Uncovering the Chemosensory System of a Subterranean Termite, Odontotermes formosanus (Shiraki) (Isoptera: Termitidae): Revealing the Chemosensory Genes and Gene Expression Patterns
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling of Odontotermes formosanus
2.2. Isolation of Total RNA from Odontotermes formosanus Samples
2.3. Preparation of Library for Transcriptome Sequencing, De Novo Assembly, and Functional Annotation
2.3.1. Bioinformatics Analysis
2.3.2. Data Quality Control
2.3.3. Transcriptome Data Assembly
2.4. Screening and Validation of Transcripts Encoding Putative Chemosensory Genes
2.5. Sequence Analysis and Phylogenetic Tree Construction
2.6. Sequence Confirmation and qRT-PCR Validation
2.7. Statistical Analysis
3. Results
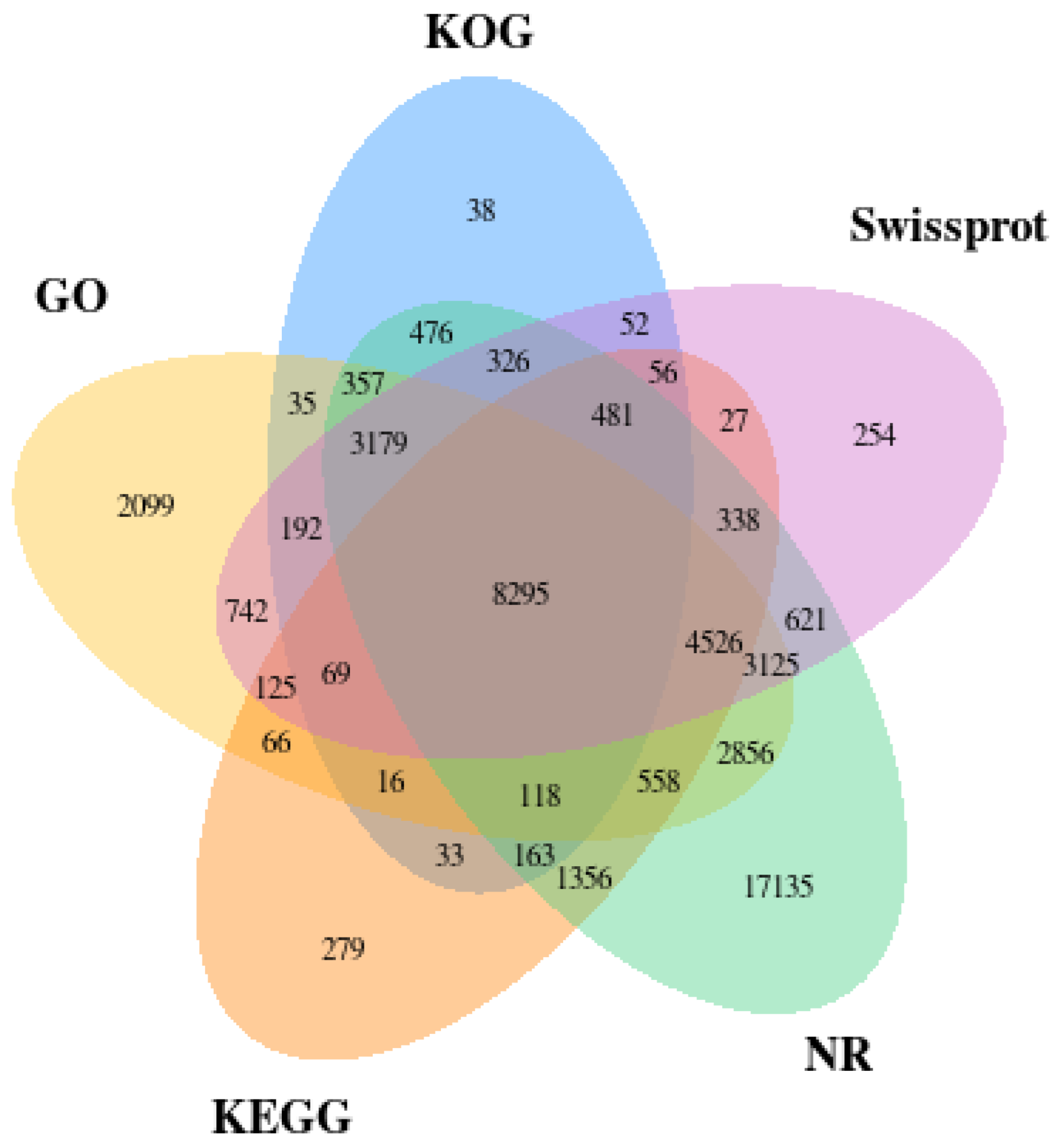
3.1. Overview of MGI Platform Sequencing and Unigene Assembly
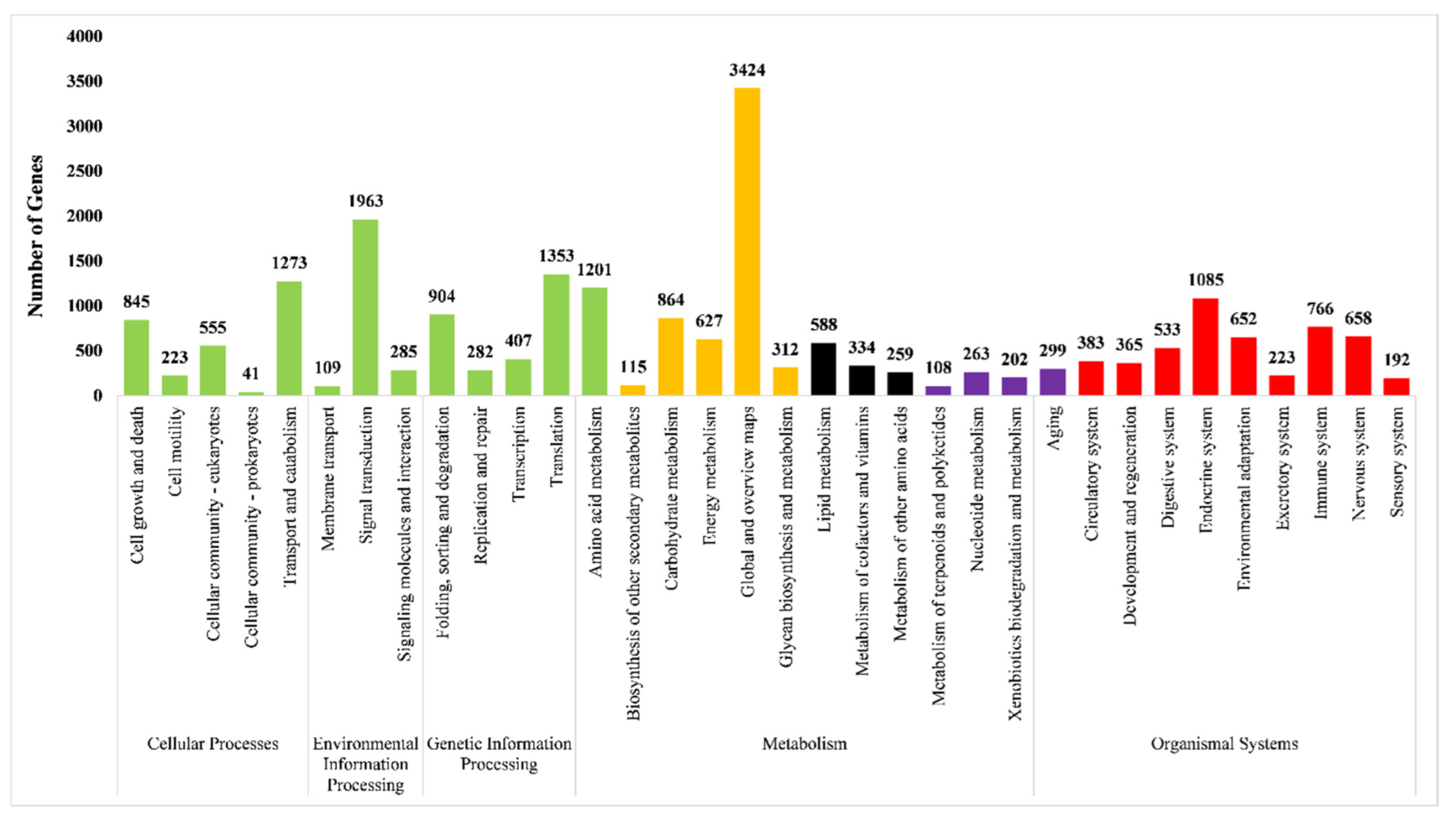
Functional Annotation
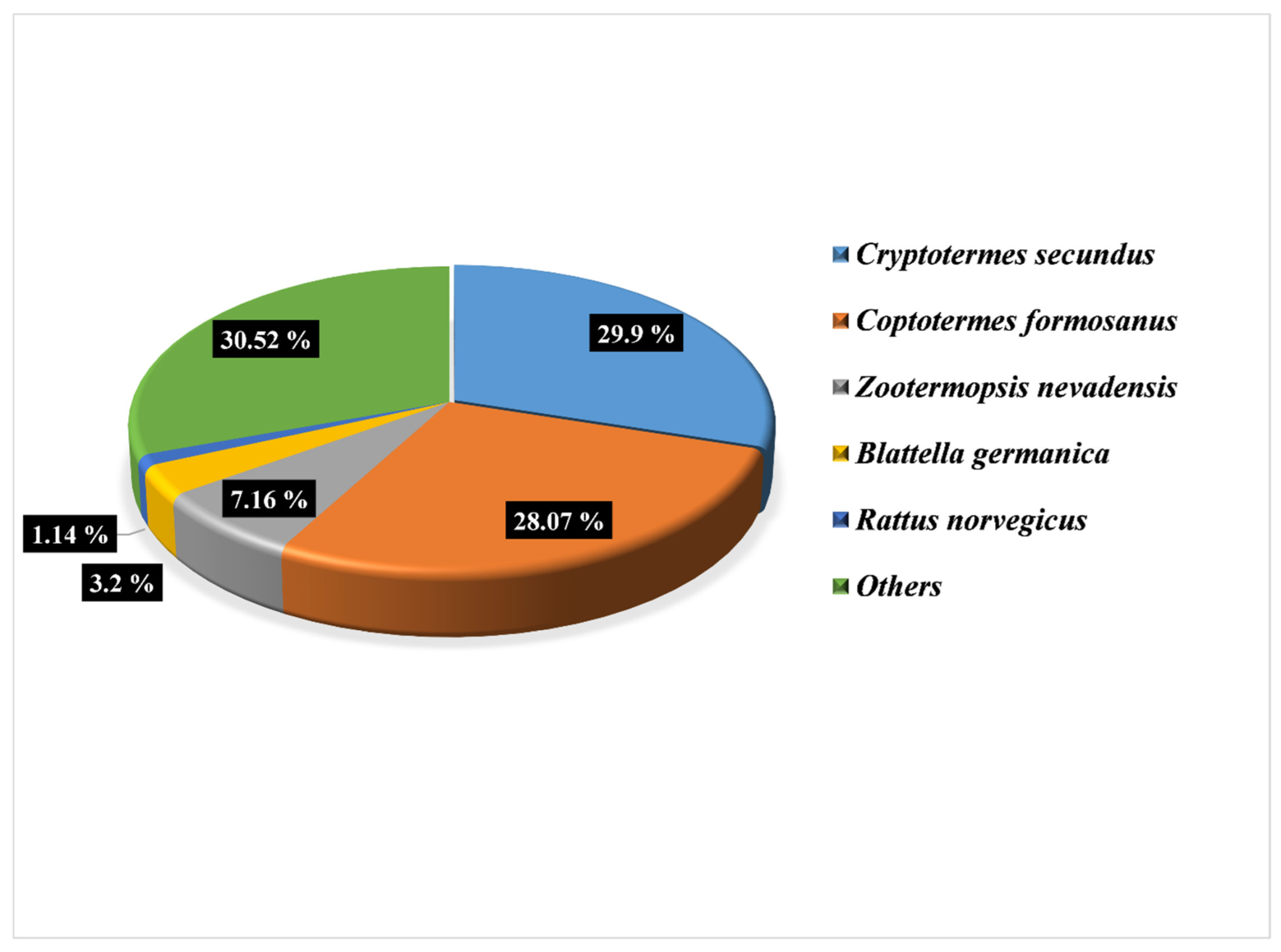
3.2. Identification of the Candidate Chemosensory Gene Families in O. formosanus
3.2.1. Candidate OBPs and CSP
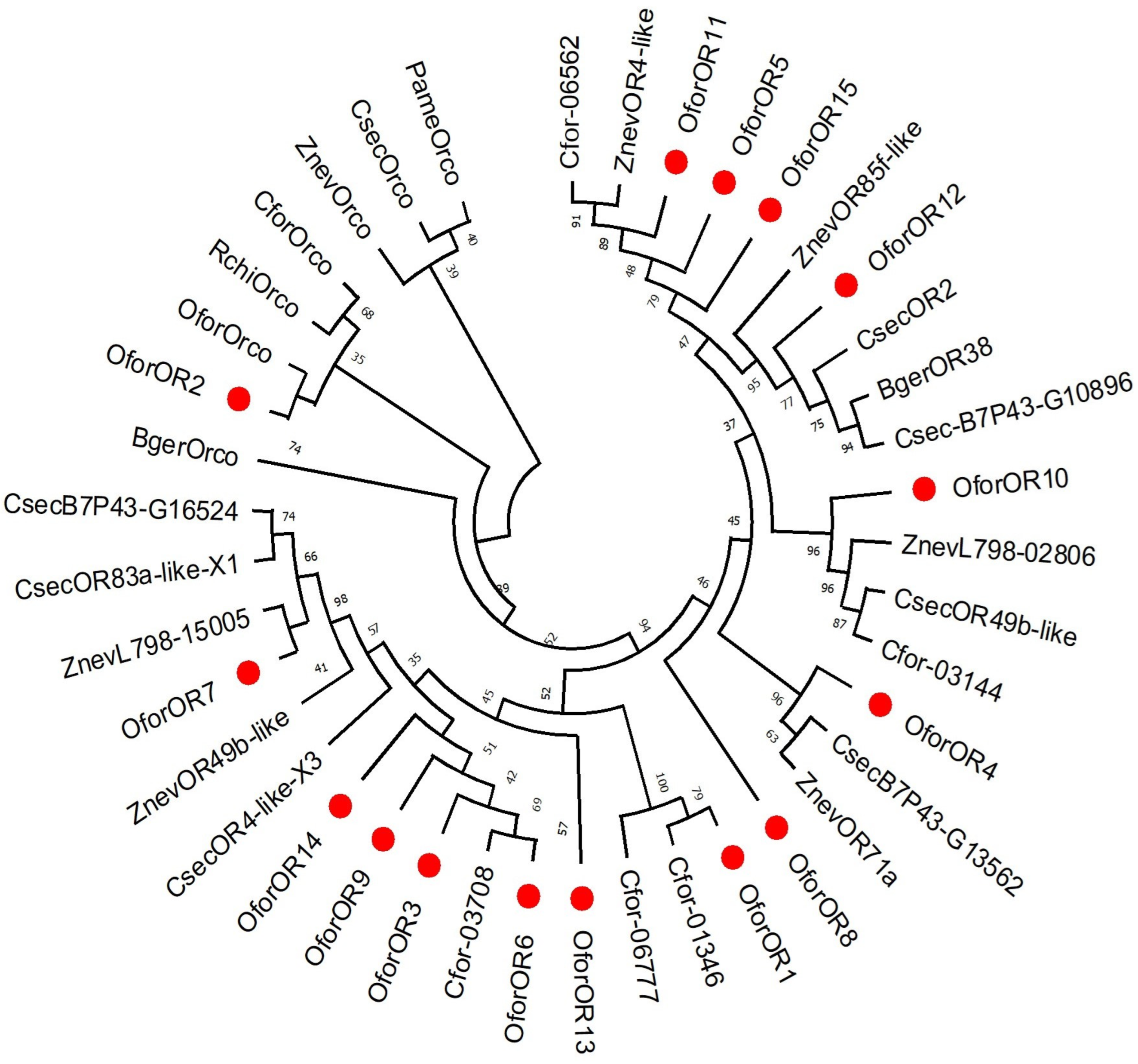
3.2.2. Candidate ORs
3.2.3. Candidate GRs
3.2.4. Candidate SNMPs
3.3. Expression Patterns of the Putative Genes from the Chemosensory Families
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, X.; Ji, B.; Liu, S.; Cao, D.; Yang, J.; Liu, J.; Ji, S.; Soleymaninejadian, E.; Wang, H. Research progress inanatomic structures of digestive system and symbiotes in termites. J. Nanjing For. Univ. 2015, 58, 155–161. [Google Scholar]
- Wang, Y.; Ji, B.; Liu, S.; Xu, L.; Jin, M.; Wang, Y. Induction Feeding Activity of Fungus Garden of Odontotermes formosanus (Isoptera: Termitidae) to Foragers. Sci. Silvae Sin. 2018, 54, 117–123. [Google Scholar]
- Xu, L.; Xiong, J.; Ji, B.; Liu, S.; Wang, Y.; Jin, M.; Zhang, L. Studies on the anatomical structures of labial gland in Odontotermes formosanus. J. Environ. Entomol. 2019, 41, 914–921. [Google Scholar]
- Feng, K.; Lu, X.; Luo, J.; Tang, F. Studies on Sunialpha 5FL against termites—Laboratory tests of soil treatment. Pesticides 1995, 34, 18–20. [Google Scholar]
- Huang, F.S.; Zhu, S.M.; Ping, Z.M.; He, X.S.; Li, G.X.; Gao, D.R. Fauna Sinica Insecta (Vol. 17): Isoptera; Science Press: Beijing, China, 2000; ISBN 703007100X. [Google Scholar]
- Huang, Q.-Y.; Lei, C.-L.; Xue, D. Field evaluation of a fipronil bait against subterranean termite Odontotermes formosanus (Isoptera: Termitidae). J. Econ. Entomol. 2006, 99, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Lei, C.; Xue, D. Food choice of the underground termite, Odontotermes formosanus. Sci. Silvae Sin. 2005, 5, 91–95. [Google Scholar] [CrossRef]
- Kirton, L.G.; Cheng, S. Ring-barking and root debarking of dipterocarp saplings by termites in an enrichment planting site in Malaysia. J. Trop. For. Sci. 2007, 19, 67–72. [Google Scholar]
- Ninkovic, V.; Markovic, D.; Rensing, M. Plant volatiles as cues and signals in plant communication. Plant. Cell Environ. 2021, 44, 1030–1043. [Google Scholar] [CrossRef] [PubMed]
- Kulmuni, J.; Wurm, Y.; Pamilo, P. Comparative genomics of chemosensory protein genes reveals rapid evolution and positive selection in ant-specific duplicates. Heredity 2013, 110, 538–547. [Google Scholar] [CrossRef]
- Lahav, S.; Soroker, V.; Hefetz, A.; Vander Meer, R.K. Direct behavioral evidence for hydrocarbons as ant recognition discriminators. Naturwissenschaften 1999, 86, 246–249. [Google Scholar] [CrossRef]
- Croset, V.; Rytz, R.; Cummins, S.F.; Budd, A.; Brawand, D.; Kaessmann, H.; Gibson, T.J.; Benton, R. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010, 6, e1001064. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Niimura, Y.; Nozawa, M. The evolution of animal chemosensory receptor gene repertoires: Roles of chance and necessity. Nat. Rev. Genet. 2008, 9, 951–963. [Google Scholar] [CrossRef]
- Clyne, P.J.; Warr, C.G.; Freeman, M.R.; Lessing, D.; Kim, J.; Carlson, J.R. A novel family of divergent seven-transmembrane proteins: Candidate odorant receptors in Drosophila. Neuron 1999, 22, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Vosshall, L.B.; Amrein, H.; Morozov, P.S.; Rzhetsky, A.; Axel, R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 1999, 96, 725–736. [Google Scholar] [CrossRef] [PubMed]
- DeGennaro, M.; McBride, C.S.; Seeholzer, L.; Nakagawa, T.; Dennis, E.J.; Goldman, C.; Jasinskiene, N.; James, A.A.; Vosshall, L.B. Orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature 2013, 498, 487–491. [Google Scholar] [CrossRef]
- Larsson, M.C.; Domingos, A.I.; Jones, W.D.; Chiappe, M.E.; Amrein, H.; Vosshall, L.B. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 2004, 43, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Rogers, M.E.; Sun, M.; Lerner, M.R.; Vogt, R.G. SNMP-1, a novel membrane protein of olfactory neurons of the silk moth Antheraea polyphemus with homology to the CD36 family of membrane proteins. J. Biol. Chem. 1997, 272, 14792–14799. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-J.; Xu, W.; Chen, Q.-M.; Sun, L.-N.; Anderson, A.; Xia, Q.-Y.; Papanicolaou, A. A phylogenomics approach to characterizing sensory neuron membrane proteins (SNMPs) in Lepidoptera. Insect Biochem. Mol. Biol. 2020, 118, 103313. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-J.; Li, G.-C.; Zhu, J.-Y.; Liu, N.-Y. Genome-based analysis reveals a novel SNMP group of the Coleoptera and chemosensory receptors in Rhaphuma horsfieldi. Genomics 2020, 112, 2713–2728. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Zhou, J.J.; Ban, L.P.; Calvello, M. Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 2006, 63, 1658–1676. [Google Scholar] [CrossRef]
- Angeli, S.; Ceron, F.; Scaloni, A.; Monti, M.; Monteforti, G.; Minnocci, A.; Petacchi, R.; Pelosi, P. Purification, structural characterization, cloning and immunocytochemical localization of chemoreception proteins from Schistocerca gregaria. Eur. J. Biochem. 1999, 262, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Leal, W.S.; Nikonova, L.; Peng, G. Disulfide structure of the pheromone binding protein from the silkworm moth, Bombyx mori. FEBS Lett. 1999, 464, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Scaloni, A.; Monti, M.; Angeli, S.; Pelosi, P. Structural analysis and disulfide-bridge pairing of two odorant-binding proteins from Bombyx mori. Biochem. Biophys. Res. Commun. 1999, 266, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Lagarde, A.; Spinelli, S.; Tegoni, M.; He, X.; Field, L.; Zhou, J.-J.; Cambillau, C. The crystal structure of odorant binding protein 7 from Anopheles gambiae exhibits an outstanding adaptability of its binding site. J. Mol. Biol. 2011, 414, 401–412. [Google Scholar] [CrossRef] [PubMed]
- Spinelli, S.; Lagarde, A.; Iovinella, I.; Legrand, P.; Tegoni, M.; Pelosi, P.; Cambillau, C. Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules. Insect Biochem. Mol. Biol. 2012, 42, 41–50. [Google Scholar] [CrossRef]
- Xu, P.X.; Zwiebel, L.J.; Smith, D.P. Identification of a distinct family of genes encoding atypical odorant-binding proteins in the malaria vector mosquito, Anopheles gambiae. Insect Mol. Biol. 2003, 12, 549–560. [Google Scholar] [CrossRef]
- Zhou, X.; Slone, J.D.; Rokas, A.; Berger, S.L.; Liebig, J.; Ray, A.; Reinberg, D.; Zwiebel, L.J. Phylogenetic and transcriptomic analysis of chemosensory receptors in a pair of divergent ant species reveals sex-specific signatures of odor coding. PLOS Genet. 2012, 8, e1002930. [Google Scholar] [CrossRef]
- Sánchez-Gracia, A.; Vieira, F.G.; Rozas, J. Molecular evolution of the major chemosensory gene families in insects. Heredity 2009, 103, 208–216. [Google Scholar] [CrossRef]
- Yang, B.; Liu, Y.; Wang, B.; Wang, G. Olfaction-based behaviorally manipulated technology of pest insects research progress, opportunities and challenges. Bull. Nat. Nat. Sci. Found. Chin 2020, 34, 441–446. [Google Scholar]
- Gao, Q.; Chess, A. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics 1999, 60, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Oi, F.M.; Scharf, M.E. Social exploitation of hexamerin: RNAi reveals a major caste-regulatory factor in termites. Proc. Natl. Acad. Sci. USA 2006, 103, 4499–4504. [Google Scholar] [CrossRef]
- Husseneder, C.; McGregor, C.; Lang, R.P.; Collier, R.; Delatte, J. Transcriptome profiling of female alates and egg-laying queens of the Formosan subterranean termite. Comp. Biochem. Physiol. Part D Genom. Proteom. 2012, 7, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Scharf, M.E.; Wu-Scharf, D.; Zhou, X.; Pittendrigh, B.R.; Bennett, G.W. Gene expression profiles among immature and adult reproductive castes of the termite Reticulitermes flavipes. Insect Mol. Biol. 2005, 14, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Korb, J.; Weil, T.; Hoffmann, K.; Foster, K.R.; Rehli, M. A gene necessary for reproductive suppression in termites. Science 2009, 324, 758. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, D.; Lax, A.R.; Henrissat, B.; Coutinho, P.; Katiya, N.; Nierman, W.C.; Fedorova, N. Carbohydrate-active enzymes revealed in Coptotermes formosanus (Isoptera: Rhinotermitidae) transcriptome. Insect Mol. Biol. 2012, 21, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Leonardo, F.C.; da Cunha, A.F.; da Silva, M.J.; Carazzolle, M.F.; Costa-Leonardo, A.M.; Costa, F.F.; Pereira, G.A. Analysis of the workers head transcriptome of the Asian subterranean termite, Coptotermes gestroi. Bull. Entomol. Res. 2011, 101, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Miura, T.; Kamikouchi, A.; Sawata, M.; Takeuchi, H.; Natori, S.; Kubo, T.; Matsumoto, T. Soldier caste-specific gene expression in the mandibular glands of Hodotermopsis japonica (Isoptera: Termopsidae). Proc. Natl. Acad. Sci. USA 1999, 96, 13874–13879. [Google Scholar] [CrossRef]
- Robertson, H.M.; Wanner, K.W. The chemoreceptor superfamily in the honey bee, Apis mellifera: Expansion of the odorant, but not gustatory, receptor family. Genome Res. 2006, 16, 1395–1403. [Google Scholar] [CrossRef]
- Zhou, X.; Rokas, A.; Berger, S.L.; Liebig, J.; Ray, A.; Zwiebel, L.J. Chemoreceptor evolution in hymenoptera and its implications for the evolution of eusociality. Genome Biol. Evol. 2015, 7, 2407–2416. [Google Scholar] [CrossRef]
- Engsontia, P.; Sangket, U.; Chotigeat, W.; Satasook, C. Molecular evolution of the odorant and gustatory receptor genes in lepidopteran insects: Implications for their adaptation and speciation. J. Mol. Evol. 2014, 79, 21–39. [Google Scholar] [CrossRef]
- Terrapon, N.; Li, C.; Robertson, H.M.; Ji, L.; Meng, X.; Booth, W.; Chen, Z.; Childers, C.P.; Glastad, K.M.; Gokhale, K.; et al. Molecular traces of alternative social organization in a termite genome. Nat. Commun. 2014, 5, 3636. [Google Scholar] [CrossRef]
- De Fouchier, A.; Walker, W.B., III; Montagné, N. Functional evolution of Lepidoptera olfactory receptors revealed by deorphanization of a moth repertoire. Nat. Commun. 2017, 8, 15709. [Google Scholar] [CrossRef]
- Auer, T.O.; Khallaf, M.A.; Silbering, A.F.; Zappia, G.; Ellis, K.; Álvarez-Ocaña, R.; Arguello, J.R.; Hansson, B.S.; Jefferis, G.S.X.E.; Caron, S.J.C. Olfactory receptor and circuit evolution promote host specialization. Nature 2020, 579, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Obiero, G.F.; Pauli, T.; Geuverink, E.; Veenendaal, R.; Niehuis, O.; Große-Wilde, E. Chemoreceptor diversity in apoid wasps and its reduction during the evolution of the pollen-collecting lifestyle of bees (Hymenoptera: Apoidea). Genome Biol. Evol. 2021, 13, evaa269. [Google Scholar] [CrossRef]
- Shigenobu, S.; Hayashi, Y.; Watanabe, D.; Tokuda, G.; Hojo, M.Y.; Toga, K.; Saiki, R.; Yaguchi, H.; Masuoka, Y.; Suzuki, R.; et al. Genomic and transcriptomic analyses of the subterranean termite Reticulitermes speratus: Gene duplication facilitates social evolution. Proc. Natl. Acad. Sci. USA 2022, 119, e2110361119. [Google Scholar] [CrossRef]
- Harrison, M.C.; Jongepier, E.; Robertson, H.M.; Arning, N.; Bitard-Feildel, T.; Chao, H.; Childers, C.P.; Dinh, H.; Doddapaneni, H.; Dugan, S.; et al. Hemimetabolous genomes reveal molecular basis of termite eusociality. Nat. Ecol. Evol. 2018, 2, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Mitaka, Y.; Kobayashi, K.; Mikheyev, A.; Tin, M.M.Y.; Watanabe, Y.; Matsuura, K. Caste-specific and sex-specific expression of chemoreceptor genes in a termite. PLoS ONE 2016, 11, e0146125. [Google Scholar] [CrossRef] [PubMed]
- Johny, J.; Diallo, S.; Lukšan, O.; Shewale, M.; Kalinová, B.; Hanus, R.; Große-Wilde, E. Conserved orthology in termite chemosensory gene families. Front. Ecol. Evol. 2023, 10, 1065947. [Google Scholar] [CrossRef]
- Suzuki, R.H.; Hanada, T.; Hayashi, Y.; Shigenobu, S.; Maekawa, K.; Hojo, M.K. Gene expression profiles of chemosensory genes of termite soldier and worker antennae. Insect Mol. Biol. 2023, 32, 424–435. [Google Scholar] [CrossRef]
- Liu, Y.Z.; Tang, G.Q.; Pan, Y.Z.; Chen, L.D.; He, Y.Z. Observations on the reproductive caste of Odontotermes formosanus (Shiraki): Larval development and naptial flight. Acta Entomol. Sin. 1985, 28, 111–114. [Google Scholar]
- Cheng, M.; Mo, J.; Deng, T.; Mao, W.; Li, D. Biology and ecology of Odontotermes formosanus in China. Sociobiology 2007, 50, 45–61. [Google Scholar]
- Huang, Q.-Y.; Chen, Y.; Li, J.-H.; Lei, C.-L. Intercolony agonism in the subterranean termite Odontotermes formosanus (Isoptera: Termitidae). Sociobiology 2007, 50, 867–880. [Google Scholar]
- Huang, Q.; Sun, P.; Zhou, X.; Lei, C. Characterization of head transcriptome and analysis of gene expression involved in caste differentiation and aggression in Odontotermes formosanus (Shiraki). PLoS ONE 2012, 7, e50383. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Deng, Y.Y.; Li, J.Q.; Wu, S.F.; Zhu, Y.P.; Chen, Y.W.; He, F.C. Integrated nr database in protein annotation system and its localization. Comput. Eng. 2006, 32, 71–74. [Google Scholar]
- Conesa, A.; Götz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008, 2008, 619832. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef]
- Jacobs, S.P.; Liggins, A.P.; Zhou, J.J.; Pickett, J.A.; Jin, X.; Field, L.M. OS-D-like genes and their expression in aphids (Hemiptera: Aphididae). Insect Mol. Biol. 2005, 14, 423–432. [Google Scholar] [CrossRef]
- Gao, Y.; Huang, Q.; Xu, H. Silencing orco impaired the ability to perceive trail pheromones and affected locomotion behavior in two termite species. J. Econ. Entomol. 2020, 113, 2941–2949. [Google Scholar] [CrossRef] [PubMed]
- Hekmat-Scafe, D.S.; Scafe, C.R.; McKinney, A.J.; Tanouye, M.A. Genome-Wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 2002, 12, 1357–1369. [Google Scholar] [CrossRef] [PubMed]
- Robertson, H.M.; Baits, R.L.; Walden, K.K.O.; Wada-Katsumata, A.; Schal, C. Enormous expansion of the chemosensory gene repertoire in the omnivorous German cockroach Blattella germanica. J. Exp. Zool. Part B Mol. Dev. Evol. 2018, 330, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Atkinson, R.; Jones, D.N.M.; Smith, D.P. Drosophila OBP LUSH Is required for activity of pheromone-sensitive neurons. Neuron 2005, 45, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Biessmann, H.; Andronopoulou, E.; Biessmann, M.R.; Douris, V.; Dimitratos, S.D.; Eliopoulos, E.; Guerin, P.M.; Iatrou, K.; Justice, R.W.; Kröber, T.; et al. The Anopheles gambiae odorant binding protein 1 (AgamOBP1) mediates indole recognition in the antennae of female mosquitoes. PLoS ONE 2010, 5, e9471. [Google Scholar] [CrossRef]
- Swarup, S.; Williams, T.I.; Anholt, R.R.H. Functional dissection of Odorant binding protein genes in Drosophila melanogaster. Genes Brain Behav. 2011, 10, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.F.; de Biasio, F.; Qiao, H.L.; Iovinella, I.; Yang, S.X.; Ling, Y.; Riviello, L.; Battaglia, D.; Falabella, P.; Yang, X.L.; et al. Two odorant-binding proteins mediate the behavioural response of aphids to the alarm pheromone (E)-ß-farnesene and structural analogues. PLoS ONE 2012, 7, e32759. [Google Scholar] [CrossRef] [PubMed]
- Ullah, R.M.K.; Waris, M.I.; Qureshi, S.R.; Rasool, F.; Duan, S.-G.; Zaka, S.M.; Atiq, M.N.; Wang, M.-Q. Silencing of an odorant binding protein (SaveOBP10) involved in the behavioural shift of the wheat aphid Sitobion avenae (Fabricius). Insect Mol. Biol. 2022, 31, 568–584. [Google Scholar] [CrossRef]
- Tegoni, M.; Campanacci, V.; Cambillau, C. Structural aspects of sexual attraction and chemical communication in insects. Trends Biochem. Sci. 2004, 29, 257–264. [Google Scholar] [CrossRef] [PubMed]
- Gong, D.P.; Zhang, H.J.; Zhao, P.; Xia, Q.Y.; Xiang, Z.H. The odorant binding protein gene family from the genome of silkworm, Bombyx mori. BMC Genom. 2009, 10, 332. [Google Scholar] [CrossRef]
- Yang, G.; Winberg, G.; Ren, H.; Zhang, S. Expression, purification and functional analysis of an odorant binding protein AaegOBP22 from Aedes aegypti. Protein Expr. Purif. 2011, 75, 165–171. [Google Scholar] [CrossRef]
- Pelosi, P.; Iovinella, I.; Zhu, J.; Wang, G.; Dani, F.R. Beyond chemoreception: Diverse tasks of soluble olfactory proteins in insects. Biol. Rev. 2018, 93, 184–200. [Google Scholar] [CrossRef]
- Saba, N.U.; Ye, C.; Zhang, W.; Wu, T.; Wang, Y.; Zhang, X.; Song, Z.; Xing, L.; Su, X. The antennal sensilla and expression patterns of olfactory genes in the lower termite Reticulitermes aculabialis (Isoptera: Rhinotermitidae). J. Insect Sci. 2022, 22, 11. [Google Scholar] [CrossRef]
- Pikielny, C.W.; Hasan, G.; Rouyer, F.; Rosbash, M. Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron 1994, 12, 35–49. [Google Scholar] [CrossRef]
- McKenna, M.P.; Hekmat-Scafe, D.S.; Gaines, P.; Carlson, J.R. Putative Drosophila pheromone-binding proteins expressed in a subregion of the olfactory system. J. Biol. Chem. 1994, 269, 16340–16347. [Google Scholar] [CrossRef] [PubMed]
- Sabatier, L.; Jouanguy, E.; Dostert, C.; Zachary, D.; Dimarcq, J.-L.; Bulet, P.; Imler, J.-L. Pherokine-2 and-3: Two Drosophila molecules related to pheromone/odor-binding proteins induced by viral and bacterial infections. Eur. J. Biochem. 2003, 270, 3398–3407. [Google Scholar] [CrossRef]
- Xin, Z.; Huang, D.; Zhao, D.; Li, J.; Wei, X.; Xiao, J. Genome-Wide analysis of chemosensory protein genes (CSPs) family in Fig wasps (Hymenoptera, Chalcidoidea). Genes 2020, 11, 1149. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, K.; Breer, H. Chemosensory proteins of Locusta migratoria (Orthoptera: Acrididae). Insect Biochem. Mol. Biol. 2000, 30, 233–241. [Google Scholar] [CrossRef]
- Maleszka, R.; Stange, G. Molecular cloning, by a novel approach, of a cDNA encoding a putative olfactory protein in the labial palps of the moth Cactoblastis cactorum. Gene 1997, 202, 39–43. [Google Scholar] [CrossRef]
- Sun, Y.; Huang, L.; Pelosi, P.; Wang, C. Expression in antennae and reproductive organs suggests a dual role of an odorant-binding protein in two sibling Helicoverpa species. PLoS ONE 2012, 7, e30040. [Google Scholar] [CrossRef] [PubMed]
- Mameli, M.; Tuccini, A.; Mazza, M.; Petacchi, R.; Pelosi, P. Soluble Proteins in chemosensory organs of phasmids. Insect Biochem. Mol. Biol. 1996, 26, 875–882. [Google Scholar] [CrossRef]
- Dietrich, K.; Breer, H. Identity and expression pattern of chemosensory proteins in Heliothis virescens (Lepidoptera, Noctuidae). Insect Biochem. Mol. Biol. 2001, 31, 1173–1181. [Google Scholar] [CrossRef]
- Jacquin-joly, E.; Vogt, R.G.; François, M.; Meillour, P.N. Functional and expression pattern analysis of chemosensory proteins expressed in antennae and pheromonal gland of Mamestra brassicae. Chem. Senses 2001, 26, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-N.; Qian, J.-L.; Xu, J.-W.; Zhu, X.-Y.; Li, M.-Y.; Xu, X.-X.; Liu, C.-X.; Xue, T.; Sun, L. Identification of chemosensory genes based on the transcriptomic analysis of six different chemosensory organs in Spodoptera exigua. Front. Physiol. 2018, 9, 432. [Google Scholar] [CrossRef]
- Zhong, J.; Wang, Y.; Lu, Y.; Ma, X.; Zhang, Q.; Wang, X.; Zhang, Q.; Sun, M. Identification and expression analysis of chemosensory genes in the antennal transcriptome of Chrysanthemum Aphid Macrosiphoniella sanborni. Insects 2022, 13, 597. [Google Scholar] [CrossRef] [PubMed]
- Robertson, H.M.; Warr, C.G.; Carlson, J.R. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2003, 100, 14537–14542. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.R.; Smith, C.D.; Robertson, H.M.; Helmkampf, M.; Zimin, A.; Yandell, M.; Holt, C.; Hu, H.; Abouheif, E.; Benton, R.; et al. Draft genome of the red harvester ant Pogonomyrmex barbatus. Proc. Natl. Acad. Sci. USA 2011, 108, 5667–5672. [Google Scholar] [CrossRef]
- Smith, C.D.; Zimin, A.; Holt, C.; Abouheif, E.; Benton, R.; Cash, E.; Croset, V.; Currie, C.R.; Elhaik, E.; Elsik, C.G.; et al. Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile). Proc. Natl. Acad. Sci. USA 2011, 108, 5673–5678. [Google Scholar] [CrossRef]
- Sanes, J.R.; Hildebrand, J.G. Structure and development of antennae in a moth, Manduca sexta. Dev. Biol. 1976, 51, 282–299. [Google Scholar] [CrossRef]
- Hallem, E.A.; Dahanukar, A.; Carlson, J.R. Insect odor and taste receptors. Annu. Rev. Entomol. 2006, 51, 113–135. [Google Scholar] [CrossRef]
- Sato, K.; Tohara, K. Insect seven-transmembrane olfactory receptor complex is an odor-gated ion channel. Folia Pharmacol. Jpn. 2009, 134, 248–253. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, N.-Y.; Xu, W.; Dong, S.-L.; Zhu, J.-Y.; Xu, Y.-X.; Anderson, A. Genome-wide analysis of ionotropic receptor gene repertoire in Lepidoptera with an emphasis on its functions of Helicoverpa armigera. Insect Biochem. Mol. Biol. 2018, 99, 37–53. [Google Scholar] [CrossRef] [PubMed]
- Fleischer, J.; Pregitzer, P.; Breer, H.; Krieger, J. Access to the odor world: Olfactory receptors and their role for signal transduction in insects. Cell. Mol. Life Sci. 2018, 75, 485–508. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Yu, S.; Merchant, A.; Lei, C.; Zhou, X.; Huang, Q. Downregulation of Orco and 5-HTT alters nestmate discrimination in the subterranean termite Odontotermes formosanus (Shiraki). Front. Physiol. 2019, 10, 714. [Google Scholar] [CrossRef]
- Clyne, P.J.; Warr, C.G.; Carlson, J.R. Candidate taste receptors in Drosophila. Science 2000, 287, 1830–1834. [Google Scholar] [CrossRef] [PubMed]
- Scott, K.; Brady, R.J.; Cravchik, A.; Morozov, P.; Rzhetsky, A.; Zuker, C.; Axel, R. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell 2001, 104, 661–673. [Google Scholar] [CrossRef]
- Agnihotri, A.R.; Roy, A.A.; Joshi, R.S. Gustatory receptors in Lepidoptera: Chemosensation and beyond. Insect Mol. Biol. 2016, 25, 519–529. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, Y.-R.; Zhou, W.-W.; Liang, Q.-M.; Yuan, X.; Cheng, J.; Zhu, Z.-R.; Gong, Z.-J. Identification and characterization of two sensory neuron membrane proteins from Cnaphalocrocis medinalis (Lepidoptera: Pyralidae). Arch. Insect Biochem. Physiol. 2013, 82, 29–42. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, J.; Liu, Y.; Wang, G.; Dong, S. Expression of SNMP1 and SNMP2 genes in antennal sensilla of Spodoptera exigua (Hübner). Arch. Insect Biochem. Physiol. 2014, 85, 114–126. [Google Scholar] [CrossRef]
- Jin, X.; Ha, T.S.; Smith, D.P. SNMP is a signaling component required for pheromone sensitivity in Drosophila. Proc. Natl. Acad. Sci. USA 2008, 105, 10996–11001. [Google Scholar] [CrossRef]
- Vogt, R.G.; Miller, N.E.; Litvack, R.; Fandino, R.A.; Sparks, J.; Staples, J.; Friedman, R.; Dickens, J.C. The insect SNMP gene family. Insect Biochem. Mol. Biol. 2009, 39, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Benton, R.; Vannice, K.S.; Vosshall, L.B. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature 2007, 450, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Pregitzer, P.; Greschista, M.; Breer, H.; Krieger, J. The sensory neurone membrane protein SNMP1 contributes to the sensitivity of a pheromone detection system. Insect Mol. Biol. 2014, 23, 733–742. [Google Scholar] [CrossRef]
- Younas, A.; Waris, M.I.; Chang, X.Q.; Shaaban, M.; Abdelnabby, H.; Ul Qamar, M.T.; Wang, M.Q. A chemosensory protein MsepCSP5 involved in chemoreception of oriental armyworm Mythimna separata. Int. J. Biol. Sci. 2018, 14, 1935–1949. [Google Scholar] [CrossRef] [PubMed]
- Younas, A.; Waris, M.I.; Ul Qamar, M.T.; Shaaban, M.; Prager, S.M.; Wang, M.Q. Functional analysis of the chemosensory protein MsepCSP8 from the oriental armyworm Mythimna separata. Front. Physiol. 2018, 9, 872. [Google Scholar] [CrossRef]
- Waris, M.I.; Younas, A.; Adeel, M.M.; Duan, S.G.; Quershi, S.R.; Ullah, R.M.K.; Wang, M.Q. The role of chemosensory protein 10 in the detection of behaviorally active compounds in brown planthopper, Nilaparvata lugens. Insect Sci. 2020, 27, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Zhang, Z.N. Comparative study on the antennal sensilla of various forms of Myzus persicae. Acta Entomol. Sin. 2000, 43, 131–136. [Google Scholar]
- Zhang, F.; Li, X.; Zhang, Y.; Cheng, D. Morphological examination of winged and wingless forms of the grain aphid Sitobion miscanthi. Plant Prot. 2015, 41, 56–62. [Google Scholar]
- Shao, S.; Yang, Z.; Chen, H.; Wang, C.; Wu, H.; Jiang, B.; Chen, X. Comparison and analysis of the antenna sensilla of morphs of the aphid Schlechtendalia chinensis (bell)(Hemiptera; Pemphigidae). J. Environ. Entomol. 2020, 42, 1510–1517. [Google Scholar]
- Liu, T.; Wang, Y.; Tian, Y.; Zhang, J.; Zhao, J.; Guo, A. The receptor channel formed by ppk25, ppk29 and ppk23 can sense the Drosophila female pheromone 7,11-heptacosadiene. Genes Brain Behav. 2020, 19, e12529. [Google Scholar] [CrossRef]
- Latorre-Estivalis, J.M.; Almeida, F.C.; Pontes, G.; Dopazo, H.; Barrozo, R.B.; Lorenzo, M.G. Evolution of the insect PPK gene family. Genome Biol. Evol. 2021, 13, evab185. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, A.; Chandel, A.; Turner, H.; Wang, S.; Liman, E.R.; Montell, C. Requirement for an Otopetrin-like protein for acid taste in Drosophila. Proc. Natl. Acad. Sci. USA 2021, 118, e2110641118. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y.-H.; Cooper, A.J.; Teng, B.; Chang, R.B.; Artiga, D.J.; Turner, H.N.; Mulhall, E.M.; Ye, W.; Smith, A.D.; Liman, E.R. An evolutionarily conserved gene family encodes proton-selective ion channels. Science 2018, 359, 1047–1050. [Google Scholar] [CrossRef] [PubMed]
- Hughes, I.; Binkley, J.; Hurle, B.; Green, E.D.; Sidow, A.; Ornitz, D.M. Identification of the Otopetrin Domain, a conserved domain in vertebrate otopetrins and invertebrate otopetrin-like family members. BMC Evol. Biol. 2008, 8, 41. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.-J.; Guo, D.; Wang, L.-X.; Niu, C.-D.; Wu, S.-F.; Zhang, Y.V.; Gao, C.-F. Function of transient receptor potential-like channel in insect egg laying. Front. Mol. Neurosci. 2022, 15, 823563. [Google Scholar] [CrossRef]
- Sharkey, C.R.; Fujimoto, M.S.; Lord, N.P.; Shin, S.; McKenna, D.D.; Suvorov, A.; Martin, G.J.; Bybee, S.M. Overcoming the loss of blue sensitivity through opsin duplication in the largest animal group, beetles. Sci. Rep. 2017, 7, 8. [Google Scholar] [CrossRef]
- Carulli, J.P.; Chen, D.M.; Stark, W.S.; Hartl, D.L. Phylogeny and physiology of Drosophila opsins. J. Mol. Evol. 1994, 38, 250–262. [Google Scholar] [CrossRef] [PubMed]
- Bao, R.; Friedrich, M. Molecular evolution of the Drosophila retinome: Exceptional gene gain in the higher Diptera. Mol. Biol. Evol. 2009, 26, 1273–1287. [Google Scholar] [CrossRef]
- Sakai, K.; Tsutsui, K.; Yamashita, T.; Iwabe, N.; Takahashi, K.; Wada, A.; Shichida, Y. Drosophila melanogaster rhodopsin Rh7 is a UV-to-visible light sensor with an extraordinarily broad absorption spectrum. Sci. Rep. 2017, 7, 7349. [Google Scholar] [CrossRef]


| Parameters | Odontotermes formosanus |
|---|---|
| Total reads | 39,251,722 |
| Clean reads | 39,036,281 |
| Total bases | 5,927,010,022 |
| Clean bases | 5,872,123,432 |
| Q20 (%) | 98.08 |
| Q30 (%) | 94.14 |
| GC (%) | 41.27 |
| Total number of unigenes | 138,762 |
| Total length of unigenes | 121,676,404 |
| Average length of unigenes | 876.87 |
| Median length of unigenes | 431 |
| Maximum length of unigenes | 27,662 |
| Minimum length of unigenes | 201 |
| Total number of Trinity reads | 234,541 |
| Total length of Trinity reads | 311,356,994 |
| Average length of Trinity reads | 1327.52 |
| Median length of Trinity reads | 588 |
| Maximum length of Trinity reads | 27,662 |
| Minimum length of Trinity reads | 185 |
| Gene Name | GenBank Accession Number | Length (aa) | Signal Peptide | Sequence * (Yes/No) | Domain Incomplete ** | Conserved Domain | Blastp Match | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | Accession Number | Score | QC (%) | E-Value | Identity (%) | |||||||
| OforOBP1 | OR651388 | 106 | 1-18 | Yes | C | PBP_GOBP super family | Cryptotermes secundus | XP_023722114.1 | 118 | 80 | 2 × 10−311 | 64.71 |
| OforOBP2 | OR651389 | 155 | 1-23 | Yes | - | PBP_GOBP super family | Cryptotermes secundus | XP_023719059.1 | 194 | 98 | 1 × 10−60 | 59.87 |
| OforOBP3 | OR651390 | 139 | 1-18 | Yes | - | PBP_GOBP super family | Zootermopsis nevadensis | XP_021937238.1 | 234 | 100 | 2 × 10−76 | 79.14 |
| OforOBP4 | OR651391 | 138 | 1-19 | Yes | - | PBP_GOBP super family | Cryptotermes secundus | XP_023725124.1 | 201 | 99 | 1 × 10−63 | 64.96 |
| OforOBP5 | OR651392 | 146 | 1-24 | Yes | - | PBP_GOBP super family | Cryptotermes secundus | XP_023722798.1 | 175 | 84 | 4 × 10−53 | 63.41 |
| OforOBP6 | OR651393 | 72 | - | Yes | C | PBP_GOBP super family | Cryptotermes secundus | XP_023722772.1 | 123 | 100 | 3 × 10−32 | 79.17 |
| OforOBP7 | OR651394 | 160 | 1-21 | Yes | - | PBP_GOBP super family | Zootermopsis nevadensis | XP_021924927.1 | 248 | 100 | 2 × 10−81 | 75 |
| OforOBP8 | OR651395 | 170 | 1-20 | Yes | - | PBP_GOBP super family | Cryptotermes secundus | XP_023719061.1 | 190 | 82 | 2 × 10−58 | 59.57 |
| OforOBP9 | OR651396 | 210 | 1-19 | Yes | - | PBP_GOBP super family | Cryptotermes secundus | XP_023704937.1 | 176 | 100 | 1 × 10−51 | 40.65 |
| OforOBP10 | OR651397 | 144 | 1-20 | Yes | - | PBP_GOBP super family | Zootermopsis nevadensis | XP_021928285.1 | 167 | 96 | 3 × 10−50 | 51.80 |
| OforOBP11 | OR651398 | 151 | 1-24 | Yes | - | PBP_GOBP super family | Zootermopsis nevadensis | XP_021937236.1 | 272 | 100 | 3 × 10−91 | 82.78 |
| OforOBP12 | OR651399 | 146 | 1-24 | Yes | - | PBP_GOBP super family | Cryptotermes secundus | XP_023722796.1 | 193 | 99 | 4 × 10−60 | 62.07 |
| OforOBP13 | OR651400 | 58 | - | NO | NC | PBP_GOBP super family | Coptotermes formosanus | GFG37024.1 | 107 | 100 | 4 × 10−28 | 86.21 |
| OforCSP1 | OR651283 | 52 | - | Yes | N | OS-D super family | Plodia interpunctella | XP_053615073.1 | 52 | 94 | 3 × 10−06 | 44.90 |
| OforOR1 | OR651429 | 307 | - | Yes | NC | 7tm_6 super family | Coptotermes formosanus | GFG35287.1 | 515 | 99 | 4 × 10−179 | 80.59 |
| OforOR2 | OR651430 | 346 | - | Yes | N | 7tm_6 super family | Odontotermes formosanus | QZA87370.1 | 718 | 100 | 0.00 | 100 |
| OforOR3 | OR651431 | 417 | - | Yes | - | 7tm_6 super family | Zootermopsis nevadensis | KDR19176.1 | 479 | 96 | 2 × 10−164 | 57.32 |
| OforOR4 | OR651432 | 111 | - | Yes | N | 7tm_6 super family | Cryptotermes secundus | PNF21445.1 | 141 | 80 | 1 × 10−39 | 74.16 |
| OforOR5 | OR651433 | 479 | - | Yes | - | 7tm_6 super family | Coptotermes formosanus | GFG30512.1 | 605 | 99 | 0.0 | 59.41 |
| OforOR6 | OR651434 | 413 | - | NO | - | 7tm_6 super family | Zootermopsis nevadensis | KDR19177.1 | 508 | 95 | 7 × 10−176 | 59.90 |
| OforOR7 | OR651435 | 286 | - | Yes | N | 7tm_6 super family | Zootermopsis nevadensis | KDR10322.1 | 485 | 99 | 4 × 10−169 | 80.35 |
| OforOR8 | OR651436 | 110 | - | Yes | N | 7tm_6 super family | Zootermopsis nevadensis | XP_021932574.1 | 109 | 88 | 5 × 10−28 | 51 |
| OforOR9 | OR651437 | 275 | - | Yes | N | 7tm_6 super family | Coptotermes formosanus | GFG34088.1 | 390 | 91 | 1 × 10−133 | 71.31 |
| OforOR10 | OR651438 | 174 | - | Yes | N | 7tm_6 super family | Zootermopsis nevadensis | XP_021915284.1 | 243 | 98 | 5 × 10−76 | 68.60 |
| OforOR11 | OR651439 | 121 | - | Yes | N | 7tm_6 super family | Coptotermes formosanus | GFG28618.1 | 192 | 85 | 3 × 10−56 | 83.65 |
| OforOR12 | OR651440 | 124 | - | Yes | N | 7tm_6 super family | Cryptotermes secundus | XP_033607852.1 | 217 | 100 | 1 × 10−69 | 81.45 |
| OforOR13 | OR651441 | 96 | - | NO | N | 7tm_6 super family | Cryptotermes secundus | XP_033606671.1 | 112 | 100 | 2 × 10−28 | 54.17 |
| OforOR14 | OR651442 | 80 | - | NO | N | 7tm_6 super family | Coptotermes formosanus | GFG41080.1 | 129 | 100 | 1 × 10−35 | 73.75 |
| OforOR15 | OR651443 | 157 | - | Yes | N | 7tm_6 super family | Coptotermes formosanus | GFG30511.1 | 218 | 89 | 1 × 10−51 | 62.86 |
| OforGR1 | OR651376 | 273 | - | Yes | N | 7tm_7 super family | Cryptotermes secundus | PNF40292.1 | 404 | 87 | 6 × 10−137 | 81.25 |
| OforGR2 | OR651377 | 290 | - | Yes | N | 7tm_7 super family | Cryptotermes secundus | XP_023711366.1 | 447 | 100 | 6 × 10−154 | 73.79 |
| OforGR3 | OR651378 | 308 | 1-20 | Yes | N | 7tm_7 super family | Cryptotermes secundus | XP_023708030.1 | 460 | 100 | 1 × 10−158 | 70.78 |
| OforGR4 | OR651379 | 348 | - | NO | - | 7tm_7 super family | Cryptotermes secundus | XP_023704213.2 | 492 | 99 | 1 × 10−170 | 69.71 |
| OforGR5 | OR651380 | 248 | - | Yes | N | 7tm_7 super family | Zootermopsis nevadensis | XP_021929494.1 | 368 | 95 | 2 × 10−123 | 74.15 |
| OforGR6 | OR651381 | 116 | - | NO | N | 7tm_7 super family | Zootermopsis nevadensis | XP_021920101.1 | 138 | 87 | 7 × 10−39 | 66.67 |
| OforGR7 | OR651382 | 82 | - | Yes | N | 7tm_7 super family | Cryptotermes secundus | PNF27532.1 | 112 | 98 | 7 × 10−28 | 67.90 |
| OforGR8 | OR651383 | 117 | - | NO | C | 7tm_7 super family | Cryptotermes secundus | XP_023708030.1 | 80.5 | 56 | 1 × 10−14 | 60.29 |
| OforGR9 | OR651384 | 59 | - | Yes | N | 7tm_7 super family | Zootermopsis nevadensis | XP_021915038.1 | 79.7 | 96 | 1 × 10−15 | 68.42 |
| OforSNMP1 | OR651358 | 496 | - | Yes | - | CD36 super family | Zootermopsis nevadensis | XP_021913553.1 | 1461 | 100 | 0.00 | 89.11 |
| OforSNMP2 | OR651359 | 515 | - | Yes | - | CD36 super family | Zootermopsis nevadensis | XP_021913531.1 | 921 | 100 | 0.00 | 83.50 |
| OforSNMP3 | OR651360 | 190 | - | NO | C | CD36 super family | Coptotermes formosanus | GFG35002.1 | 295 | 99 | 2 × 10−94 | 72.49 |
| OforSNMP4 | OR651361 | 118 | - | NO | N | CD36 super family | Blattella germanica | AMA98193.1 | 133 | 98 | 7 × 10−36 | 49.14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaleem Ullah, R.M.; Jia, B.; Liang, S.; Sikandar, A.; Gao, F.; Wu, H. Uncovering the Chemosensory System of a Subterranean Termite, Odontotermes formosanus (Shiraki) (Isoptera: Termitidae): Revealing the Chemosensory Genes and Gene Expression Patterns. Insects 2023, 14, 883. https://doi.org/10.3390/insects14110883
Kaleem Ullah RM, Jia B, Liang S, Sikandar A, Gao F, Wu H. Uncovering the Chemosensory System of a Subterranean Termite, Odontotermes formosanus (Shiraki) (Isoptera: Termitidae): Revealing the Chemosensory Genes and Gene Expression Patterns. Insects. 2023; 14(11):883. https://doi.org/10.3390/insects14110883
Chicago/Turabian StyleKaleem Ullah, Rana Muhammad, Bao Jia, Sheng Liang, Aatika Sikandar, Fukun Gao, and Haiyan Wu. 2023. "Uncovering the Chemosensory System of a Subterranean Termite, Odontotermes formosanus (Shiraki) (Isoptera: Termitidae): Revealing the Chemosensory Genes and Gene Expression Patterns" Insects 14, no. 11: 883. https://doi.org/10.3390/insects14110883
APA StyleKaleem Ullah, R. M., Jia, B., Liang, S., Sikandar, A., Gao, F., & Wu, H. (2023). Uncovering the Chemosensory System of a Subterranean Termite, Odontotermes formosanus (Shiraki) (Isoptera: Termitidae): Revealing the Chemosensory Genes and Gene Expression Patterns. Insects, 14(11), 883. https://doi.org/10.3390/insects14110883









