Comprehensive Transcriptome Analysis in the Testis of the Silkworm, Bombyx mori
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. RNA-seq Data of B. mori Tissues
2.2. RNA Extraction and RNA Sequencing
2.3. Morphological Observation of B. mori Testis
2.4. RNA Sequencing (RNA-seq) Experiment
2.5. Statistical Calculations for Tissue-Specific and Region-Enriched Gene Calculations
2.6. Comparative Analysis of Total Genes and Transcripts Between D. melanogaster and B. mori
2.7. GO Enrichment Analysis
3. Results and Discussion
3.1. Comparative Analysis of Gene Expression among B. mori Tissues
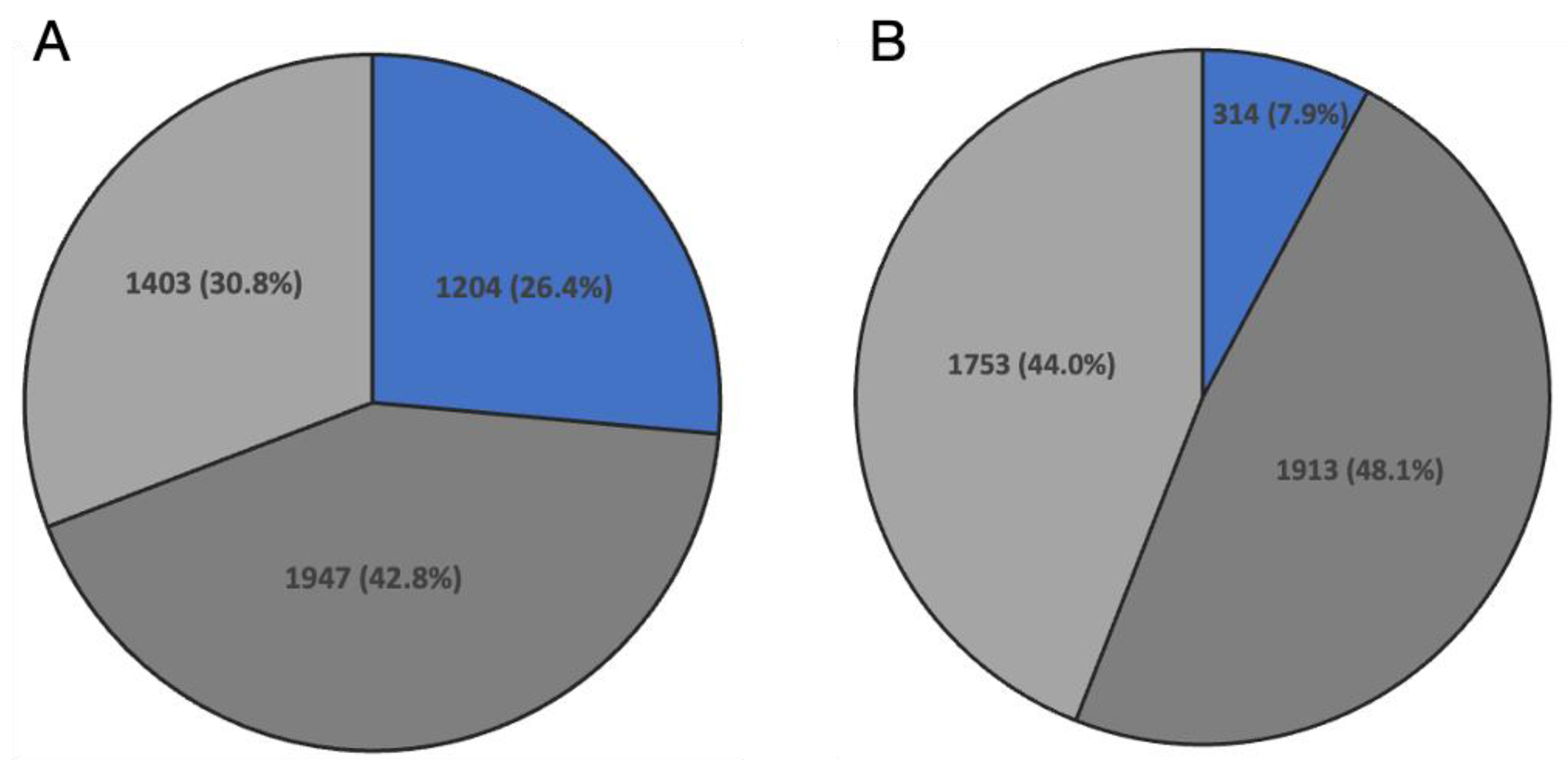
3.2. Transcriptome Analysis of Silkworm, B. mori, and Testis between Different Regions
3.3. Comparative Analysis of Gene Expression in Histologically and Functionally Homologous Regions of B. mori and D. melanogaster Testis
3.4. Gene Ontology (GO) Enrichment Analysis of Differentially Expressed Genes (DEGs) in B. mori Testis
3.4.1. GO Enrichment Analysis of SR-Specific Enriched Genes
3.4.2. GO Enrichment Analysis of DR-Specific Enriched Genes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Phillips, B.T.; Gassei, K.; Orwig, K.E. Spermatogonial stem cell regulation and spermatogenesis. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 1663–1678. [Google Scholar] [CrossRef]
- De Kretser, D.M.; Loveland, K.L.; Meinhardt, A.; Simorangkir, D.; Wreford, N. Spermatogenesis. Hum. Reprod. 1998, 13, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Toyama, K. On the Spermatogenesis of the Silkworm. Bull. Coll. Agric. Tokyo Imp. Univ. 1894, 2, 125–188. [Google Scholar]
- Sakae, K.; Yamashiki, N. Characterization of the apical cell in the testicular follicle of the silkworm Bombyx mori L. from the embryonic to adult stage. J. Insect Biotechnol. Sericol. 2018, 24, 17–24. [Google Scholar] [CrossRef]
- Miya, K. The Reproductive System and Gametogenesis of Bombyx mori—An Ultrastructural Point of View. II.; Tokai University Press: Hadano-shi, Japan, 2015; pp. 3–21. [Google Scholar]
- Sado, T. Spermatogenesis of the Silkworm and Its Bearing on Radiation Induced Sterility. II. J. Fac. Agric. Kyushu Univ. 1963, 12, 387–404. [Google Scholar] [CrossRef]
- Katsuno, S. Studies on Eupyrene and Apyrene Spermatozoa in the Silkworm, Bombyx mori L. (Lepidoptera: Bombycidae): I. The Intratesticular Behaviour of the Spermatozoa at Various Stages from the 5th-Instar to the Adult. Appl. Entomol. Zool. 1977, 12, 142–153. [Google Scholar] [CrossRef]
- Mita, K.; Nenoi, M.; Morimyo, M.; Tsuji, H.; Ichimura, S.; Sawai, M.; Hamana, K. Expression of the Bombzyx mori β-tubulin-encoding gene in testis. Gene 1995, 162, 329–330. [Google Scholar] [PubMed]
- Kusakabe, T.; Kawaguchi, Y.; Maeda, T.; Koga, K. Role of Interaction between Two Silkworm RecA Homologs in Homologous DNA Pairing. Arch. Biochem. Biophys. 2001, 388, 39–44. [Google Scholar] [CrossRef]
- Miyagawa, Y.; Lee, J.M.; Maeda, T.; Koga, K.; Kawaguchi, Y.; Kusakabe, T. Differential expression of a Bombyx mori AHA1 homologue during spermatogenesis. Insect Mol. Biol. 2005, 14, 245–253. [Google Scholar] [CrossRef]
- Yang, D.; Xu, J.; Chen, K.; Liu, Y.; Yang, X.; Tang, L.; Luo, X.; Liu, Z.; Li, M.; Walters, J.R.; et al. BmPMFBP1 regulates the development of eupyrene sperm in the silkworm, Bombyx mori. PLoS Genet. 2022, 18, e1010131. [Google Scholar] [CrossRef]
- Sakai, H.; Oshima, H.; Yuri, K.; Gotoh, H.; Daimon, T.; Yaginuma, T.; Sahara, K.; Niimi, T. Dimorphic sperm formation by Sex-Lethal. Proc. Natl. Acad. Sci. USA 2019, 116, 10412–10417. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, Y.; Yang, X.; Liu, Z.; Luo, X.; Xu, J.; Huang, Y. Dysfunction of dimorphic sperm impairs male fertility in the Silkworm. Cell Discov. 2020, 6, 60. [Google Scholar] [CrossRef] [PubMed]
- Fabian, L.; Brill, J.A. Drosophila Spermiogenesis Big Things Come from Little Packages. Spermatogenesis 2012, 2, 197–212. [Google Scholar] [CrossRef] [PubMed]
- Siddall, N.A.; Hime, G.R. A Drosophila toolkit for defining gene function in spermatogenesis. Reproduction 2017, 153, R121–R132. [Google Scholar] [CrossRef]
- Witt, E.; Benjamin, S.; Svetec, N.; Zhao, L. Testis single-cell RNA-seq reveals the dynamics of de novo gene transcription and germline mutational bias in Drosophila. Elife 2019, 8, e47138. [Google Scholar] [CrossRef]
- Witt, E.; Shao, Z.; Hu, C.; Krause, H.M.; Zhao, L. Single-cell RNA-sequencing reveals pre-meiotic X-chromosome dosage compensation in Drosophila Testis. PLoS Genet. 2021, 17, e1009728. [Google Scholar] [CrossRef]
- Vedelek, V.; Bodai, L.; Grézal, G.; Kovács, B.; Boros, I.M.; Laurinyecz, B.; Sinka, R. Analysis of Drosophila melanogaster testis transcriptome. BMC Genom. 2018, 19, 697. [Google Scholar] [CrossRef]
- Mita, K.; Kasahara, M.; Sasaki, S.; Nagayasu, Y.; Yamada, T.; Kanamori, H.; Namiki, N.; Kitagawa, M.; Yamashita, H.; Yasukochi, Y.; et al. The Genome Sequence of Silkworm, Bombyx mori. DNA Res. 2004, 11, 27–35. [Google Scholar] [CrossRef]
- Xia, Q.; Zhou, Z.; Lu, C.; Cheng, D.; Dai, F.; Li, B.; Zhao, P.; Zha, X.; Cheng, T.; Chai, C.; et al. A Draft Sequence for the Genome of the Domesticated Silkworm (Bombyx mori). Science 2004, 306, 1937–1940. [Google Scholar] [CrossRef]
- Li, Y.; Wang, G.; Tian, J.; Liu, H.; Yang, H.; Yi, Y.; Wang, J.; Shi, X.; Jiang, F.; Yao, B.; et al. Transcriptome Analysis of the Silkworm (Bombyx mori) by High-Throughput RNA Sequencing. PLoS ONE 2012, 7, e43713. [Google Scholar] [CrossRef]
- Yokoi, K.; Tsubota, T.; Jouraku, A.; Sezutsu, H.; Bono, H. Reference Transcriptome Data in Silkworm Bombyx mori. Insects 2021, 12, 519. [Google Scholar] [CrossRef] [PubMed]
- Kamitani, M.; Kashima, M.; Tezuka, A.; Nagano, A.J. Lasy-Seq: A high-throughput library preparation method for RNA-Seq and its application in the analysis of plant responses to fluctuating temperatures. Sci. Rep. 2019, 9, 7091. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D.; Floor, S.N. Differential analyses for RNA-Seq: Transcript-level estimates improve gene-level inferences. F1000Research 2016, 4, 1521. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhu, Z.; Guan, Z.; Liu, G.; Wang, Y.; Zhang, Z. SGID: A comprehensive and interactive database of the silkworm. Database 2019, 2019, baz134. [Google Scholar] [CrossRef]
- Klopfenstein, D.V.; Zhang, L.; Pedersen, B.S.; Ramírez, F.; Vesztrocy, A.W.; Naldi, A.; Mungall, C.J.; Yunes, J.M.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef]
- Xia, B.; Yan, Y.; Baron, M.; Wagner, F.; Barkley, D.; Chiodin, M.; Kim, S.Y.; Keefe, D.L.; Alukal, J.P.; Boeke, J.D.; et al. Widespread Transcriptional Scanning in the Testis Modulates Gene Evolution Rates. Cell 2020, 180, 248–262.e21. [Google Scholar] [CrossRef]
- Kaessmann, H. Origins, evolution, and phenotypic impact of new genes. Genome Res. 2010, 20, 1313. [Google Scholar] [CrossRef]
- Long, M.; Betrán, E.; Thornton, K.; Wang, W. The origin of new genes: Glimpses from the young and old. Nat. Rev. Genet. 2003, 4, 865–875. [Google Scholar] [CrossRef]
- Zhao, L.; Saelao, P.; Jones, C.D.; Begun, D.J. Origin and Spread of de Novo Genes in Drosophila melanogaster Populations. Science 2014, 343, 769–772. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Zhang, Y.E.; Chen, J.Y.; Liu, C.J.; Zhou, W.Z.; Li, Y.; Zhang, M.; Zhang, R.; Wei, L.; Li, C.Y. Hominoid-Specific de Novo Protein-Coding Genes Originating from Long Non-Coding RNAs. PLoS Genet. 2012, 8, e1002942. [Google Scholar] [CrossRef]
- Cridland, J.M.; Majane, A.C.; Zhao, L.; Begun, D.J. Population biology of accessory gland-expressed de Novo genes in Drosophila melanogaster. Genetics 2022, 220, iyab207. [Google Scholar] [CrossRef] [PubMed]
- Oud, M.S.; Smits, R.M.; Smith, H.E.; Mastrorosa, F.K.; Holt, G.S.; Houston, B.J.; de Vries, P.F.; Alobaidi, B.K.S.; Batty, L.E.; Ismail, H.; et al. A de novo paradigm for male infertility. Nat. Commun. 2022, 13, 154. [Google Scholar] [CrossRef] [PubMed]
- Rivard, E.L.; Ludwig, A.G.; Patel, P.H.; Grandchamp, A.; Arnold, S.E.; Berger, A.; Scott, E.M.; Kelly, B.J.; Mascha, G.C.; Bornberg-Bauer, E.; et al. A putative de Novo evolved gene required for spermatid chromatin condensation in Drosophila melanogaster. PLoS Genet. 2021, 17, e1009787. [Google Scholar] [CrossRef]
- Robertson, M.J.; Kent, K.; Tharp, N.; Nozawa, K.; Dean, L.; Mathew, M.; Grimm, S.L.; Yu, Z.; Légaré, C.; Fujihara, Y.; et al. Large-scale discovery of male reproductive tract-specific genes through analysis of RNA-seq datasets. BMC Biol. 2020, 18, 103. [Google Scholar] [CrossRef]
- Mon, H.; Sato, M.; Lee, J.M.; Kusakabe, T. Construction of gene co-expression networks in cultured silkworm cells and identification of previously uncharacterized lepidopteran-specific genes required for chromosome dynamics. Insect Biochem. Mol. Biol. 2022, 151, 103875. [Google Scholar] [CrossRef]
- Vogt, N.; Koch, I.; Schwarz, H.; Schnorrer, F.; Nüsslein-Volhard, C. The γTuRC components Grip75 and Grip128 have an essential microtubule-anchoring function in the Drosophila germline. Development 2006, 133, 3963–3972. [Google Scholar] [CrossRef]
- Rastelli, L.; Kuroda, M.I. An analysis of maleless and Histone H4 acetylation in Drosophila melanogaster spermatogenesis. Mech. Dev. 1998, 71, 107–117. [Google Scholar] [CrossRef]
- Prozzillo, Y.; Fattorini, G.; Ferreri, D.; Leo, M.; Dimitri, P.; Messina, G. Knockdown of DOM/Tip60 Complex Subunits Impairs Male Meiosis of Drosophila melanogaster. Cells 2023, 12, 1348. [Google Scholar] [CrossRef]
- Leatherman, J.L.; Dinardo, S. Zfh-1 Controls Somatic Stem Cell Self-Renewal in the Drosophila Testis and Nonautonomously Influences Germline Stem Cell Self-Renewal. Cell Stem Cell 2008, 3, 44–45. [Google Scholar] [CrossRef] [PubMed]
- Morgan, M.; Kumar, L.; Li, Y.; Baptissart, M. Post-transcriptional regulation in spermatogenesis: All RNA pathways lead to healthy sperm. Cell. Mol. Life Sci. 2021, 78, 8049–8071. [Google Scholar] [CrossRef]
- Haynes, S.R.; Cooper, M.T.; Pype, S.; Stolow, D.T. Involvement of a Tissue-Specific RNA Recognition Motif Protein in Drosophila Spermatogenesis. Mol. Cell. Biol. 1997, 17, 2708–2715. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, X.; Li, X.; Guo, J.; Zhang, P.; Zeng, W. The roles of microRNAs in regulation of mammalian spermatogenesis. J. Anim. Sci. Biotechnol. 2017, 8, 35. [Google Scholar] [CrossRef]
- Lim, C.; Tarayrah, L.; Chen, X. Transcriptional regulation during Drosophila spermatogenesis. Spermatogenesis 2012, 2, 158. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Luan, X.; Zheng, Q.; Qiao, C.; Chen, W.; Wang, M.; Yan, Y.; Xie, B.; Shen, C.; He, Z.; et al. Precursor RNA processing 3 is required for male fertility, and germline stem cell self-renewal and differentiation via regulating spliceosome function in Drosophila testes. Sci. Rep. 2019, 9, 9988. [Google Scholar] [CrossRef]
- Gan, Q.; Chepelev, I.; Wei, G.; Tarayrah, L.; Cui, K.; Zhao, K.; Chen, X. Dynamic regulation of alternative splicing and chromatin structure in Drosophila gonads revealed by RNA-seq. Cell Res. 2010, 20, 763–783. [Google Scholar] [CrossRef]
- Liao, S.E.; Ai, Y.; Fukunaga, R. An RNA-binding protein Blanks plays important roles in defining small RNA and mRNA profiles in Drosophila testes. Heliyon 2018, 4, e00706. [Google Scholar] [CrossRef]
- Giansanti, M.G.; Belloni, G.; Gatti, M. Rab11 Is Required for Membrane Trafficking and Actomyosin Ring Constriction in Meiotic Cytokinesis of Drosophila Males. Mol. Biol. Cell 2007, 18, 5034–5047. [Google Scholar] [CrossRef] [PubMed]
- Basu, J.; Bousbaa, H.; Logarinho, E.; Li, Z.X.; Williams, B.C.; Lopes, C.; Sunkel, C.E.; Goldberg, M.L. Mutations in the Essential Spindle Checkpoint Gene Bub1 Cause Chromosome Missegregation and Fail to Block Apoptosis in Drosophila. J. Cell Biol. 1999, 146, 13. [Google Scholar] [CrossRef]
- Adashev, V.E.; Bazylev, S.S.; Potashnikova, D.M.; Godneeva, B.K.; Shatskikh, A.S.; Olenkina, O.M.; Olenina, L.V.; Kotov, A.A. Comparative transcriptional analysis uncovers molecular processes in early and mature somatic cyst cells of Drosophila testes. Eur. J. Cell Biol. 2022, 101, 151246. [Google Scholar] [CrossRef] [PubMed]
- de Cuevas, M.; Matunis, E.L. The stem cell niche: Lessons from the Drosophila testis. Development 2011, 138, 2861–2869. [Google Scholar] [CrossRef]
- Losick, V.P.; Morris, L.X.; Fox, D.T.; Spradling, A. Drosophila Stem Cell Niches: A decade of discovery suggests a unified view of stem cell regulation. Dev. Cell 2011, 21, 159–171. [Google Scholar] [CrossRef]
- Chen, C.; Inaba, M.; Venkei, Z.G.; Yamashita, Y.M. Klp10A, a stem cell centrosome-enriched kinesin, balances asymmetries in Drosophila male germline stem cell division. Elife 2016, 5, e20977. [Google Scholar] [CrossRef]
- Zhang, J.; Luo, J.; Chen, J.; Dai, J.; Montell, C. The Role of Y Chromosome Genes in Male Fertility in Drosophila melanogaster. Genetics 2020, 215, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Agarwal, A. Spermatogenesis: An Overview. In Sperm Chromatin; Zini, A., Agarwal, A., Eds.; Springer: New York, NY, USA, 2011; pp. 19–44. [Google Scholar] [CrossRef]
- Suede, S.H.; Malik, A.; Sapra, A. Histology, Spermatogenesis. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2023. [Google Scholar]
- Li, M.G.; Serr, M.; Newman, E.A.; Hays, T.S. The Drosophila Tctex-1 Light Chain Is Dispensable for Essential Cytoplasmic Dynein Functions but Is Required during Spermatid Differentiation. Mol. Biol. Cell 2004, 15, 3005. [Google Scholar] [CrossRef] [PubMed]
- Williams, B.C.; Riedy, M.F.; Williams, E.V.; Gatti, M.; Goldberg, M.L. The Drosophila kinesin-like protein KLP3A is a midbody component required for central spindle assembly and initiation of cytokinesis. J. Cell Biol. 1995, 129, 709–723. [Google Scholar] [CrossRef]
- Couderc, J.L.; Richard, G.; Vachias, C.; Mirouse, V. Drosophila LKB1 is required for the assembly of the polarized actin structure that allows spermatid individualization. PLoS ONE 2017, 12, e0182279. [Google Scholar] [CrossRef] [PubMed]
- Tang, E.I.; Mruk, D.D.; Cheng, C.Y. Regulation of microtubule (MT)-based cytoskeleton in the seminiferous epithelium during spermatogenesis. Semin. Cell Dev. Biol. 2016, 59, 35. [Google Scholar] [CrossRef]
- Wang, L.; Yan, M.; Wu, S.; Mao, B.; Wong, C.K.C.; Ge, R.; Sun, F.; Cheng, C.Y. Microtubule Cytoskeleton and Spermatogenesis—Lesson from Studies of Toxicant Models. Toxicol. Sci. 2020, 177, 305. [Google Scholar] [CrossRef] [PubMed]
- Yamashiki, N.; Kawamura, N. Behaviors of nucleus, basal bodies and microtubules during eupyrene and apyrene spermiogenesis in the silkworm, Bombyx mori (Lepidoptera). Dev. Growth Differ. 1997, 39, 715–722. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, Y.; Yamashiki, N. Microtubule Dynamics and Distribution of γ-Tubulin in Male Germ Cells of Bombyx mori (Lepidoptera). J. Insect Biotechnol. Sericol. 2007, 76, 113–120. [Google Scholar]
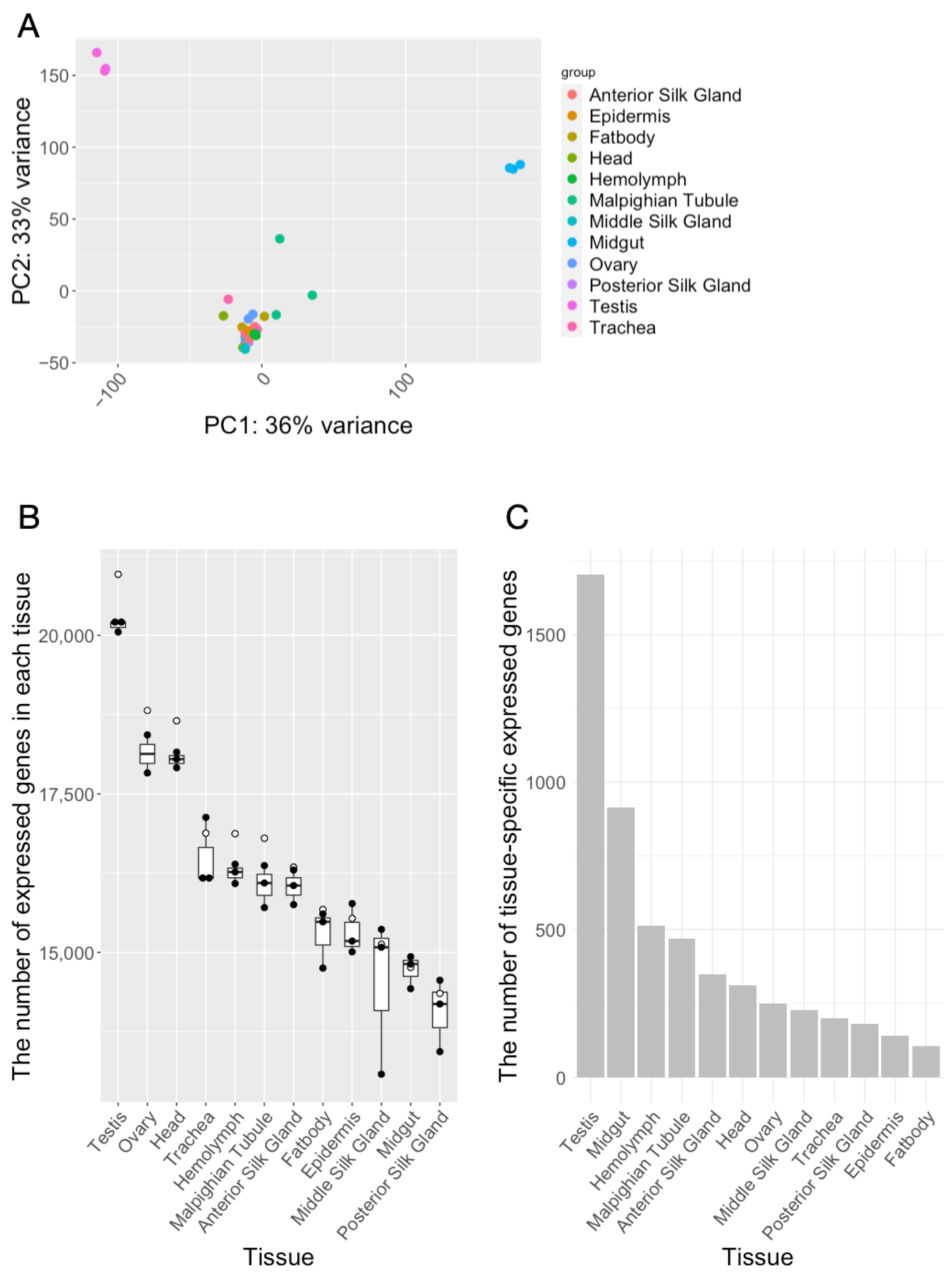



| Tissue | Expressed Genes | Tissue-Specific Expressed Genes |
|---|---|---|
| Anterior Silk Gland | 16,346 | 349 |
| Epidermis | 15,537 | 139 |
| Fat body | 15,677 | 105 |
| Head | 18,653 | 310 |
| Hemolymph | 16,873 | 513 |
| Malpighian Tubule | 16,801 | 470 |
| Middle Silk Gland | 15,130 | 227 |
| Midgut | 14,763 | 915 |
| Ovary | 18,816 | 248 |
| Posterior Silk Gland | 14,354 | 179 |
| Testis | 20,962 | 1705 |
| Trachea | 16,881 | 198 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kakino, K.; Mon, H.; Ebihara, T.; Hino, M.; Masuda, A.; Lee, J.M.; Kusakabe, T. Comprehensive Transcriptome Analysis in the Testis of the Silkworm, Bombyx mori. Insects 2023, 14, 684. https://doi.org/10.3390/insects14080684
Kakino K, Mon H, Ebihara T, Hino M, Masuda A, Lee JM, Kusakabe T. Comprehensive Transcriptome Analysis in the Testis of the Silkworm, Bombyx mori. Insects. 2023; 14(8):684. https://doi.org/10.3390/insects14080684
Chicago/Turabian StyleKakino, Kohei, Hiroaki Mon, Takeru Ebihara, Masato Hino, Akitsu Masuda, Jae Man Lee, and Takahiro Kusakabe. 2023. "Comprehensive Transcriptome Analysis in the Testis of the Silkworm, Bombyx mori" Insects 14, no. 8: 684. https://doi.org/10.3390/insects14080684
APA StyleKakino, K., Mon, H., Ebihara, T., Hino, M., Masuda, A., Lee, J. M., & Kusakabe, T. (2023). Comprehensive Transcriptome Analysis in the Testis of the Silkworm, Bombyx mori. Insects, 14(8), 684. https://doi.org/10.3390/insects14080684






