; Shatters, R.G. Asian citrus psyllid adults inoculate huanglongbing bacterium more efficiently than nymphs when this bacterium is acquired by early instar nymphs. Sci. Rep. 2020, 10, 18244. [Google Scholar] [CrossRef] [PubMed]
- Grafton-Cardwell, E.E.; Stelinski, L.L.; Stansly, P.A. Biology and Management of Asian Citrus Psyllid, Vector of the Huanglongbing Pathogens. Annu. Rev. Entomol. 2013, 58, 413–432. [Google Scholar] [CrossRef] [PubMed]
- Hall, D.G. Biology, History and World Status of Diaphorina citri. In I Taller Internacional sobre Huanglongbing de los cítricos (Candidatus Liberibacter spp) y el psílido asiático de los cítricos (Diaphorina citri); USDA-ARS, U.S. Horticultural Research Laboratory: Fort Pierce, FL, USA, 2008; pp. 1–11. [Google Scholar]
- Qureshi, J.A.; Stansly, P.A. Dormant season foliar sprays of broad-spectrum insecticides: An effective component of integrated management for Diaphorina citri (Hemiptera: Psyllidae) in citrus orchards. Crop Prot. 2010, 29, 860–866. [Google Scholar] [CrossRef]
- Qureshi, J.A.; Stansly, P.A. Integrated Approaches for Managing the Asian Citrus Psyllid Diaphorina citri (Homoptera: Psyllidae) in Florida. Proc. Fla. State Hortic. Soc. 2007, 120, 110–115. [Google Scholar]
- Rogers, M.E.; Shawer, D.B. Effectiveness of Several Soil-applied Systemic Insecticides for Managing the Asian Citrus Psyllid, Diaphorina citri Kuwayama (Homoptera: Psyllidae). Proc. Fla. State Hortic. Soc. 2007, 120, 116–119. [Google Scholar]
- Tiwari, S.; Mann, R.S.; Rogers, M.E.; Stelinski, L.L. Insecticide resistance in field populations of Asian citrus psyllid in Florida. Pest Manag. Sci. 2011, 67, 1258–1268. [Google Scholar] [CrossRef]
- Kanga, L.H.B.; Eason, J.; Haseeb, M.; Qureshi, J.; Stansly, P. Monitoring for insecticide resistance in Asian Citrus Psyllid (Hemiptera: Psyllidae) populations in Florida. J. Econ. Entomol. 2016, 109, 832–836. [Google Scholar] [CrossRef]
- Stansly, P.A.; Arevalo, H.A.; Qureshi, J.A.; Jones, M.M.; Hendricks, K.; Roberts, P.D.; Roka, F.M. Vector control and foliar nutrition to maintain economic sustainability of bearing citrus in Florida groves affected by huanglongbing. Pest Manag. Sci. 2014, 70, 415–426. [Google Scholar] [CrossRef]
- Tansey, J.A.; Vanaclocha, P.; Monzo, C.; Jones, M.; Stansly, P.A. Costs and benefits of insecticide and foliar nutrient applications to huanglongbing-infected citrus trees. Pest Manag. Sci. 2017, 73, 904–916. [Google Scholar] [CrossRef]
- Monzó, C.; Stansly, P.A. Thresholds for vector control and compatibility with beneficial fauna in citrus with high incidence of huanglongbing. Acta Hortic. 2015, 1065, 1137–1144. [Google Scholar] [CrossRef]
- Tsakas, S.; Marmaras, V.J. Insect immunity and its signalling: An overview. Invertebr. Surviv. J. 2010, 7, 228–238. [Google Scholar]
- De Gregorio, E.; Spellman, P.T.; Tzou, P.; Rubin, G.M.; Lemaitre, B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 2002, 21, 2568–2579. [Google Scholar] [CrossRef]
- Gottar, M.; Gobert, V.; Michel, T.; Belvin, M.; Duyk, G.; Hoffmann, J.A.; Ferrandon, D.; Royet, J. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 2002, 416, 640–644. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, V.; Reichhart, J.M. The immune response of Drosophila melanogaster. Immunol. Rev. 2004, 198, 59–71. [Google Scholar] [CrossRef]
- Zhang, C.; Yan, S.Q.; Shen, B.B.; Ali, S.; Wang, X.M.; Jin, F.L.; Cuthbertson, A.G.S.; Qiu, B.L. RNAi knock-down of the Bemisia tabaci Toll gene (BtToll) increases mortality after challenge with destruxin A. Mol. Immunol. 2017, 88, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Zambon, R.A.; Nandakumar, M.; Vakharia, V.W.; Wu, L.P. The Toll pathway is important for an antiviral response in Drosophila. Proc. Natl. Acad. Sci. USA 2005, 102, 7257–7262. [Google Scholar] [CrossRef] [Green Version]
- Gillespie, J.P.; Kanost, M.R.; Trenczek, T. Biological mediators of insect immunity. Annu. Rev. Entomol. 1997, 42, 611–643. [Google Scholar] [CrossRef]
- Pili-Floury, S.; Leulier, F.; Takahashi, K.; Saigo, K.; Samain, E.; Ueda, R.; Lemaitre, B. In Vivo RNA Interference Analysis Reveals an Unexpected Role for GNBP1 in the Defense against Gram-positive Bacterial Infection in Drosophila Adults. J. Biol. Chem. 2004, 279, 12848–12853. [Google Scholar] [CrossRef]
- Wang, Y.; Sumathipala, N.; Rayaprolu, S.; Jiang, H. Recognition of microbial molecular patterns and stimulation of prophenoloxidase activation by a β-1,3-glucanase-related protein in Manduca sexta larval plasma. Insect Biochem. Mol. Biol. 2011, 41, 322–331. [Google Scholar] [CrossRef]
- Dziarski, R.; Gupta, D. The peptidoglycan recognition proteins (PGRPs). Genome Biol. 2006, 7, 232. [Google Scholar] [CrossRef]
- Lemaitre, B.; Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743. [Google Scholar] [CrossRef] [PubMed]
- Silverman, N.; Maniatis, T. NF-κB signaling pathways in mammalian and insect innate immunity. Genes Dev. 2001, 15, 2321–2342. [Google Scholar] [CrossRef]
- Bangham, J.; Jiggins, F.; Lemaitre, B. Insect Immunity: The Post-Genomic Era. Immunity 2006, 25, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Müller, U.; Vogel, P.; Alber, G.; Schaub, G. The innate immune system of mammals and insects. Contrib. Microbiol. 2008, 15, 21–44. [Google Scholar] [CrossRef]
- He, Y.J.; Lu, G.; Qi, Y.H.; Zhang, Y.; Zhang, X.D.; Huang, H.J.; Zhuo, J.C.; Sun, Z.T.; Yan, F.; Chen, J.P.; et al. Activation of Toll Immune Pathway in an Insect Vector Induced by a Plant Virus. Front. Immunol. 2021, 11, 613957. [Google Scholar] [CrossRef]
- Bang, I.S. JAK/STAT signaling in insect innate immunity. Entomol. Res. 2019, 49, 339–353. [Google Scholar] [CrossRef]
- Govind, S. Innate immunity in Drosophila: Pathogens and pathways. Insect Sci. 2008, 15, 29. [Google Scholar] [CrossRef] [PubMed]
- Kojour, M.A.M.; Han, Y.S.; Hun, Y.J.; Jo, Y.H. An overview of insect innate immunity. Entomol. Res. 2020, 50, 282–291. [Google Scholar] [CrossRef]
- Akira, S.; Takeda, K.; Kaisho, T. Toll-like receptors: Critical proeins linking innate and acquired immunity. Nat. Immunol. 2001, 2, 675–680. [Google Scholar] [CrossRef]
- Stokes, B.A.; Yadav, S.; Shokal, U.; Smith, L.C.; Eleftherianos, I. Bacterial and fungal pattern recognition receptors in homologous innate signaling pathways of insects and mammals. Front. Microbiol. 2015, 6, 19. [Google Scholar] [CrossRef]
- Kanost, M.R.; Jiang, H.; Yu, X.Q. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev. 2004, 198, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Buchon, N.; Silverman, N.; Cherry, S. Immunity in Drosophila melanogaster-from microbial recognition to whole-organism physiology. Nat. Rev. Immunol. 2014, 14, 796–810. [Google Scholar] [CrossRef] [PubMed]
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645. [Google Scholar] [CrossRef] [PubMed]
- Ferrandon, D.; Imler, J.L.; Hetru, C.; Hoffmann, J.A. The Drosophila systemic immune response: Sensing and signalling during bacterial and fungal infections. Nat. Rev. Immunol. 2007, 7, 862–874. [Google Scholar] [CrossRef]
- Cao, X.; He, Y.; Hu, Y.; Wang, Y.; Chen, Y.R.; Bryant, B.; Clem, R.J.; Schwartz, L.M.; Blissard, G.; Jiang, H. The immune signaling pathways of Manduca sexta. Insect Biochem. Mol. Biol. 2015, 62, 64–74. [Google Scholar] [CrossRef]
- Arp, A.P.; Hunter, W.B.; Pelz-Stelinski, K.S. Annotation of the Asian citrus psyllid genome reveals a reduced innate immune system. Front. Physiol. 2016, 7, 570. [Google Scholar] [CrossRef] [Green Version]
- Gerardo, N.M.; Altincicek, B.; Anselme, C.; Atamian, H.; Barribeau, S.M.; De Vos, M.; Duncan, E.J.; Evans, J.D.; Gabaldón, T.; Ghanim, M.; et al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 2010, 11, R21. [Google Scholar] [CrossRef]
- Kim, J.H.; Min, J.S.; Kang, J.S.; Kwon, D.H.; Yoon, K.S.; Strycharz, J.; Koh, Y.H.; Pittendrigh, B.R.; Clark, J.M.; Lee, S.H. Comparison of the humoral and cellular immune responses between body and head lice following bacterial challenge. Insect Biochem. Mol. Biol. 2011, 41, 332–339. [Google Scholar] [CrossRef]
- Mesquita, R.D.; Vionette-Amaral, R.J.; Lowenberger, C.; Rivera-Pomar, R.; Monteiro, F.A.; Minx, P.; Spieth, J.; Carvalho, A.B.; Panzera, F.; Lawson, D.; et al. Genome of Rhodnius prolixus, an insect vector of Chagas disease, reveals unique adaptations to hematophagy and parasite infection. Proc. Natl. Acad. Sci. USA 2015, 112, 14936–14941. [Google Scholar] [CrossRef]
- Arp, A.P.; Martini, X.; Pelz-Stelinski, K.S. Innate immune system capabilities of the Asian citrus psyllid, Diaphorina citri. J. Invertebr. Pathol. 2017, 148, 94–101. [Google Scholar] [CrossRef]
- Moreira, L.A.; Iturbe-Ormaetxe, I.; Jeffery, J.A.; Lu, G.; Pyke, A.T.; Hedges, L.M.; Rocha, B.C.; Hall-Mendelin, S.; Day, A.; Riegler, M.; et al. A Wolbachia Symbiont in Aedes aegypti Limits Infection with Dengue, Chikungunya, and Plasmodium. Cell 2009, 139, 1268–1278. [Google Scholar] [CrossRef] [PubMed]
- Nakabachi, A.; Ueoka, R.; Oshima, K.; Teta, R.; Mangoni, A.; Gurgui, M.; Oldham, N.J.; Van Echten-Deckert, G.; Okamura, K.; Yamamoto, K.; et al. Defensive bacteriome symbiont with a drastically reduced genome. Curr. Biol. 2013, 23, 1478–1484. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, J.S.; Johnson, R.S.; Hoki, J.S.; Kruse, A.; Mahoney, J.; Hilf, M.E.; Hunter, W.B.; Hall, D.G.; Schroeder, F.C.; MacCoss, M.J.; et al. Metabolic interplay between the asian citrus psyllid and its profftella symbiont: An achilles’ heel of the citrus greening insect vector. PLoS ONE 2015, 10, e0140826. [Google Scholar] [CrossRef]
- Tatineni, S.; Sagaram, U.S.; Gowda, S.; Robertson, C.J.; Dawson, W.O.; Iwanami, T.; Wang, N. In planta distribution of “Candidatus Liberibacter asiaticus” as revealed by polymerase chain reaction (PCR) and real-time PCR. Phytopathology 2008, 98, 592–599. [Google Scholar] [CrossRef] [PubMed]
- Goto, A.; Blandin, S.; Royet, J.; Reichhart, J.M.; Levashina, E.A. Silencing of Toll pathway components by direct injection of double-stranded RNA into Drosophila adult flies. Nucleic Acids Res. 2003, 31, 6619–6623. [Google Scholar] [CrossRef]
- Bingsohn, L.; Knorr, E.; Billion, A.; Narva, K.E.; Vilcinskas, A. Knockdown of genes in the Toll pathway reveals new lethal RNA interference targets for insect pest control. Insect Mol. Biol. 2017, 26, 92–102. [Google Scholar] [CrossRef]
- Tauszig-Delamasure, S.; Bilak, H.; Capovilla, M.; Hoffmann, J.A.; Imler, J.L. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat. Immunol. 2002, 3, 91–97. [Google Scholar] [CrossRef]
- Clemmons, A.W.; Wasserman, S.A. Combinatorial effects of transposable elements on gene expression and phenotypic robustness in Drosophila melanogaster development. G3 2013, 3, 1531–1538. [Google Scholar] [CrossRef]
- Parthier, C.; Stelter, M.; Ursel, C.; Fandrich, U.; Lilie, H.; Breithaupt, C.; Stubbs, M.T. Structure of the Toll-Spätzle complex, a molecular hub in Drosophila development and innate immunity. Proc. Natl. Acad. Sci. USA 2014, 111, 6281–6286. [Google Scholar] [CrossRef]
- Altschul, S.F.; Wootton, J.C.; Gertz, E.M.; Agarwala, R.; Morgulis, A.; Schäffer, A.A.; Yu, Y.K. Protein database searches using compositionally adjusted substitution matrices. FEBS J. 2005, 272, 5101–5109. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs; Oxford University Press: Oxford, UK, 1997; Volume 25. [Google Scholar]
- Flores-Gonzalez, M.; Hosmani, P.S.; Fernandez-Pozo, N.; Mann, M.; Humann, J.L.; Main, D.; Heck, M.; Brown, S.; Mueller, L.A.; Saha, S. Citrusgreening.org: An open access and integrated systems biology portal for the Huanglongbing (HLB) disease complex. BioRxiv 2019, 868364. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 1992, 8, 275–282. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Nehela, Y.; Killiny, N. Infection with phytopathogenic bacterium inhibits melatonin biosynthesis, decreases longevity of its vector, and suppresses the free radical-defense. J. Pineal Res. 2018, 65, e12511. [Google Scholar] [CrossRef]
- Santos-Ortega, Y.; Killiny, N. Silencing of sucrose hydrolase causes nymph mortality and disturbs adult osmotic homeostasis in Diaphorina citri (Hemiptera: Liviidae). Insect Biochem. Mol. Biol. 2018, 101, 131–143. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Rutschmann, S.; Kilinc, A.; Ferrandon, D. Cutting Edge: The Toll Pathway Is Required for Resistance to Gram-Positive Bacterial Infections in Drosophila. J. Immunol. 2002, 168, 1542–1546. [Google Scholar] [CrossRef]
- Lemaitre, B.; Nicolas, E.; Michaut, L.; Reichhart, J.M.; Hoffmann, J.A. The dorsoventral regulatory gene cassette spatzle/Toll/Cactus controls the potent antifungal response in Drosophila adults. Cell 1996, 86, 973–983. [Google Scholar] [CrossRef]
- Lemaitre, B.; Kromer-Metzger, E.; Michaut, L.; Nicolas, E.; Meister, M.; Georgel, P.; Reichhart, J.M.; Hoffmann, J.A. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc. Natl. Acad. Sci. USA 1995, 92, 9465–9469. [Google Scholar] [CrossRef]
- Tanji, T.; Hu, X.; Weber, A.N.R.; Ip, Y.T. Toll and IMD Pathways Synergistically Activate an Innate Immune Response in Drosophila melanogaster. Mol. Cell. Biol. 2007, 27, 4578–4588. [Google Scholar] [CrossRef]
- Valanne, S.; Wang, J.-H.; Rämet, M. The Drosophila Toll Signaling Pathway. J. Immunol. 2011, 186, 649–656. [Google Scholar] [CrossRef]
- Lindsay, S.A.; Wasserman, S.A. Conventional and non-conventional Drosophila Toll signaling. Dev. Comp. Immunol. 2014, 42, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Towb, P.; Sun, H.; Wasserman, S.A. Tube is an IRAK-4 homolog in a toll pathway adapted for development and immunity. J. Innate Immun. 2009, 1, 309–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanji, T.; Ip, Y.T. Regulators of the Toll and Imd pathways in the Drosophila innate immune response. Trends Immunol. 2005, 26, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Qiu, P.; Pan, P.C.; Govind, S. A role for the Drosophila Toll/Cactus pathway in larval hematopoiesis. Development 1998, 125, 1909–1920. [Google Scholar] [CrossRef]
- Ganesan, S.; Aggarwal, K.; Paquette, N.; Silverman, N. NF-κB/Rel Proteins and the Humoral Immune Responses of Drosophila melanogaster. Curr. Top. Microbiol. Immunol. 2010, 349. [Google Scholar] [CrossRef]
- Hanks, S.K.; Quinn, A.M.; Hunter, T. The Protein Kinase Family: Conserved Features and Deduced PHylogeny of the Catalytic Domains. Science 1988, 241, 42–52. [Google Scholar] [CrossRef]
- Ferreira, Á.G.; Naylor, H.; Esteves, S.S.; Pais, I.S.; Martins, N.E.; Teixeira, L. The Toll-Dorsal Pathway Is Required for Resistance to Viral Oral Infection in Drosophila. PLoS Pathog. 2014, 10, e1004507. [Google Scholar] [CrossRef] [PubMed]
- Berni, M.; Fontenele, M.R.; Tobias-Santos, V.; Caceres-Rodrigues, A.; Mury, F.B.; Vionette-do-Amaral, R.; Masuda, H.; Sorgine, M.; da Fonseca, R.N.; Araujo, H. Toll signals regulate dorsal-ventral patterning and anterior-posterior placement of the embryo in the hemipteran Rhodnius prolixus. Evodevo 2014, 5, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Gene | Primers | Nucleotide Sequence 5′-3′ | Length (bp) |
|---|---|---|---|
| Cact | Dc-Cact F | GAGCTAGCACAAGTGATGGG | 147 |
| Dc-Cact R | CAACACTAGCTGTCGCATGA | ||
| Dif/Di | Dc-Dif F | AGCGTATCTGCCCAAGGATA | 120 |
| Dc_Dif R | GCAGACACATGGGACAAAGT | ||
| TI | Dc-TI F | TTGTCGGTATCCTGACCCTT | 112 |
| Dc-TI R | CGATCGGTCCTAATCGGTTG | ||
| Myd88 | Dc-Myd88 F | TTGAAGCCATCAGGCCAAAG | 147 |
| Dc-Myd88 R | CACAGTTGGGATTTGGAGGG | ||
| Spz | Dc-Spz F | GGTGCGTCCAGGAATACATC | 122 |
| Dc-Spz R | GGGTTCAAAGTTGTGTTGCG | ||
| pll | Dc-pll F | CCAGAGCAGAGCTGTGTAGCA | 161 |
| Dc-pll R | CTGAGCACCCCACTTGGACA | ||
| tub | Dc-tub F | AACCGGAATCTCCCGGTGAC | 143 |
| Dc-Tub R | ACCATCAGTAGCGCGTTCCA | ||
| actin | Dc-Actin F | CCCTGGACTTTGAACAGGAA | 170 |
| Dc-Actin R | CTCGTGGATACCGCAAGATT | ||
| Alpha-tubulin | Dc-Tubulin F | CTTTCCAACACCACCGCTAT | 142 |
| Dc-Tubulin R | GGTCTTCCCTCGCCTCTG |
| Drosophila melanogaster (Accession #) | Diaphorina citri | |||||||
|---|---|---|---|---|---|---|---|---|
| Annotation in D. citri | Accession # | aa | Max Score | Total Score | Query Cover % | E Value | Identity % | |
| Cactus (AAA85908.1). | NF-kappa-B inhibitor Cactus | XP_008487783.1 | 304 | 122 | 122 | 34 | 1 × 10−31 | 37.57 |
| * Dorsal related immunity factor (AAA28465.1) | embryonic polarity protein Dorsal-like isoform X3 | XP_017305229.1 | 541 | 211 | 211 | 47 | 9 × 10−61 | 39.44 |
| embryonic polarity protein Dorsal-like isoform X2 | XP_026675878.1 | 667 | 208 | 208 | 44 | 2 × 10−58 | 41.14 | |
| embryonic polarity protein Dorsal-like isoform X1 | XP_017305228.1 | 691 | 208 | 208 | 44 | 2 × 10−58 | 41.14 | |
| Toll (AAA28941.1) | protein Toll-like | XP_026684268.1 | 857 | 283 | 283 | 68 | 4 × 10−81 | 29.54 |
| protein Toll-like | XP_026685794.1 | 1109 | 269 | 403 | 76 | 4 × 10−75 | 30.8 | |
| protein Toll-like | XP_008479067.1 | 169 | 63.2 | 63.2 | 11 | 2 × 10−11 | 24.81 | |
| Pelle (AAA28750.1) | serine/threonine-protein kinase pelle-like isoform X3 | XP_026680533.1 | 701 | 266 | 266 | 90 | 2 × 10−81 | 38 |
| serine/threonine-protein kinase pelle-like isoform X2 | XP_026680532.1 | 703 | 266 | 266 | 90 | 2 × 10−81 | 38 | |
| serine/threonine-protein kinase pelle-like isoform X1 | XP_026680531.1 | 708 | 266 | 266 | 90 | 2 × 10−81 | 38 | |
| ** Myd88 (AAL56570.1) | myd88-RA. (AHRD V3.11 *** Myeloid differentiation primary response protein MyD88 tr | DcitrP049290.1.1 | 396 | 92.0 | 5 × 10−20 | 28.06 | ||
| Tube (P22812.4) | protein Tube, partial | XP_008486711.1 | 229 | 67.0 | 67.0 | 33 | 8 × 10−13 | 29.49 |
| interleukin-1 receptor-associated kinase 4-like | XP_008485884.1 | 455 | 67.0 | 67.0 | 33 | 5 × 10−12 | 29.49 | |
| ** Spaetzle (NP_733194.1) | spaetzle5-PA partial | DcitrP089585.1.5 | 296 | - | - | - | 2 × 10−18 | 30.61 |
| spaetzle5-PA partial | DcitrP089585.1.4 | 296 | - | - | - | 2 × 10−18 | 30.61 | |
| spaetzle5-PA partial | DcitrP089585.1.3 | 296 | - | - | - | 2 × 10−18 | 30.61 | |
| spaetzle5-PA partial | DcitrP089585.1.2 | 296 | - | - | - | 2 × 10−18 | 30.61 | |
| spaetzle5-PA partial | DcitrP089585.1.1 | 296 | - | - | - | 2 × 10−18 | 30.61 | |
| * Dorsal (Di; NP_724054.1) | embryonic polarity protein Dorsal-like isoform X2 | XP_026675878.1 | 667 | 289 | 384 | 52 | 1 × 10−85 | 47.38 |
| embryonic polarity protein Dorsal-like isoform X1 | XP_017305228.1 | 691 | 289 | 384 | 53 | 2 × 10−85 | 46.85 | |
| embryonic polarity protein Dorsal-like isoform X3 | XP_017305229.1 | 541 | 280 | 280 | 32 | 7 × 10−84 | 46.58 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rashidi, M.; Killiny, N. In Silico Characterization and Gene Expression Analysis of Toll Signaling Pathway-Related Genes in Diaphorina citri. Insects 2022, 13, 783. https://doi.org/10.3390/insects13090783
Rashidi M, Killiny N. In Silico Characterization and Gene Expression Analysis of Toll Signaling Pathway-Related Genes in Diaphorina citri. Insects. 2022; 13(9):783. https://doi.org/10.3390/insects13090783
Chicago/Turabian StyleRashidi, Mahnaz, and Nabil Killiny. 2022. "In Silico Characterization and Gene Expression Analysis of Toll Signaling Pathway-Related Genes in Diaphorina citri" Insects 13, no. 9: 783. https://doi.org/10.3390/insects13090783
APA StyleRashidi, M., & Killiny, N. (2022). In Silico Characterization and Gene Expression Analysis of Toll Signaling Pathway-Related Genes in Diaphorina citri. Insects, 13(9), 783. https://doi.org/10.3390/insects13090783







