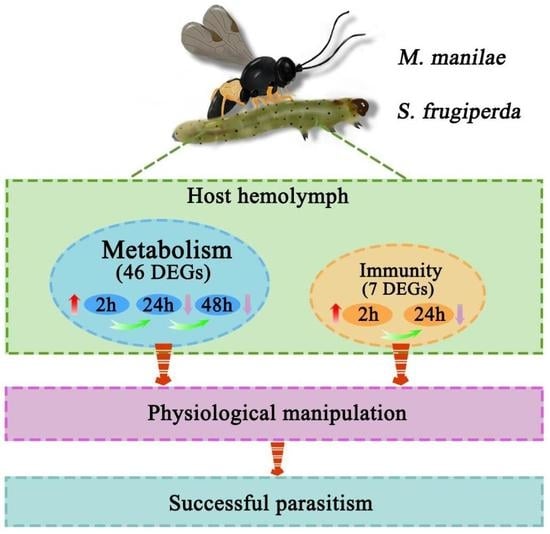
Host Transcriptome Analysis of Spodoptera frugiperda Larvae Parasitized by Microplitis manilae
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insects
2.2. Sample Collection
2.3. RNA Extraction and Transcriptome Sequencing
2.4. Transcriptomic Data Analysis
2.5. DEGs Identification
2.6. qPCR Analysis
3. Results
3.1. Analyses of Host Larvae Transcriptome after M. manilae Parasitization
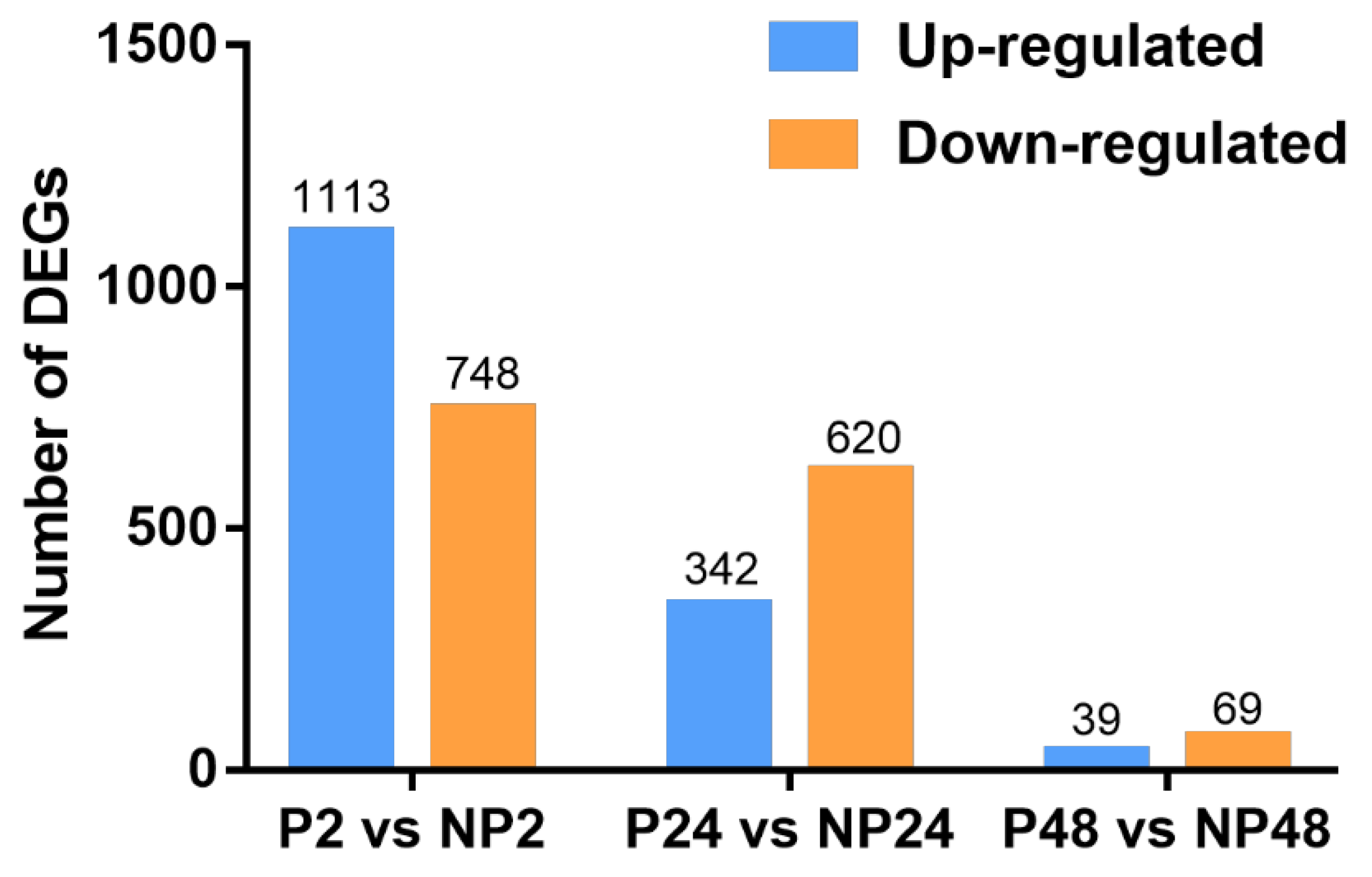
3.2. Identification of DEGs
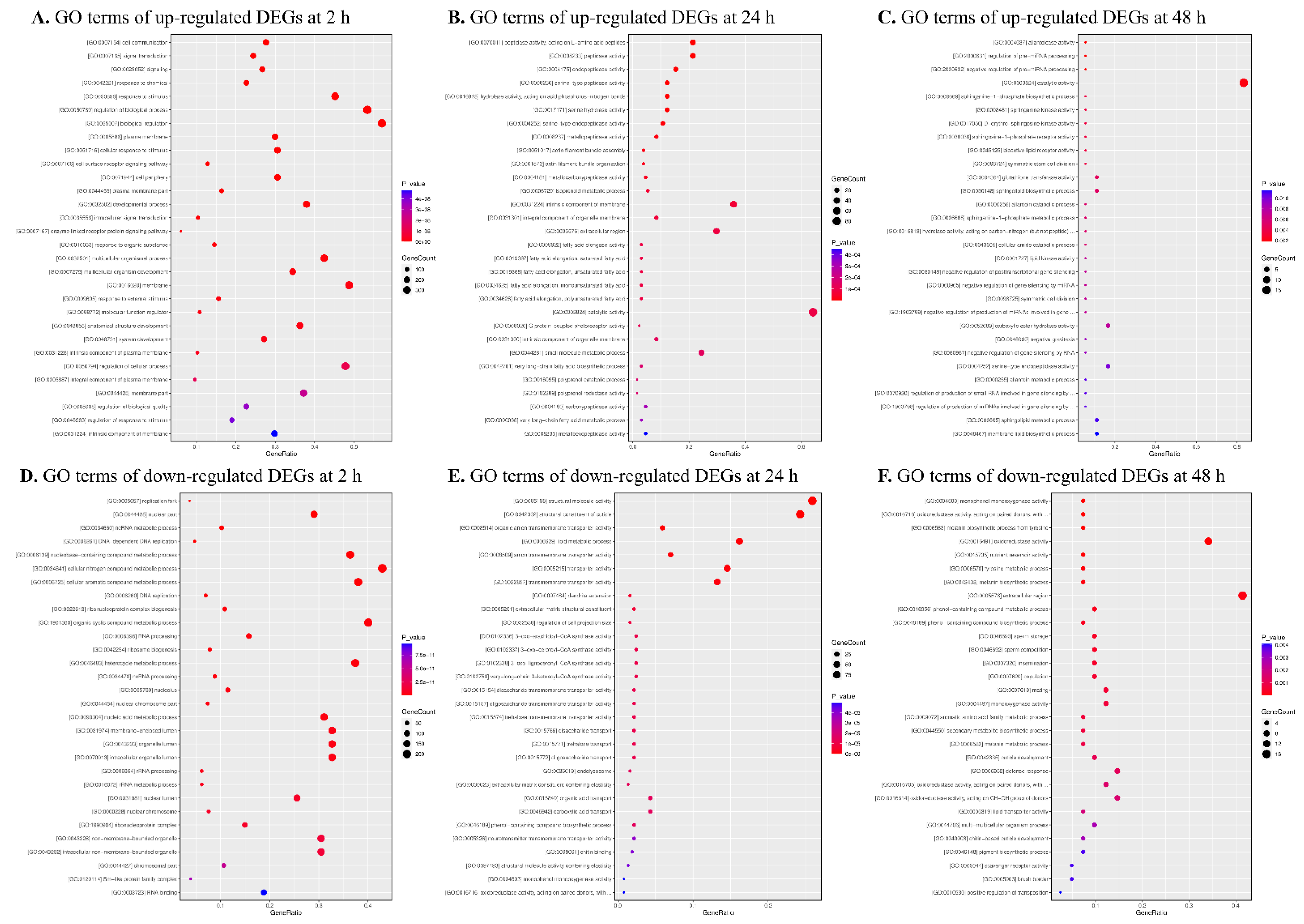
3.3. Functional Characterization of DEGs
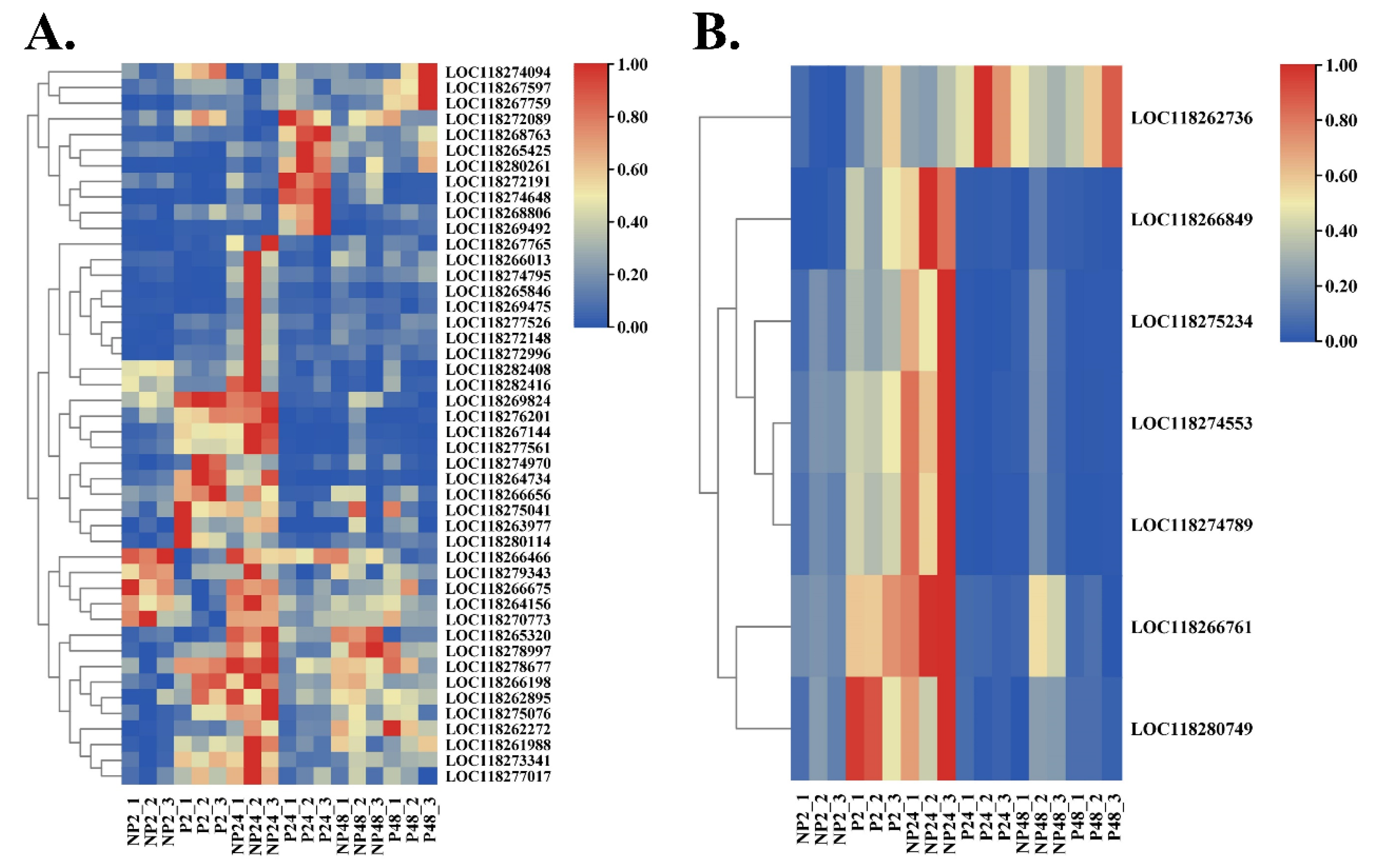
3.4. Identification of Common DEGs after M. manilae Parasitization
3.5. Common Metabolism and Immune-Related DEGs
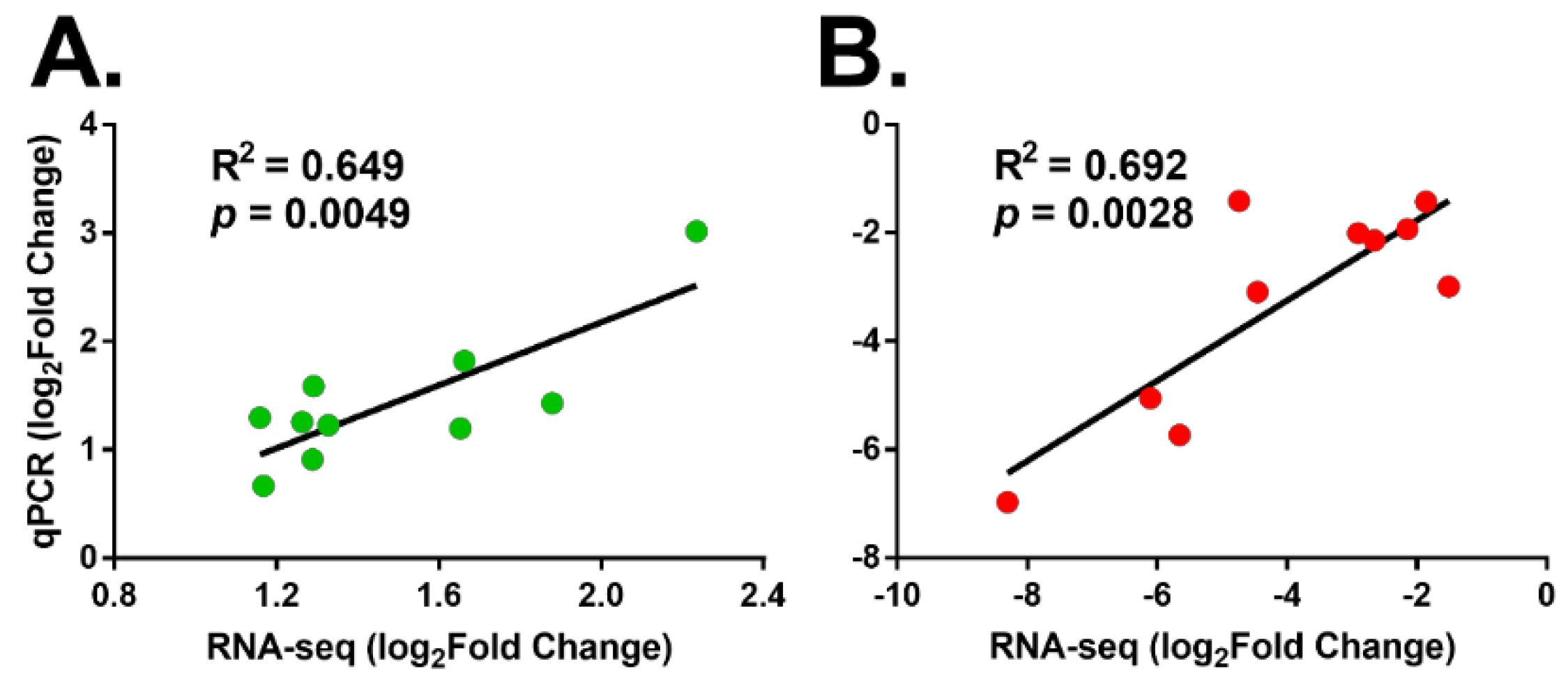
3.6. Verification of the Expression Profile of DEGs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Capinera, J.L. Fall Armyworm, Spodoptera frugiperda (J.E. Smith) (Insecta: Lepidoptera: Noctuidae). Univ. Fla. 2002, 2002, 1–7. [Google Scholar] [CrossRef]
- Wan, J.; Huang, C.; Li, C.y.; Zhou, H.x.; Ren, Y.l.; Li, Z.y.; Xing, L.s.; Zhang, B.; Qiao, X.; Liu, B.; et al. Biology, invasion and management of the agricultural invader: Fall armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae). J. Integr. Agric. 2020, 19, 2–19. [Google Scholar] [CrossRef]
- Maruthadurai, R.; Ramesh, R. Occurrence, damage pattern and biology of fall armyworm, Spodoptera frugiperda (JE smith)(Lepidoptera: Noctuidae) on fodder crops and green amaranth in Goa, India. Phytoparasitica 2020, 48, 15–23. [Google Scholar] [CrossRef]
- Van Driesche, R.; Bellows, T.S., Jr. Biological Control; Springer Science & Business Media: Berlin, Germany, 2012. [Google Scholar]
- Hassell, M.; Waage, J. Host-parasitoid population interactions. Annu. Rev. Entomol. 1984, 29, 89–114. [Google Scholar] [CrossRef]
- Yang, L.; Li, F.; Wu, S. A review of the parasitoid wasps of the fall armyworm Spodoptera frugiperda (Lepidoptera: Noctuidae) and their regulations on host immune responses. Chin. J. Biol. Control 2020, 36, 496–506. [Google Scholar]
- Harvey, J.A.; Poelman, E.H.; Tanaka, T. Intrinsic inter-and intraspecific competition in parasitoid wasps. Annu. Rev. Entomol. 2013, 58, 333–351. [Google Scholar] [CrossRef] [Green Version]
- Beckage, N.E.; Gelman, D.B. Wasp parasitoid disruption of host development: Implications for new biologically based strategies for insect control. Annu. Rev. Entomol. 2004, 49, 299–330. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Qiu, L.M.; Fang, Q.; Stanley, D.W.; Ye, G.Y. Cellular and humoral immune interactions between Drosophila and its parasitoids. Insect Sci. 2020, 11, 1208–1227. [Google Scholar] [CrossRef]
- Ye, X.Q.; Shi, M.; Huang, J.H.; Chen, X.X. Parasitoid polydnaviruses and immune interaction with secondary hosts. Dev. Comp. Immunol. 2018, 83, 124–129. [Google Scholar] [CrossRef]
- Asgari, S.; Rivers, D.B. Venom proteins from endoparasitoid wasps and their role in host-parasite interactions. Annu. Rev. Entomol. 2011, 56, 313–335. [Google Scholar] [CrossRef]
- Teng, Z.W.; Wu, H.Z.; Ye, X.H.; Xiong, S.J.; Xu, G.; Wang, F.; Fang, Q.; Ye, G.Y. An ovarian protein involved in passive avoidance of an endoparasitoid to evade its host immune response. J. Proteome Res. 2019, 18, 2695–2705. [Google Scholar] [CrossRef]
- Wang, Z.Z.; Ye, X.Q.; Shi, M.; Li, F.; Wang, Z.H.; Zhou, Y.N.; Gu, Q.J.; Wu, X.T.; Yin, C.L.; Guo, D.H.; et al. Parasitic insect-derived miRNAs modulate host development. Nat. Commun. 2018, 9, 2205. [Google Scholar] [CrossRef] [Green Version]
- Pennacchio, F.; Caccia, S.; Digilio, M.C. Host regulation and nutritional exploitation by parasitic wasps. Curr. Opin. Insect Sci. 2014, 6, 74–79. [Google Scholar] [CrossRef]
- Wang, B.B.; Ren, C.D.; Yang, L.; Fang, Q.; Song, Q.S.; Ye, G.Y. Venom alpha-amylase of the endoparasitic wasp Pteromalus puparum influences host metabolism. Pest Manag. Sci. 2020, 76, 2180–2189. [Google Scholar] [CrossRef]
- Wertheim, B.; Kraaijeveld, A.R.; Schuster, E.; Blanc, E.; Hopkins, M.; Pletcher, S.D.; Strand, M.R.; Partridge, L.; Godfray, H.C. Genome-wide gene expression in response to parasitoid attack in Drosophila. Genome Biol. 2005, 6, R94. [Google Scholar] [CrossRef] [Green Version]
- Schlenke, T.A.; Morales, J.; Govind, S.; Clark, A.G. Contrasting infection strategies in generalist and specialist wasp parasitoids of Drosophila melanogaster. PLoS Pathog. 2007, 3, e158. [Google Scholar] [CrossRef]
- Salazar-Jaramillo, L.; Jalvingh, K.M.; De Haan, A.; Kraaijeveld, K.; Buermans, H.; Wertheim, B. Inter- and intra-species variation in genome-wide gene expression of Drosophila in response to parasitoid wasp attack. BMC Genom. 2017, 18, 331. [Google Scholar] [CrossRef] [Green Version]
- Danneels, E.L.; Formesyn, E.M.; Hahn, D.A.; Denlinger, D.L.; Cardoen, D.; Wenseleers, T.; Schoofs, L.; De Graaf, D.C. Early changes in the pupal transcriptome of the flesh fly Sarcophagha crassipalpis to parasitization by the ectoparasitic wasp, Nasonia vitripennis. Insect Biochem. Mol. Biol. 2013, 43, 1189–1200. [Google Scholar] [CrossRef]
- Chevignon, G.; Cambier, S.; Da Silva, C.; Poulain, J.; Drezen, J.M.; Huguet, E.; Moreau, S.J. Transcriptomic response of Manduca sexta immune tissues to parasitization by the bracovirus associated wasp Cotesia congregata. Insect Biochem. Mol. Biol. 2015, 62, 86–99. [Google Scholar] [CrossRef]
- Sheng, S.; Wang, J.; Chu, J.; Ding, J.; Liu, Z.X.; Jiang, D.; Liang, X.; Shao, Z.; Wang, J.; Wu, F.A. Analysis of the Glyphodes pyloalis larvae immune transcriptome in response to parasitization by its endoparasitoid, Aulacococentrum confusum. Comp. Biochem. Physiol. D Genom. Proteom. 2021, 38, 100803. [Google Scholar] [CrossRef]
- Zhou, S.; Lu, Y.; Wang, Y.; Chen, J.; Pang, L.; Zhang, Q.; Sheng, Y.; Liu, Z.; Shi, M.; Chen, X.; et al. Comparative transcriptome analysis reveals a potential mechanism for host nutritional manipulation after parasitization by Leptopilina boulardi. Comp. Biochem. Physiol. D Genom. Proteom. 2021, 39, 100862. [Google Scholar] [CrossRef]
- Rajapakse, R.H.; Ashley, T.R.; Waddill, V.H. Biology and host acceptance of Microplitis manilae (Hymenoptera: Braconidae) raised on fall armyworm larvae Spodoptera frugiperda (Lepidoptera: Noctuidae). Fla. Entomol. 1985, 68, 653–657. [Google Scholar] [CrossRef]
- Yang, L.; Xing, B.; Wang, L.; Yuan, L.; Manzoor, M.; Li, F.; Wu, S. Identification of serine protease, serine protease homolog and prophenoloxidase genes in Spodoptera frugiperda (Lepidoptera: Noctuidae). J. Asia Pac. Entomol. 2021, 24, 1144–1152. [Google Scholar] [CrossRef]
- Yamaguchi, K.; Matsumoto, H.; Ochiai, M.; Tsuzuki, S.; Hayakawa, Y. Enhanced expression of stress-responsive cytokine-like gene retards insect larval growth. Insect Biochem. Mol. Biol. 2012, 42, 183–192. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Roberts, A.; Pachter, L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods 2013, 10, 71–73. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta (Delta CT)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Tang, Q.Y.; Zhang, C.X. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 2013, 20, 254–260. [Google Scholar] [CrossRef]
- Forbes, A.A.; Bagley, R.K.; Beer, M.A.; Hippee, A.C.; Widmayer, H.A. Quantifying the unquantifiable: Why Hymenoptera, not Coleoptera, is the most speciose animal order. BMC Ecol. 2018, 18, 11. [Google Scholar] [CrossRef] [Green Version]
- Martinson, E.O.; Wheeler, D.; Wright, J.; Mrinalini; Siebert, A.L.; Werren, J.H. Nasonia vitripennis venom causes targeted gene expression changes in its fly host. Mol. Ecol. 2014, 23, 5918–5930. [Google Scholar] [CrossRef] [Green Version]
- Song, W.; Veenstra, J.A.; Perrimon, N. Control of lipid metabolism by tachykinin in Drosophila. Cell Rep. 2014, 9, 40–47. [Google Scholar] [CrossRef] [Green Version]
- Canavoso, L.E.; Frede, S.; Rubiolo, E.R. Metabolic pathways for dietary lipids in the midgut of hematophagous Panstrongylus megistus (Hemiptera: Reduviidae). Insect Biochem. Mol. Biol. 2004, 34, 845–854. [Google Scholar] [CrossRef]
- Visser, B.; Ellers, J. Lack of lipogenesis in parasitoids: A review of physiological mechanisms and evolutionary implications. J. Insect Physiol. 2008, 54, 1315–1322. [Google Scholar] [CrossRef]
- Visser, B.; Roelofs, D.; Hahn, D.A.; Teal, P.E.; Marien, J.; Ellers, J. Transcriptional changes associated with lack of lipid synthesis in parasitoids. Genome Biol. Evol. 2012, 4, 864–874. [Google Scholar] [CrossRef] [Green Version]
- Visser, B.; Le Lann, C.; den Blanken, F.J.; Harvey, J.A.; van Alphen, J.J.; Ellers, J. Loss of lipid synthesis as an evolutionary consequence of a parasitic lifestyle. Proc. Natl. Acad. Sci. USA 2010, 107, 8677–8682. [Google Scholar] [CrossRef] [Green Version]
- Lammers, M.; Kraaijeveld, K.; Mariën, J.; Ellers, J. Gene expression changes associated with the evolutionary loss of a metabolic trait: Lack of lipogenesis in parasitoids. BMC Genom. 2019, 20, 309. [Google Scholar] [CrossRef]
- Rivers, D.B.; Denlinger, D.L. Venom-induced alterations in fly lipid metabolism and its impact on larval development of the ectoparasitoid Nasonia vitripennis (Walker)(Hymenoptera: Pteromalidae). J. Invertebr. Pathol. 1995, 66, 104–110. [Google Scholar] [CrossRef]
- Jiang, J.-X.; Ji, X.-Y.; Yin, Y.-Y.; Wan, N.-F. The effect of nucleopolyhedrovirus infection and/or parasitism by Microplitis pallidipes on hemolymph proteins, sugars, and lipids in Spodoptera exigua larvae. BioControl 2013, 58, 777–788. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, X.; Wang, Z.; Chen, T.; Zhou, S.; Chen, J.; Pang, L.; Ye, X.; Shi, M.; Huang, J.; et al. Symbiotic bracovirus of a parasite manipulates host lipid metabolism via tachykinin signaling. PLoS Pathog. 2021, 17, e1009365. [Google Scholar] [CrossRef]
- Yang, L.; Qiu, L.; Fang, Q.; Wu, S.; Ye, G. A venom protein of ectoparasitoid Pachycrepoideus vindemiae, PvG6PDH, contributes to parasitism by inhibiting host glucose-6-phosphate metabolism. Insect Sci. 2022, 29, 399–410. [Google Scholar] [CrossRef]
- Yu, H.; Lin, L.; Zhang, Z.; Zhang, H.; Hu, H. Targeting NF-κB pathway for the therapy of diseases: Mechanism and clinical study. Signal Transduct. Target. Ther. 2020, 5, 209. [Google Scholar] [CrossRef]
- Danneels, E.L.; Formesyn, E.M.; De Graaf, D.C. Exploring the potential of venom from Nasonia vitripennis as therapeutic agent with high-throughput screening tools. Toxins 2015, 7, 2051–2070. [Google Scholar] [CrossRef] [Green Version]
- Saba, E.; Shafeeq, T.; Irfan, M.; Lee, Y.Y.; Kwon, H.-W.; Seo, M.G.; Park, S.-J.; Lee, K.-Y.; Rhee, M.H. Anti-inflammatory activity of crude venom isolated from parasitoid wasp, bracon hebetor say. Mediat. Inflamm. 2017, 2017, 6978194. [Google Scholar] [CrossRef] [Green Version]
- Danneels, E.L.; Gerlo, S.; Heyninck, K.; Van Craenenbroeck, K.; De Bosscher, K.; Haegeman, G.; de Graaf, D.C. How the venom from the ectoparasitoid wasp Nasonia vitripennis exhibits anti-inflammatory properties on mammalian cell lines. PLoS ONE 2014, 9, e96825. [Google Scholar] [CrossRef] [Green Version]
- Jo, Y.H.; Lee, J.H.; Patnaik, B.B.; Keshavarz, M.; Lee, Y.S.; Han, Y.S. Autophagy in Tenebrio molitor immunity: Conserved antimicrobial functions in insect defenses. Front. Immunol. 2021, 12, 667664. [Google Scholar] [CrossRef]
- Ying, S.-H.; Feng, M.-G. Insight into vital role of autophagy in sustaining biological control potential of fungal pathogens against pest insects and nematodes. Virulence 2019, 10, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Romanelli, D.; Casati, B.; Franzetti, E.; Tettamanti, G. A molecular view of autophagy in Lepidoptera. BioMed Res. Int. 2014, 2014, 902315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, L.; Wan, B.; Wang, B.B.; Liu, M.M.; Fang, Q.; Song, Q.S.; Ye, G.Y. The pupal ectoparasitoid Pachycrepoideus vindemmiae regulates cellular and humoral immunity of host Drosophila melanogaster. Front. Physiol. 2019, 10, 1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, S.; Pardini, R. Mechanisms for regulating oxygen toxicity in phytophagous insects. Free Radic. Biol. Med. 1990, 8, 401–413. [Google Scholar] [CrossRef]
- Carton, Y.; Poirié, M.; Nappi, A.J. Insect immune resistance to parasitoids. Insect Sci. 2008, 15, 67–87. [Google Scholar] [CrossRef]
- Zhang, Z.; Inomata, N.; Yamazaki, T.; Kishino, H. Evolutionary history and mode of the amylase multigene family in Drosophila. J. Mol. Evol. 2003, 57, 702–709. [Google Scholar] [CrossRef]
- Lemaitre, B.; Miguel-Aliaga, I. The digestive tract of Drosophila melanogaster. Annu. Rev. Genet. 2013, 47, 377–404. [Google Scholar] [CrossRef] [Green Version]
- Tang, H. Regulation and function of the melanization reaction in Drosophila. Fly 2014, 3, 105–111. [Google Scholar] [CrossRef]
- Armstrong, P.B. The contribution of proteinase inhibitors to immune defense. Trends Immunol. 2001, 22, 47–52. [Google Scholar] [CrossRef]
- Gregorio, E.D.; Han, S.J.; Lee, W.J.; Baek, M.J.; Osaki, T.; Kawabata, S.I.; Lee, B.L.; Iwanaga, S.; Lemaitre, B.; Brey, P.T. An immune-responsive serpin regulates the melanization cascade in Drosophila. Dev. Cell 2002, 3, 581–592. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Qiu, L.M.; Fang, Q.; Ye, G.Y. A venom protein, Kazal-type serine protease inhibitor, of ectoparasitoid Pachycrepoideus vindemiae inhibits the hemolymph melanization of host Drosophila melanogaster. Arch. Insect Biochem. Physiol. 2020, 105, e21736. [Google Scholar] [CrossRef]
- Qian, C.; Fang, Q.; Wang, L.; Ye, G.Y. Molecular cloning and functional studies of two kazal-type serine protease inhibitors specifically expressed by Nasonia vitripennis venom apparatus. Toxins 2015, 7, 2888–2905. [Google Scholar] [CrossRef] [Green Version]
- Yan, Z.C.; Fang, Q.; Liu, Y.; Xiao, S.; Yang, L.; Wang, F.; An, C.J.; Werren, J.H.; Ye, G.Y. A venom serpin splicing isoform of the endoparasitoid wasp Pteromalus puparum suppresses host prophenoloxidase cascade by forming complexes with host hemolymph proteinases. J. Biol. Chem. 2017, 292, 1038–1051. [Google Scholar] [CrossRef] [Green Version]
- Colinet, D.; Dubuffet, A.; Cazes, D.; Moreau, S.; Drezen, J.M.; Poirie, M. A serpin from the parasitoid wasp Leptopilina boulardi targets the Drosophila phenoloxidase cascade. Dev. Comp. Immunol. 2009, 33, 681–689. [Google Scholar] [CrossRef]
- Dudzic, J.P.; Kondo, S.; Ueda, R.; Bergman, C.M.; Lemaitre, B. Drosophila innate immunity: Regional and functional specialization of prophenoloxidases. BMC Biol. 2015, 13, 81. [Google Scholar] [CrossRef] [Green Version]
- Nam, H.-J.; Jang, I.-H.; Asano, T.; Lee, W.-J. Involvement of Pro-Phenoloxidase 3 in Lamellocyte-Meidated Spontaneous Melanization in Drosophila. Mol. Cells 2008, 26, 606–610. [Google Scholar]
- Barat-Houari, M.; Hilliou, F.; Jousset, F.X.; Sofer, L.; Deleury, E.; Rocher, J.; Ravallec, M.; Galibert, L.; Delobel, P.; Feyereisen, R.; et al. Gene expression profiling of Spodoptera frugiperda hemocytes and fat body using cDNA microarray reveals polydnavirus-associated variations in lepidopteran host genes transcript levels. BMC Genom. 2006, 7, 160. [Google Scholar] [CrossRef] [Green Version]
- Dai, M.; Yang, J.; Liu, X.; Gu, H.; Li, F.; Li, B.; Wei, J. Parasitism by the Tachinid Parasitoid Exorista japonica Leads to Suppression of Basal Metabolism and Activation of Immune Response in the Host Bombyx mori. Insects 2022, 13, 792. [Google Scholar] [CrossRef]
- Fang, Q.; Wang, L.; Zhu, Y.; Stanley, D.W.; Chen, X.; Hu, C.; Ye, G. Pteromalus puparum venom impairs host cellular immune responses by decreasing expression of its scavenger receptor gene. Insect Biochem. Mol. Biol. 2011, 41, 852–862. [Google Scholar] [CrossRef]
- Fang, Q.; Wang, B.-B.; Ye, X.-H.; Wang, F.; Ye, G.-Y. Venom of parasitoid Pteromalus puparum impairs host humoral antimicrobial activity by decreasing host cecropin and lysozyme gene expression. Toxins 2016, 8, 52. [Google Scholar] [CrossRef] [Green Version]
- Cerenius, L.; Kawabata, S.; Lee, B.L.; Nonaka, M.; Soderhall, K. Proteolytic cascades and their involvement in invertebrate immunity. Trends Biochem. Sci. 2010, 35, 575–583. [Google Scholar] [CrossRef]
- Cerenius, L.; Lee, B.L.; Soderhall, K. The proPO-system: Pros and cons for its role in invertebrate immunity. Trends Immunol. 2008, 29, 263–271. [Google Scholar] [CrossRef]
- Cooper, D.; Wuebbolt, C.; Heryanto, C.; Eleftherianos, I. The prophenoloxidase system in Drosophila participates in the anti-nematode immune response. Mol. Immunol. 2019, 109, 88–98. [Google Scholar] [CrossRef]
- Gu, Q.J.; Zhou, S.M.; Zhou, Y.N.; Huang, J.H.; Shi, M.; Chen, X.X. A trypsin inhibitor-like protein secreted by Cotesia vestalis teratocytes inhibits hemolymph prophenoloxidase activation of Plutella xylostella. J. Insect Physiol. 2019, 116, 41–48. [Google Scholar] [CrossRef]
- Tian, C.; Wang, L.; Ye, G.Y.; Zhu, S. Inhibition of melanization by a Nasonia defensin-like peptide: Implications for host immune suppression. J. Insect Physiol. 2010, 56, 1857–1862. [Google Scholar] [CrossRef]
- Thoetkiattikul, H.; Beck, M.H.; Strand, M.R. Inhibitor kappaB-like proteins from a polydnavirus inhibit NF-kappaB activation and suppress the insect immune response. Proc. Natl. Acad. Sci. USA 2005, 102, 11426–11431. [Google Scholar] [CrossRef] [Green Version]
- Beck, M.H.; Strand, M.R. A novel polydnavirus protein inhibits the insect prophenoloxidase activation pathway. Proc. Natl. Acad. Sci. USA 2007, 104, 19267–19272. [Google Scholar] [CrossRef]


| KEGG Pathway | Category | Description | Number of Gene | Ratio (%) | |
|---|---|---|---|---|---|
| P2 vs. NP2 | ko01100 | Metabolism | Metabolic pathways | 119 | 25.32 |
| ko01110 | Biosynthesis of secondary metabolites | 37 | 7.87 | ||
| ko00230 | Purine metabolism | 30 | 6.38 | ||
| ko00240 | Pyrimidine metabolism | 23 | 4.89 | ||
| ko03013 | Genetic Information Processing | RNA transport | 27 | 5.74 | |
| ko03040 | Spliceosome | 22 | 4.68 | ||
| ko03008 | Ribosome biogenesis in eukaryotes | 20 | 4.26 | ||
| ko03010 | Ribosome | 19 | 4.04 | ||
| ko05200 | Human Diseases | Pathways in cancer | 20 | 4.26 | |
| ko04140 | Cellular Processes | Autophagy-animal | 17 | 3.62 | |
| P24 vs. NP24 | ko01100 | Metabolism | Metabolic pathways | 50 | 38.17 |
| ko01110 | Biosynthesis of secondary metabolites | 20 | 15.27 | ||
| ko01130 | Biosynthesis of antibiotics | 10 | 7.63 | ||
| ko00561 | Glycerolipid metabolism | 6 | 4.58 | ||
| ko00564 | Glycerophospholipid metabolism | 6 | 4.58 | ||
| ko01120 | Microbial metabolism in diverse environments | 6 | 4.58 | ||
| ko00040 | Pentose and glucuronate interconversions | 5 | 3.82 | ||
| ko04146 | Cellular Processes | Peroxisome | 9 | 6.87 | |
| ko04212 | Organismal Systems | Longevity regulating pathway–worm | 6 | 4.58 | |
| ko05200 | Human Diseases | Pathways in cancer | 6 | 4.58 | |
| P48 vs. NP48 | ko01100 | Metabolism | Metabolic pathways | 8 | 9.30 |
| ko01110 | Biosynthesis of secondary metabolites | 4 | 4.65 | ||
| ko00230 | Purine metabolism | 3 | 3.49 | ||
| ko00982 | Drug metabolism-cytochrome P450 | 2 | 2.33 | ||
| ko00980 | Metabolism of xenobiotics by cytochrome P450 | 2 | 2.33 | ||
| ko00600 | Sphingolipid metabolism | 2 | 2.33 | ||
| ko00561 | Glycerolipid metabolism | 2 | 2.33 | ||
| ko00480 | Glutathione metabolism | 2 | 2.33 | ||
| ko04144 | Cellular Processes | Endocytosis | 3 | 3.49 | |
| ko05204 | Human Diseases | Chemical carcinogenesis | 2 | 2.33 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gulinuer, A.; Xing, B.; Yang, L. Host Transcriptome Analysis of Spodoptera frugiperda Larvae Parasitized by Microplitis manilae. Insects 2023, 14, 100. https://doi.org/10.3390/insects14020100
Gulinuer A, Xing B, Yang L. Host Transcriptome Analysis of Spodoptera frugiperda Larvae Parasitized by Microplitis manilae. Insects. 2023; 14(2):100. https://doi.org/10.3390/insects14020100
Chicago/Turabian StyleGulinuer, Ahamaijiang, Binglin Xing, and Lei Yang. 2023. "Host Transcriptome Analysis of Spodoptera frugiperda Larvae Parasitized by Microplitis manilae" Insects 14, no. 2: 100. https://doi.org/10.3390/insects14020100
APA StyleGulinuer, A., Xing, B., & Yang, L. (2023). Host Transcriptome Analysis of Spodoptera frugiperda Larvae Parasitized by Microplitis manilae. Insects, 14(2), 100. https://doi.org/10.3390/insects14020100