Bta06987, Encoding a Peptide of the AKH/RPCH Family: A Role of Energy Mobilization in Bemisia tabaci
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plants and B. tabaci
2.2. Gene Cloning
2.3. Characterization and Phylogenetic Analysis
2.4. The relative Expression of Bta06987 under Starvation
2.5. Double-Stranded RNA Synthesis
2.6. Efficacy of dsRNA
2.7. Carbohydrate Content and Triacylglycerol (TAG) Content Determination after RNAi
2.8. Feeding Behavior after RNAi
2.9. Statistical Analysis
3. Results
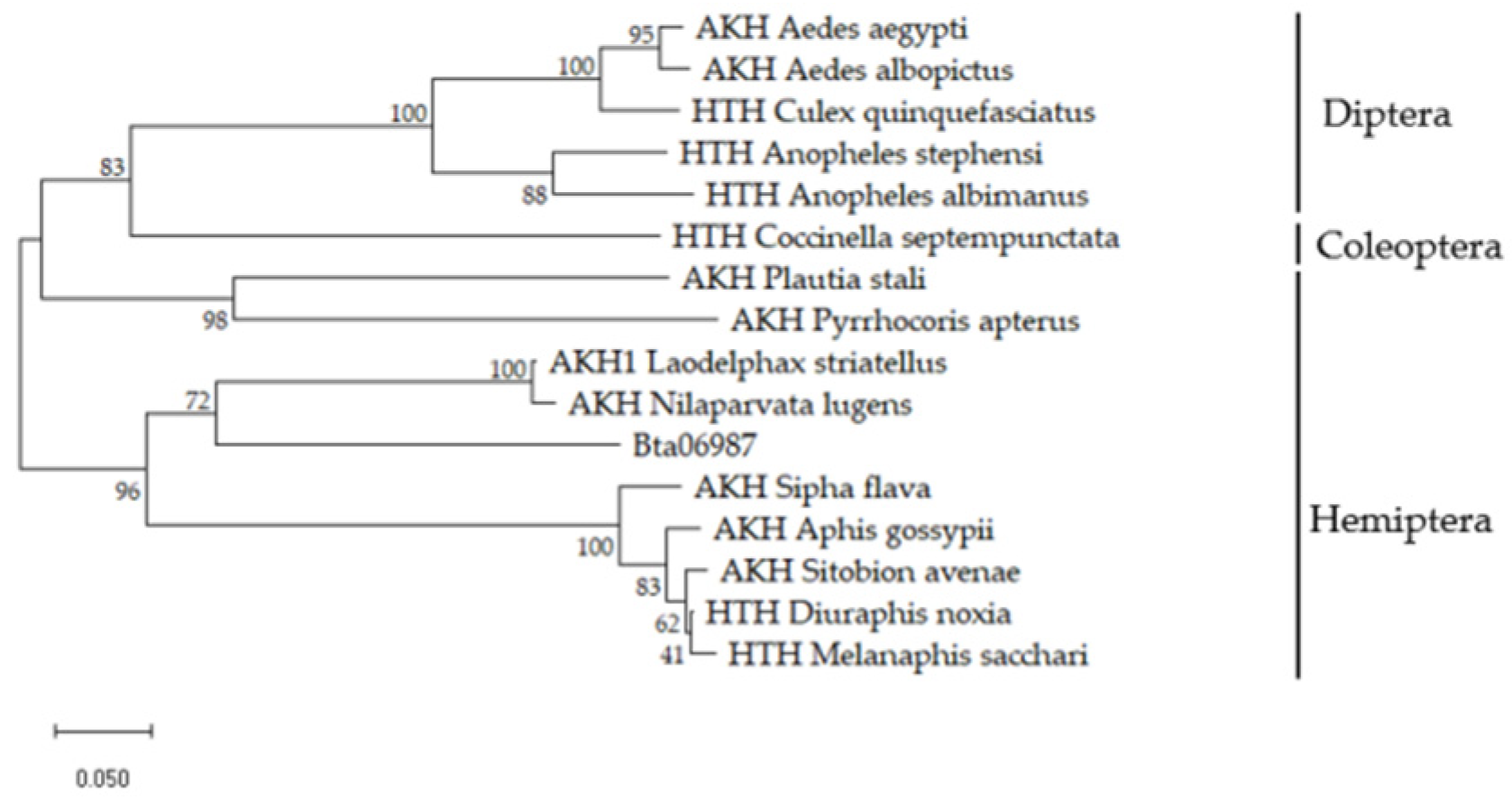
3.1. Cloning and Phylogenetic Analysis of Bta06987
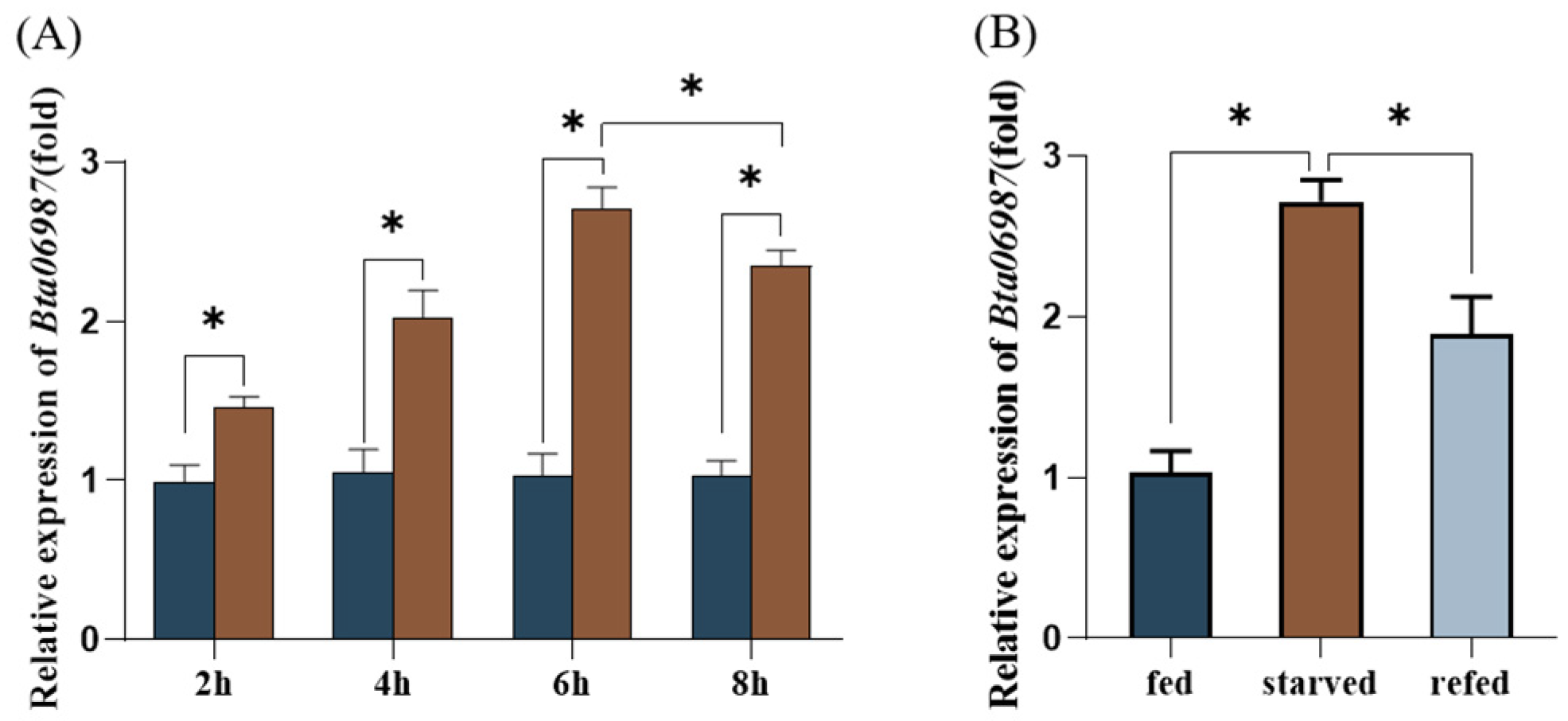
3.2. The Expression of Bta06987 under Starvation
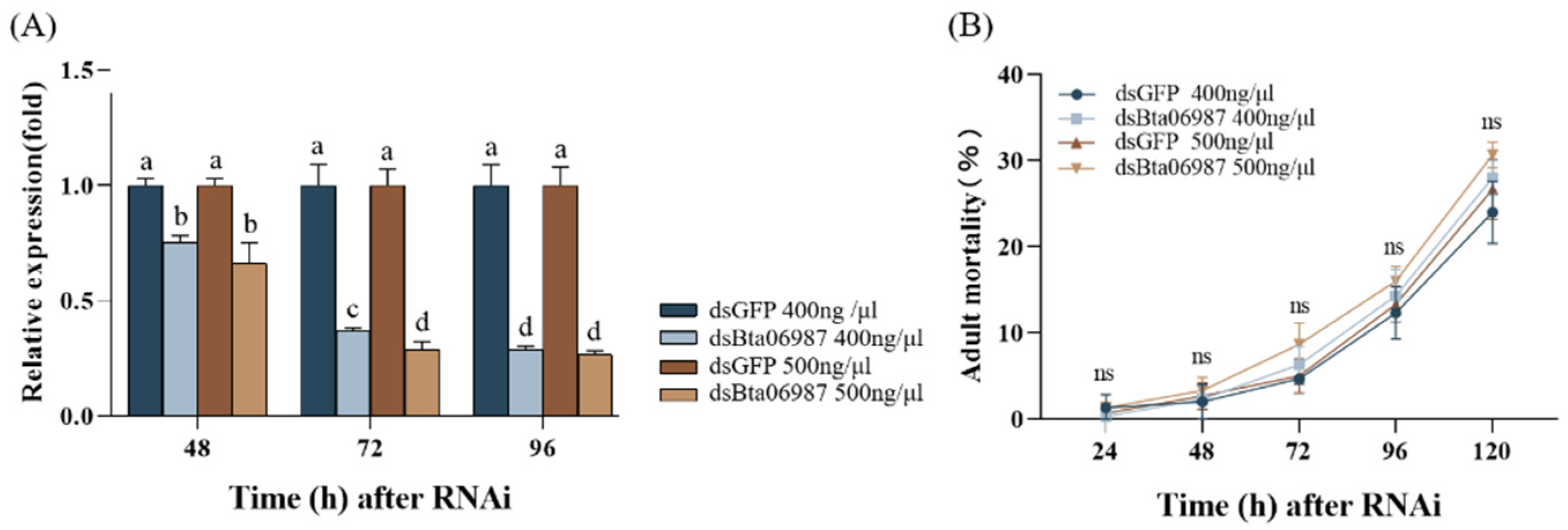
3.3. Efficacy of dsRNA
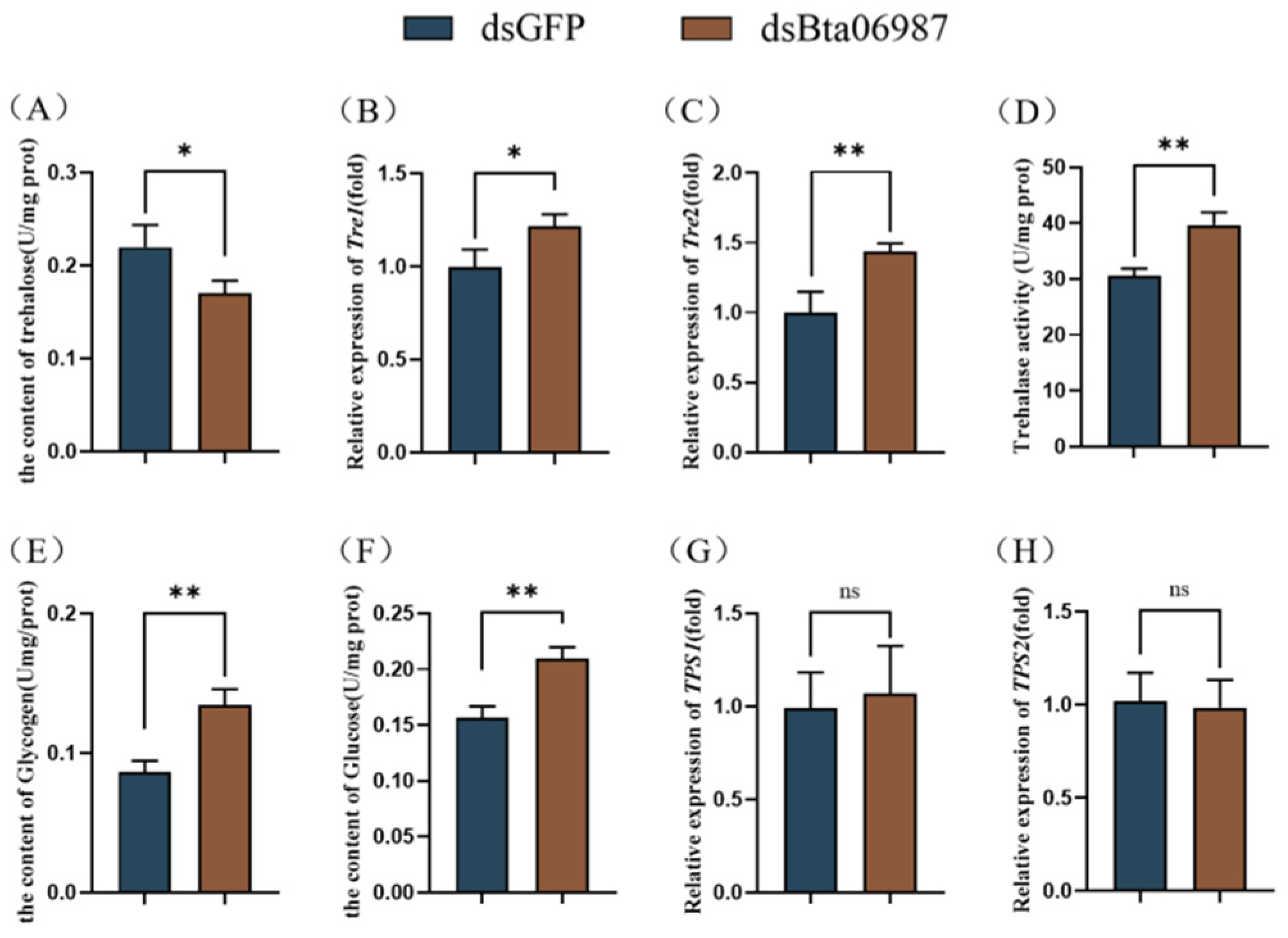
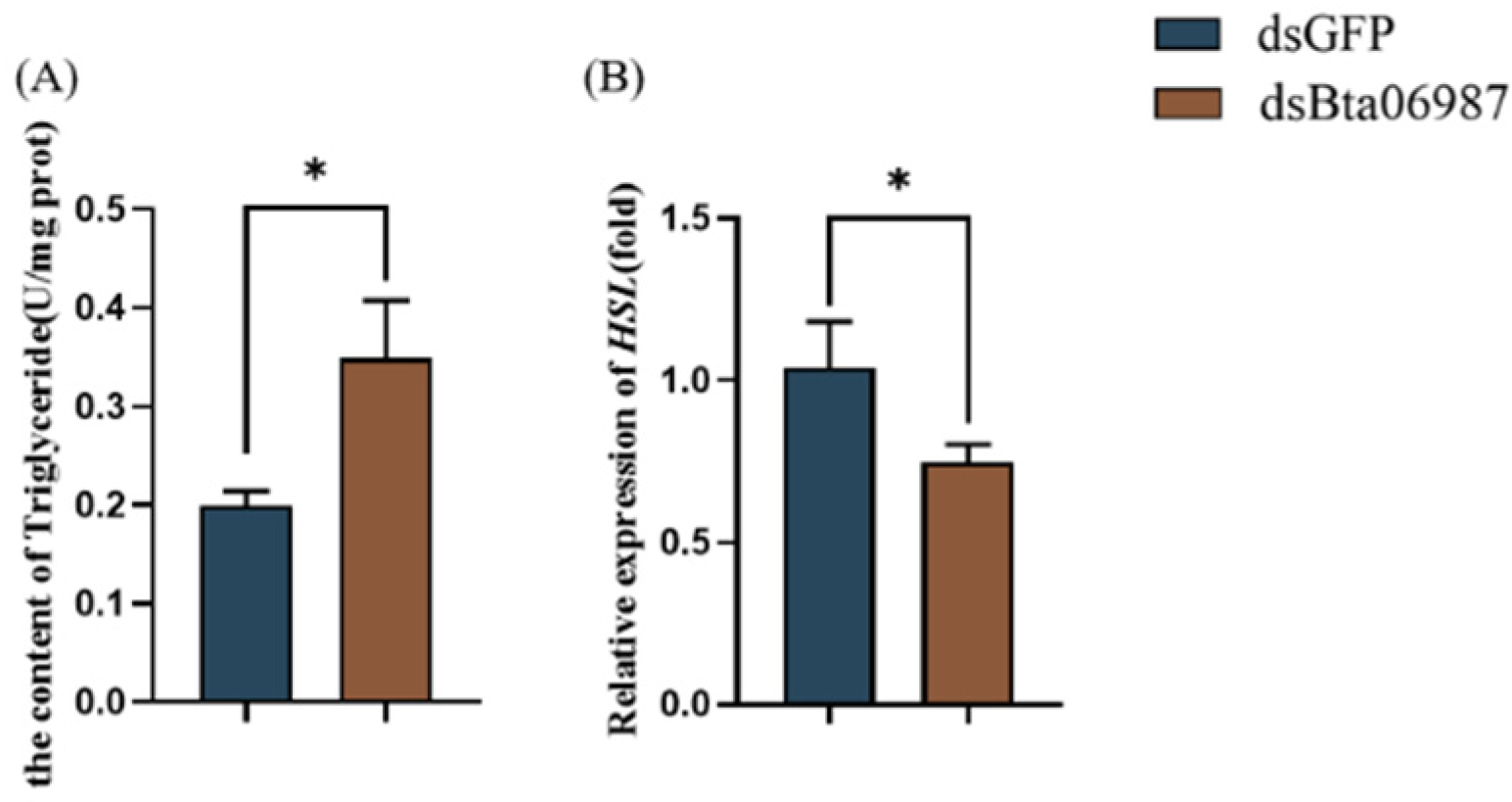
3.4. Carbohydrate Content
3.5. Triglyceride Content
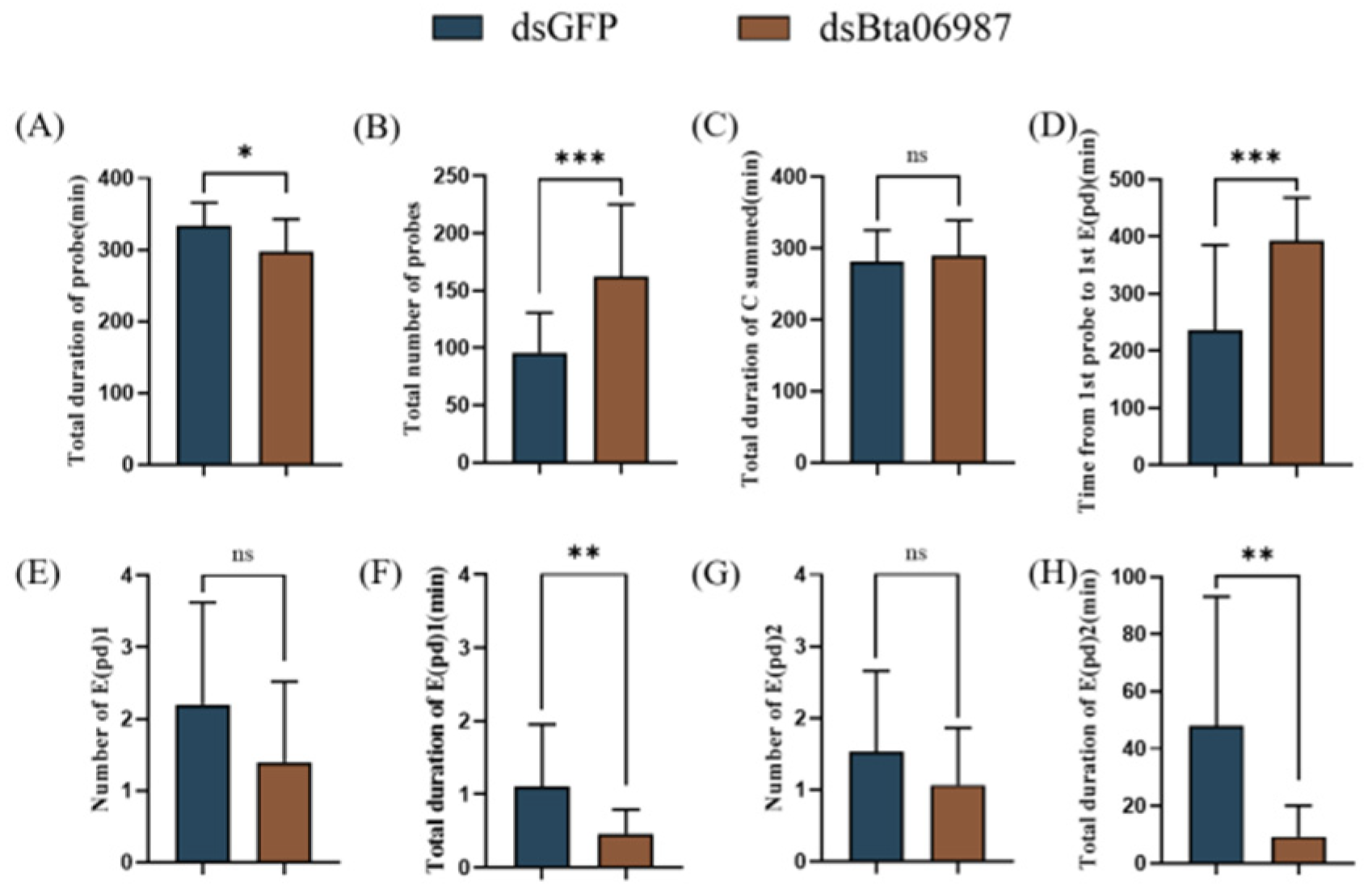
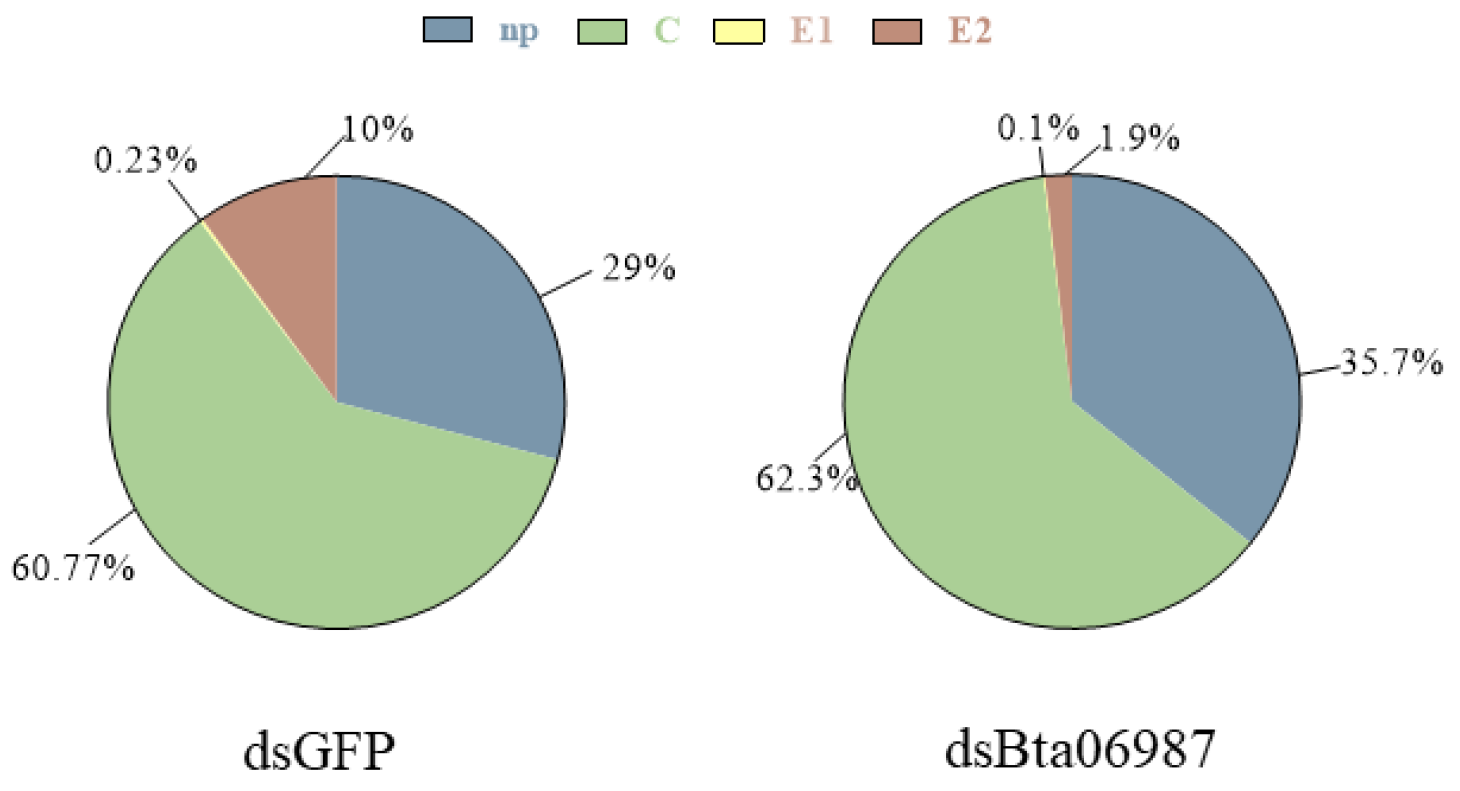
3.6. Stylet Activities
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gäde, G.; Hoffmann, K.H.; Spring, J.H. Hormonal regulation in insects: Facts, gaps, and future directions. Physiol. Rev. 1997, 77, 963–1032. [Google Scholar] [CrossRef]
- Gäde, G.; Auerswald, L. Mode of action of neuropeptides from the adipokinetic hormone family. Gen. Comp. Endocrinol. 2003, 132, 10–20. [Google Scholar] [CrossRef]
- Gäde, G.; Marco, H.G. AKH/RPCH Peptides. In Handbook of Biologically Active Peptides; Elsevier: Amsterdam, The Netherlands, 2013; pp. 185–190. [Google Scholar]
- Gäde, G.; Marco, H.G. The Adipokinetic Peptides of Hemiptera: Structure, Function, and Evolutionary Trends. Front. Insect Sci. 2022, 2, 891615. [Google Scholar] [CrossRef]
- Gäde, G. Peptides of the adipokinetic hormone/red pigment-concentrating hormone family: A new take on biodiversity. Ann. N. Y. Acad. Sci. 2009, 1163, 125–136. [Google Scholar] [CrossRef]
- Kodrík, D. Adipokinetic hormone functions that are not associated with insect flight. Physiol. Entomol. 2008, 33, 171–180. [Google Scholar] [CrossRef]
- Lorenz, M.; Gäde, G. Hormonal regulation of energy metabolism in insects as a driving force for performance. Integr. Comp. Biol. 2009, 49, 380–392. [Google Scholar] [CrossRef]
- Orchard, I. Adipokinetic hormones—An update. J. Insect Physiol. 1987, 33, 451–463. [Google Scholar] [CrossRef]
- Gäde, G. Adipokinetic Hormones and the Hormonal Control of Metabolic Activity in Hemiptera. Pestycydy 2005, 4, 49–54. [Google Scholar]
- Isabel, G.; Martin, J.R.; Chidami, S.; Veenstra, J.A.; Rosay, P. AKH-producing neuroendocrine cell ablation decreases trehalose and induces behavioral changes in Drosophila. Am. J. Regul. Integr. Comp. Physiol. 2005, 288, R531–R538. [Google Scholar] [CrossRef]
- Huang, J.-H.; Lee, H.-J. RNA interference unveils functions of the hypertrehalosemic hormone on cyclic fluctuation of hemolymph trehalose and oviposition in the virgin female Blattella germanica. J. Insect Physiol. 2011, 57, 858–864. [Google Scholar] [CrossRef]
- Hergarden, A.C.; Tayler, T.D.; Anderson, D.J. Allatostatin-A neurons inhibit feeding behavior in adult Drosophila. Proc. Natl. Acad. Sci. USA 2012, 109, 3967–3972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Qu, M.-J.; Zhang, Y.; Li, J.-W.; Liu, T.-X. Expression of neuropeptide F gene and its regulation of feeding behavior in the pea aphid, Acyrthosiphon pisum. Front. Physiol. 2018, 9, 87. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Wen, T.; Lee, G.; Park, J.H.; Cai, H.N.; Shen, P. Developmental control of foraging and social behavior by the Drosophila neuropeptide Y-like system. Neuron 2003, 39, 147–161. [Google Scholar] [CrossRef]
- Root, C.M.; Ko, K.I.; Jafari, A.; Wang, J.W. Presynaptic facilitation by neuropeptide signaling mediates odor-driven food search. Cell 2011, 145, 133–144. [Google Scholar] [CrossRef]
- Kodrík, D.; Vinokurov, K.; Tomčala, A.; Socha, R. The effect of adipokinetic hormone on midgut characteristics in Pyrrhocoris apterus L. (Heteroptera). J. Insect Physiol. 2012, 58, 194–204. [Google Scholar] [CrossRef]
- Lee, G.; Park, J.H. Hemolymph sugar homeostasis and starvation-induced hyperactivity affected by genetic manipulations of the adipokinetic hormone-encoding gene in Drosophila melanogaster. Genetics 2004, 167, 311–323. [Google Scholar] [CrossRef]
- Ahmad, M.; Khan, R.A. Field-evolved resistance of Bemisia tabaci (Hemiptera: Aleyrodidae) to carbodiimide and neonicotinoids in Pakistan. J. Econ. Entomol. 2017, 110, 1235–1242. [Google Scholar] [CrossRef]
- Horowitz, A.R.; Ghanim, M.; Roditakis, E.; Nauen, R.; Ishaaya, I. Insecticide resistance and its management in Bemisia tabaci Species. J. Pest Sci. 2020, 93, 893–910. [Google Scholar] [CrossRef]
- Mu, Y.; Shi, X.; Zhang, Z.; Zhang, Z.; Wang, T.; Wang, Y.; Wei, Y.; Zhou, X.; Xiang, M.; Liu, Y.; et al. Validamycin reduces the transmission of Tomato Chlorotic Virus by Bemisia tabaci. J. Pest Sci. 2021, 95, 1261–1272. [Google Scholar] [CrossRef]
- Mayoral, A.M.; Tjallingii, W.F.; Castañera, P. Probing behaviour of diuraphis noxia on five cereal species with different hydroxamic acid levels. Entomol. Exp. Appl. 1996, 78, 341–348. [Google Scholar] [CrossRef]
- Lu, S.; Li, J.; Bai, R.; Yan, F. EPG-Recorded Feeding Behaviors Reveal Adaptability and Competitiveness in Two Species of Bemisia Tabaci (Hemiptera: Aleyrodidae). J. Insect Behav. 2021, 34, 26–40. [Google Scholar] [CrossRef]
- De Barro, P.J.; Liu, S.S.; Boykin, L.M.; Dinsdale, A.B. Bemisia tabaci: A statement of species status. Ann. Rev. Entomol. 2011, 56, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Lalitha, S. Primer premier 5. Biotech Softw. Internet Rep. Comput. Softw. J. Sci. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Luo, Y.; Chen, Q.; Luan, J.; Chung, S.H.; Van Eck, J.; Turgeon, R.; Douglas, A.E. Towards an understanding of the molecular basis of effective RNAi against a global insect pest, the whitefly Bemisia tabaci. Insect Biochem. Mol. Biol. 2017, 88, 21–29. [Google Scholar] [CrossRef]
- Grover, S.; Jindal, V.; Banta, G.; Taning, C.N.T.; Smagghe, G.; Christiaens, O. Potential of RNA interference in the study and management of the whitefly, Bemisia tabaci. Arch. Insect Biochem. 2019, 100, e21522. [Google Scholar] [CrossRef]
- Maluta, N.K.P.; Lopes, J.R.S.; Fiallo-Olivé, E.; Navas-Castillo, J.; Lourenção, A.L. Foliar spraying of tomato plants with systemic insecticides: Effects on feeding behavior, mortality and oviposition of Bemisia tabaci (Hemiptera: Aleyrodidae) and inoculation efficiency of Tomato chlorosis virus. Insects 2020, 11, 559. [Google Scholar] [CrossRef]
- Maluta, N.; Fereres, A.; Lopes, J.R.S. Plant-mediated indirect effects of two viruses with different transmission modes on Bemisia tabaci feeding behavior and fitness. J. Pest Sci. 2019, 92, 405–416. [Google Scholar] [CrossRef]
- Steele, J.E. Occurrence of a hyperglycaemic factor in the corpus cardiacum of an insect. Nature 1961, 192, 680–681. [Google Scholar] [CrossRef]
- Unniappan, S.; Orchard, I.; Delgado, M.J. Editorial: Neuroendocrine control of energy homeostasis in non-mammalian vertebrates and invertebrates. Front. Endocrinol. 2020, 11, 404. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Shi, Y.; Lin, G.L.; Yang, C.H.; Liu, T.X. Genome-wide identification of neuropeptides and their receptor genes in Bemisia tabaci and their transcript accumulation change in response to temperature stresses. Insect Sci. 2021, 28, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Zhang, X.; Chen, X.; Li, Y.; Li, W.; Cheng, Y.; Zhou, J.; You, K.; Zhou, Q. Adipokinetic Hormone Receptor Mediates Lipid Mobilization to Regulate Starvation Resistance in the Brown Planthopper, Nilaparvata lugens. Front. Physiol. 2018, 9, 1730. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.-H.; Bradfield, J.Y.; Keeley, L.L. Age and starvation effects on hypertrehalosemic hormone-dependent gene expression of cytochrome P4504C1 in the Cockroach, Blaberus discoidalis. J. Insect Physiol. 1996, 42, 925–930. [Google Scholar] [CrossRef]
- Yu, Y.; Huang, R.; Ye, J.; Zhang, V.; Wu, C.; Cheng, G.; Jia, J.; Wang, L. Regulation of starvation-induced hyperactivity by insulin and glucagon signaling in adult Drosophila. eLife 2016, 5, e15693. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, G.; Czosnek, H. Long-term association of tomato yellow leaf curl virus with its whitefly vector Bemisia tabaci: Effect on the insect transmission capacity, longevity and fecundity. J. Gen. Virol. 1997, 78, 2683–2689. [Google Scholar] [CrossRef]
- Sheridan, M.A. Regulation of lipid metabolism in poikilothermic vertebrates. Comp. Biochem. Physiol. Part B Comp. Biochem. 1994, 107, 495–508. [Google Scholar] [CrossRef]
- Scarborough, R.M.; Jamieson, G.C.; Kalish, F.; Kramer, S.J.; McEnroe, G.A.; Miller, C.A.; Schooley, D.A. Isolation and primary structure of two peptides with cardioacceleratory and hyperglycemic activity from the corpora cardiaca of Periplaneta Americana. Proc. Natl. Acad. Sci. USA 1984, 81, 5575–5579. [Google Scholar] [CrossRef]
- Reyes-DelaTorre, A.; Peña-Rangel, M.T.; Riesgo-Escovar, J.R. Carbohydrate Metabolism in Drosophila: Reliance on the Disaccharide Trehalose. In Carbohydrates-Comprehensive Studies on Glycobiology and Glycotechnology; IntechOpen: London, UK, 2012; Volume 14, pp. 317–338. [Google Scholar]
- Elbein, A.D.; Pan, Y.T.; Pastuszak, I.; Carroll, D. New insights on trehalose: A multifunctional molecule. Glycobiology 2003, 13, 17R–27R. [Google Scholar] [CrossRef]
- Iturriaga, G.; Suárez, R.; Nova-Franco, B. Trehalose metabolism: From osmoprotection to signaling. Int. J. Mol. Sci. 2009, 10, 3793–3810. [Google Scholar] [CrossRef]
- Becker, A.; Schlöder, P.; Steele, J.E.; Wegener, G. The regulation of trehalose metabolism in insects. Experientia 1996, 52, 433–439. [Google Scholar] [CrossRef] [PubMed]
- Shukla, E.; Thorat, L.J.; Nath, B.B.; Gaikwad, S.M. Insect trehalase: Physiological significance and potential applications. Glycobiology 2015, 25, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Wang, Y.; Chen, X.; Zhang, X.; Li, W.; Cheng, Y.; Li, Y.; Zhou, J.; You, K.; Song, Y.; et al. Adipokinetic hormone receptor mediates trehalose homeostasis to promote vitellogenin uptake by oocytes in Nilaparvata lugens. Front. Physiol. 2018, 9, 1904. [Google Scholar] [CrossRef] [PubMed]
- Sanders, F.W.B.; Acharjee, A.; Walker, C.; Marney, L.; Roberts, L.D.; Imamura, F.; Jenkins, B.; Case, J.; Ray, S.; Virtue, S.; et al. Hepatic steatosis risk is partly driven by increased de novo lipogenesis following carbohydrate consumption. Genome Biol. 2018, 19, 79. [Google Scholar] [CrossRef]
- Oguri, E.; Steele, J.E. A novel function of Cockroach (Periplaneta Americana) hypertrehalosemic hormone: Translocation of lipid from hemolymph to fat body. Gen. Comp. Endocrinol. 2003, 132, 46–54. [Google Scholar] [CrossRef]
- Arrese, E.L.; Mirza, S.; Rivera, L.; Howard, A.D.; Chetty, P.S.; Soulages, J.L. Expression of lipid storage droplet protein-1 may define the role of AKH as a lipid mobilizing hormone in Manduca sexta. Insect Biochem. Mol. Biol. 2008, 38, 993–1000. [Google Scholar] [CrossRef]
- Canavoso, L.E.; Jouni, Z.E.; Karnas, K.J.; Pennington, J.E.; Wells, M.A. Fat metabolism in insects. Annu. Rev. Nutr. 2001, 21, 23–46. [Google Scholar] [CrossRef]
- Gáliková, M.; Klepsatel, P.; Xu, Y.; Kühnlein, R.P. The obesity-related adipokinetic hormone controls feeding and expression of neuropeptide regulators of Drosophila metabolism. Eur. J. Lipid Sci. Technol. 2017, 119, 1600138. [Google Scholar] [CrossRef]
- Konuma, T.; Morooka, N.; Nagasawa, H.; Nagata, S. Knockdown of the adipokinetic hormone receptor increases feeding frequency in the two-spotted Cricket Gryllus bimaculatus. Endocrinology 2012, 153, 3111–3122. [Google Scholar] [CrossRef]
- Prado, E.; Tjallingii, W.F. Aphid activities during sieve element punctures. Entomol. Exp. Appl. 1994, 72, 157–165. [Google Scholar] [CrossRef]
- Prado Maluta, N.K.; Garzo, E.; Moreno, A.; Navas-Castillo, J.; Fiallo-Olivé, E.; Spotti Lopes, J.R.; Fereres, A. Stylet penetration activities of the whitefly Bemisia tabaci associated with inoculation of the crinivirus Tomato chlorosis virus. J. Gen. Virol. 2017, 98, 1515–1520. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; de Blas, C.; Barrios, L.; Fereres, A. Correlation between whitefly (Homoptera: Aleyrodidae) feeding behavior and transmission of Tomato yellow leaf curl virus. Ann. Entomol. Soc. Am. 2009, 93, 573–579. [Google Scholar] [CrossRef]
- Chung, B.Y.; Ro, J.; Hutter, S.A.; Miller, K.M.; Guduguntla, L.S.; Kondo, S.; Pletcher, S.D. Drosophila Neuropeptide F Signaling independently regulates feeding and sleep-wake behavior. Cell Rep. 2017, 19, 2441–2450. [Google Scholar] [CrossRef]
- Wang, G.; Zhou, J.-J.; Li, Y.; Gou, Y.; Quandahor, P.; Liu, C. Trehalose and glucose levels regulate feeding behavior of the phloem-feeding insect, the Pea Aphid Acyrthosiphon pisum Harris. Sci. Rep. 2021, 11, 15864. [Google Scholar] [CrossRef] [PubMed]
| Primers | Primer Sequence (5′-3′) |
|---|---|
| For RT-PCR | |
| Bta06987-F | CTTGTCGCACAATTCTGGTGT |
| Bta06987-R | ACTTCTGAACTTCTCACAATC |
| For qPCR | |
| Bta06987-F | AGACAATCCTCCTGTCCGCTCTG |
| Bta06987-R | ACTTCTGAACTTCTCACAATC |
| THL1-F | TTGGAGGCGGAGGTGAG |
| THL1-R | GCAGTTAGTTTGGGAGGGG |
| THL2-F | ACGGATGACGGCAGAAGA |
| THL2-R | ATGGCAAAAGGAGGAAAGG |
| TPS1-F | GCCCCTTTATTTTCCACCC |
| TPS1-R | AGATTCGTACATCCTATGCCTGTT |
| TPS2-F | AATGCGGGGTTCTCGTTTG |
| TPS2-R | GGTATCGCCATCACCTTCC |
| HSL-F | ACCTCAGAGCTGTTTAGTGCC |
| HSL-R | GAGGTGCAACTTTTCCTGGC |
| Actin-R | TCTTCCAGCCATCCTTCTTG |
| Actin-F | CGGTGATTTCCTTCTGCATT |
| For dsRNA synthesis | |
| dsBta06987-F | TAATACGACTCACTATAGGGCTTGTCGCACAATTCTGGTGT |
| dsBta06987-R | TAATACGACTCACTATAGGGAGATTGCGCCTCATTCTCGATCA |
| dsGFP-F | TAATACGACTCACTATAGGGTTCAGTGGAGAGGGTGAAGGT |
| dsGFP-R | TAATACGACTCACTATAGGGTGTGTGGACAGGTAATGGTTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, X.; Liu, Y.; Zhang, Z.; Zhang, Z.; Peng, J.; Gao, Y.; Zheng, L.; Chen, J.; Du, J.; Yan, S.; et al. Bta06987, Encoding a Peptide of the AKH/RPCH Family: A Role of Energy Mobilization in Bemisia tabaci. Insects 2022, 13, 834. https://doi.org/10.3390/insects13090834
Fan X, Liu Y, Zhang Z, Zhang Z, Peng J, Gao Y, Zheng L, Chen J, Du J, Yan S, et al. Bta06987, Encoding a Peptide of the AKH/RPCH Family: A Role of Energy Mobilization in Bemisia tabaci. Insects. 2022; 13(9):834. https://doi.org/10.3390/insects13090834
Chicago/Turabian StyleFan, Xiaofan, Yong Liu, Zhuo Zhang, Zhanhong Zhang, Jing Peng, Yang Gao, Limin Zheng, Jianbin Chen, Jiao Du, Shuo Yan, and et al. 2022. "Bta06987, Encoding a Peptide of the AKH/RPCH Family: A Role of Energy Mobilization in Bemisia tabaci" Insects 13, no. 9: 834. https://doi.org/10.3390/insects13090834





