Rapid and Accurate Detection of Chrysomya megacephala (Diptera: Calliphoridae) Using Recombinase Polymerase Amplification Combined with Lateral Flow Dipstick
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and DNA Extraction
2.2. RPA Primers Design and Screening
2.3. Probe Design
2.4. RPA-LFD Detection System
2.5. Optimal Reaction Conditions for RPA-LFD Detection System
2.6. Specificity of RPA-LFD Detection System
2.7. Sensitivity of RPA-LFD Detection System
2.8. Effectiveness of RPA-LFD Detection System
2.9. RPA-LFD Detection System in Combination with Rapid DNA Extraction Methods
3. Results
3.1. Sample Collection
3.2. RPA Primers Screening
3.3. Optimal Reaction Conditions for RPA-LFD Detection System
3.4. Specificity of RPA-LFD Detection System
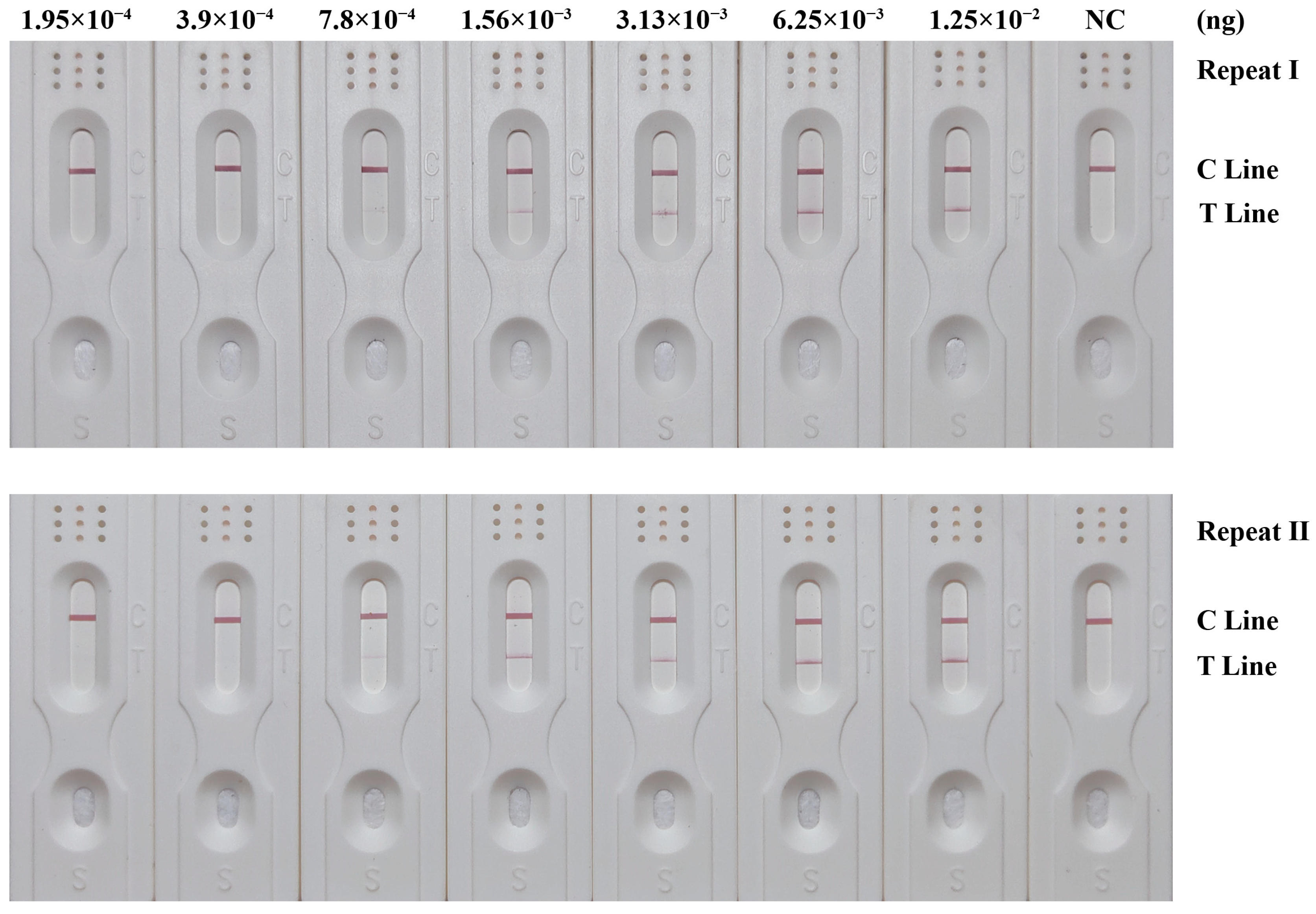
3.5. Sensitivity of RPA-LFD Detection System
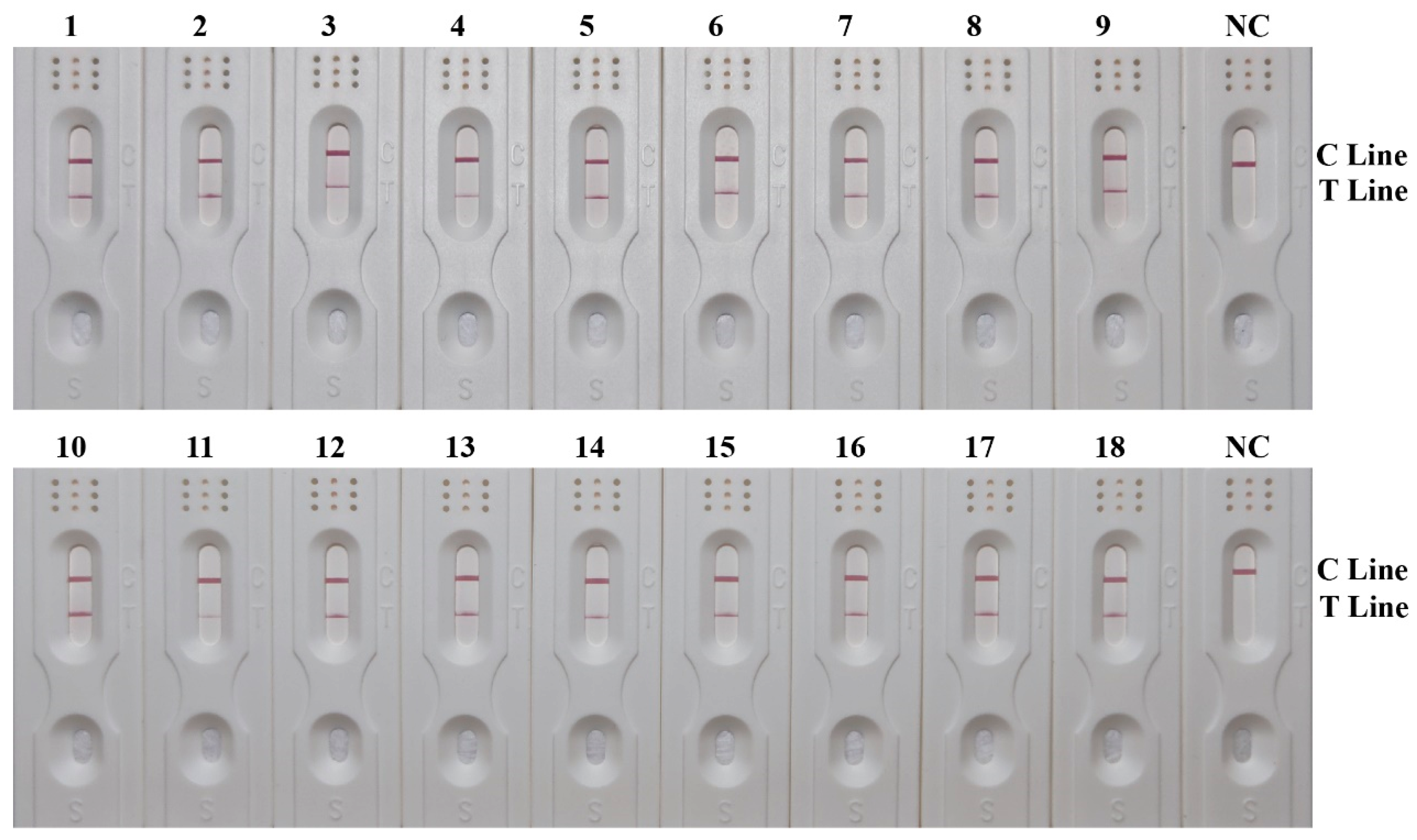
3.6. Effectiveness of RPA-LFD Detection System
3.7. RPA-LFD Detection System in Combination with Rapid DNA Extraction Methods
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, G.W.; Li, L.L.; Zhang, Y.N.; Shao, S.P.; Gao, Y.D.; Zhang, R.A.; Wang, Y.H.; Zhang, Y.A.; Guo, Y.; Kang, C.T.; et al. A global perspective of forensic entomology case reports from 1935 to 2022. Int. J. Leg. Med. 2023, 137, 1535–1553. [Google Scholar] [CrossRef] [PubMed]
- Matuszewski, S. Post-Mortem Interval Estimation Based on Insect Evidence: Current Challenges. Insects 2021, 12, 314. [Google Scholar] [CrossRef] [PubMed]
- Amendt, J.; Krettek, R.; Zehner, R. Forensic entomology. Die Naturwissenschaften 2004, 91, 51–65. [Google Scholar] [CrossRef] [PubMed]
- Amendt, J.; Richards, C.S.; Campobasso, C.P.; Zehner, R.; Hall, M.J.R. Forensic entomology: Applications and limitations. Forensic Sci. Med. Pathol. 2011, 7, 379–392. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, K.; Herrmann, E.; Amendt, J.; Verhoff, M.A.; Zehner, R. Age-dependent gene expression of Calliphora vicina pupae (Diptera: Calliphoridae) at constant and fluctuating temperatures. Int. J. Leg. Med. 2021, 135, 2625–2635. [Google Scholar] [CrossRef] [PubMed]
- Harvey, M.L.; Gaudieri, S.; Villet, M.H.; Dadour, I.R. A global study of forensically significant calliphorids: Implications for identification. Forensic Sci. Int. 2008, 177, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Y.H.; Wang, M.; Xu, W.; Zhang, Y.N.; Wang, J.F. Forensic Entomology in China and Its Challenges. Insects 2021, 12, 230. [Google Scholar] [CrossRef] [PubMed]
- Badenhorst, R.; Villet, M.H. The Uses of Chrysomya Megacephala (Fabricius, 1794) (Diptera: Calliphoridae) in Forensic Entomology. Forensic Sci. Res. 2018, 3, 2–15. [Google Scholar] [CrossRef]
- Wang, J.F.; Li, Z.G.; Chen, Y.C.; Chen, Q.S.; Yin, X.H. The succession and development of insects on pig carcasses and their significances in estimating PMI in south China. Forensic Sci. Int. 2008, 179, 11–18. [Google Scholar] [CrossRef]
- Sukontason, K.; Narongchai, P.; Kanchai, C.; Vichairat, K.; Sribanditmongkol, P.; Bhoopat, T.; Kurahashi, H.; Chockjamsai, M.; Piangjai, S.; Bunchu, N.; et al. Forensic entomology cases in Thailand: A review of cases from 2000 to 2006. Parasitol. Res. 2007, 101, 1417–1423. [Google Scholar] [CrossRef]
- Vanin, S.; Tuccia, F.; Pradelli, J.; Carta, G.; Giordani, G. Identification of Diptera Puparia in Forensic and Archeo-Funerary Contexts. Insects 2024, 15, 599. [Google Scholar] [CrossRef]
- de Carvalho, C.J.B.; de Mello-Patiu, C.A. Key to the adults of the most common forensic species of Diptera in South America. Rev. Bras. Entomol. 2008, 52, 390–406. [Google Scholar] [CrossRef]
- Chen, L.S. Necrophagous Flies in China; Guizhou Science and Technology Press: Guiyang, China, 2013. [Google Scholar]
- Gemmellaro, M.D.; Hamilton, G.C.; Ware, J.L. Review of Molecular Identification Techniques for Forensically Important Diptera. J. Med. Entomol. 2019, 56, 887–902. [Google Scholar] [CrossRef]
- Sperling, F.A.; Anderson, G.S.; Hickey, D.A. A DNA-based approach to the identification of insect species used for postmortem interval estimation. J. Forensic Sci. 1994, 39, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.D.; Stevens, J.R. Application of DNA-based methods in forensic entomology. Annu. Rev. Entomol. 2008, 53, 103–120. [Google Scholar] [CrossRef] [PubMed]
- Moore, H.E.; Adam, C.D.; Drijfhout, F.P. Identifying 1st instar larvae for three forensically important blowfly species using “fingerprint” cuticular hydrocarbon analysis. Forensic Sci. Int. 2014, 240, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Pickering, C.L.; Hands, J.R.; Fullwood, L.M.; Smith, J.A.; Baker, M.J. Rapid discrimination of maggots utilising ATR-FTIR spectroscopy. Forensic Sci. Int. 2015, 249, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Piepenburg, O.; Williams, C.H.; Stemple, D.L.; Armes, N.A. DNA detection using recombination proteins. PLoS Biol. 2006, 4, e204. [Google Scholar] [CrossRef] [PubMed]
- Crannell, Z.A.; Rohrman, B.; Richards-Kortum, R. Equipment-free incubation of recombinase polymerase amplification reactions using body heat. PLoS ONE 2014, 9, e112146. [Google Scholar] [CrossRef] [PubMed]
- Onchan, W.; Ritbamrung, O.; Changtor, P.; Pradit, W.; Chomdej, S.; Nganvongpanit, K.; Siengdee, P.; Suyasunanont, U.; Buddhachat, K. Sensitive and rapid detection of Babesia species in dogs by recombinase polymerase amplification with lateral flow dipstick (RPA-LFD). Sci. Rep. 2022, 12, 20560. [Google Scholar] [CrossRef] [PubMed]
- Daher, R.K.; Stewart, G.; Boissinot, M.; Bergeron, M.G. Recombinase Polymerase Amplification for Diagnostic Applications. Clin. Chem. 2016, 62, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Liu, Y.; Wang, Z.; Wang, H.; Zhang, X.; Lu, Q. Rapid On-Site Detection of the Bursaphelenchus xylophilus Using Recombinase Polymerase Amplification Combined with Lateral Flow Dipstick That Eliminates Interference from Primer-Dependent Artifacts. Front. Plant Sci. 2022, 13, 856109. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Macdonald, J.; von Stetten, F. Review: A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 2019, 144, 31–67. [Google Scholar] [CrossRef]
- Yao, Y.; Luo, N.; Zong, Y.; Jia, M.; Rao, Y.; Huang, H.; Jiang, H. Recombinase Polymerase Amplification Combined with Lateral Flow Dipstick Assay for the Rapid and Sensitive Detection of Pseudo-nitzschia multiseries. Int. J. Mol. Sci. 2024, 25, 1350. [Google Scholar] [CrossRef]
- Yang, Y.; Qin, X.; Wang, G.; Jin, J.; Shang, Y.; Zhang, Z. Development of an isothermoal amplification-based assay for rapid visual detection of an Orf virus. Virol. J. 2016, 13, 46. [Google Scholar] [CrossRef]
- Yang, Y.; Qin, X.; Song, Y.; Zhang, W.; Hu, G.; Dou, Y.; Li, Y.; Zhang, Z. Development of real-time and lateral flow strip reverse transcription recombinase polymerase Amplification assays for rapid detection of peste des petits ruminants virus. Virol. J. 2017, 14, 24. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Liu, J.; Liu, A.; Li, Y.; Luo, J.; Guan, G.; Yin, H. Rapid diagnosis of Theileria annulata by recombinase polymerase amplification combined with a lateral flow strip (LF-RPA) in epidemic regions. Vet. Parasitol. 2017, 237, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zheng, X.; Kan, B.; Li, W.; Zhang, W.; Jiang, T.; Lu, J.; Qin, A. Rapid detection of Burkholderia pseudomallei with a lateral flow recombinase polymerase amplification assay. PLoS ONE 2019, 14, e0213416. [Google Scholar] [CrossRef]
- Guo, Y.D.; Cai, J.F.; Meng, F.M.; Chang, Y.F.; Gu, Y.; Lan, L.M.; Liang, L.; Wen, J.F. Identification of forensically important flesh flies based on a shorter fragment of the cytochrome oxidase subunit I gene in China. Med. Vet. Entomol. 2012, 26, 307–313. [Google Scholar] [CrossRef]
- Meng, F.M.; Ren, L.P.; Wang, Z.Y.; Deng, J.Q.; Guo, Y.D.; Chen, C.; Finkelbergs, D.; Cai, J.F. Identification of Forensically Important Blow Flies (Diptera: Calliphoridae) in China Based on COI. J. Med. Entomol. 2017, 54, 1193–1200. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.D.; Zha, L.; Yan, W.T.; Li, P.; Cai, J.F.; Wu, L.X. Identification of forensically important sarcophagid flies (Diptera: Sarcophagidae) in China based on COI and period gene. Int. J. Leg. Med. 2014, 128, 221–228. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Lalitha, S. Primer Premier 5. Biotech Softw. Internet Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Priti; Jangra, S.; Baranwal, V.K.; Dietzgen, R.G.; Ghosh, A. A rapid field-based assay using recombinase polymerase amplification for identification of Thrips palmi, a vector of tospoviruses. J. Pest Sci. 2021, 94, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Jangra, S.; Ghosh, A. Rapid and zero-cost DNA extraction from soft-bodied insects for routine PCR-based applications. PLoS ONE 2022, 17, e0271312. [Google Scholar] [CrossRef]
- Li, C.J.; Sun, H.Q.; Zhao, W.X.; Wang, X.Y.; Lin, R.Z.; Yao, Y.X. Rapid assay using recombinase polymerase amplification and lateral flow dipstick for identifying Agrilus mali (Coleoptera: Buprestidae), a serious wood-boring beetle of the western Tianshan Mountains in China. J. Econ. Entomol. 2023, 116, 1969–1981. [Google Scholar] [CrossRef] [PubMed]
- Harvey, M.L.; Dadour, I.R.; Gaudieri, S. Mitochondrial DNA cytochrome oxidase I gene: Potential for distinction between immature stages of some forensically important fly species (Diptera) in western Australia. Forensic Sci. Int. 2003, 131, 134–139. [Google Scholar] [CrossRef] [PubMed]
- GilArriortua, M.; Salona Bordas, M.I.; Cainé, L.M.; Pinheiro, F.; de Pancorbo, M.M. Cytochrome b as a useful tool for the identification of blowflies of forensic interest (Diptera, Calliphoridae). Forensic Sci. Int. 2013, 228, 132–136. [Google Scholar] [CrossRef]
- Jang, H.; Shin, S.E.; Ko, K.s.; Park, S.H. SNP Typing Using Multiplex Real-Time PCR Assay for Species Identification of Forensically Important Blowflies and Fleshflies Collected in South Korea (Diptera: Calliphoridae and Sarcophagidae). BioMed Res. Int. 2019, 2019, 6762517. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Shin, S.E.; Ko, K.S.; Park, S.H. Identification of Forensically Important Calliphoridae and Sarcophagidae Species Collected in Korea Using SNaPshot Multiplex System Targeting the Cytochrome c Oxidase Subunit I Gene. BioMed Res. Int. 2018, 2018, 2953892. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Jangra, S.; Dietzgen, R.G.; Yeh, W.-B. Frontiers Approaches to the Diagnosis of Thrips (Thysanoptera): How Effective Are the Molecular and Electronic Detection Platforms? Insects 2021, 12, 920. [Google Scholar] [CrossRef]
- Arif, M.; Busot, G.Y.; Mann, R.; Rodoni, B.; Stack, J.P. Field-Deployable Recombinase Polymerase Amplification Assay for Specific, Sensitive and Rapid Detection of the US Select Agent and Toxigenic Bacterium, Rathayibacter toxicus. Biology 2021, 10, 620. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Wang, C.; Qu, W.; Xu, R.; Liu, Y.; Jia, H.; Tang, X.; Chen, S.; Li, X.; Wang, Y.; et al. A rapid identification system for vaginal fluid stains based on nested recombinant polymerase amplification and lateral flow dipstick. Int. J. Leg. Med. 2024. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Zhang, Y.; Piao, H.; Yu, D.H.; Jeong, H.J.; Yoo, G.Y.; Jo, T.H.; Hwang, J.J. Sequences of the cytochrome C oxidase subunit I (COI) gene are suitable for species identification of Korean Calliphorinae flies of forensic importance (Diptera: Calliphoridae). J. Forensic Sci. 2009, 54, 1131–1134. [Google Scholar] [CrossRef]
- Guo, Y.; Cai, J.; Chang, Y.; Li, X.; Liu, Q.; Wang, X.; Wang, X.; Zhong, M.; Wen, J.; Wang, J. Identification of forensically important sarcophagid flies (Diptera: Sarcophagidae) in China, based on COI and 16S rDNA gene sequences. J. Forensic Sci. 2011, 56, 1534–1540. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Kim, Y.H.; Ham, C.S.; Shin, S.E.; Lee, H.J.; Ko, K.S.; Choi, J.; Son, G.H.; Park, S.H. Molecular identification of forensically important calliphoridae and sarcophagidae species using ITS2 nucleotide sequences. Forensic Sci. Int. 2018, 284, 1–4. [Google Scholar] [CrossRef]
- Ye, H.; Xu, H.; Xu, J.; Liang, J.; Huang, T.; Wang, X. A novel rapid detection method for chicken adulteration based on recombinant polymerase amplification and multicomponent nuclease (MNAzyme). Microchem. J. 2024, 204, 111148. [Google Scholar] [CrossRef]
- Zhou, C.; Wang, J.; Xiang, J.; Fu, Q.; Sun, X.; Liu, L.; Ai, L.; Wang, J. Rapid detection of duck ingredient in adulterated foods by isothermal recombinase polymerase amplification assays. Food Chem. Mol. Sci. 2023, 6, 100162. [Google Scholar] [CrossRef]
- Luo, G.C.; Yi, T.T.; Jiang, B.; Guo, X.; Zhang, G.Y. Betaine-assisted recombinase polymerase assay with enhanced specificity. Anal. Biochem. 2019, 575, 36–39. [Google Scholar] [CrossRef]
- Amer, H.M.; Abd El Wahed, A.; Shalaby, M.A.; Almajhdi, F.N.; Hufert, F.T.; Weidmann, M. A new approach for diagnosis of bovine coronavirus using a reverse transcription recombinase polymerase amplification assay. J. Virol. Methods 2013, 193, 337–340. [Google Scholar] [CrossRef] [PubMed]
- Valasevich, N.; Schneider, B. Rapid detection of “Candidatus Phytoplasma mali” by recombinase polymerase amplification assays. J. Phytopathol. 2017, 165, 762–770. [Google Scholar] [CrossRef]
- Ahmed Sarah, A.; van de Sande Wendy, W.J.; Desnos-Ollivier, M.; Fahal Ahmed, H.; Mhmoud Najwa, A.; de Hoog, G.S. Application of Isothermal Amplification Techniques for Identification of Madurella mycetomatis, the Prevalent Agent of Human Mycetoma. J. Clin. Microbiol. 2015, 53, 3280–3285. [Google Scholar] [CrossRef]



| Primer | Sequence (5′–3′) | Primer Length (bp) | Amplification Length (bp) |
|---|---|---|---|
| Barcode-658 | F: GGTCAACAAATCATAAAGATATTGG | 25 | 658 |
| R: RAAACTTCAGGRTGACCAAAGAATCA | 26 |
| Assay | Primer | Sequence (5′–3′) | Primer Length (nt) | Amplification Length (bp) |
|---|---|---|---|---|
| Basic RPA | CM-RPA-1 | F: TTTACCATTTATTGTTCTAGCTGCAACTCTT | 31 | 256 |
| R: TTGAATATGAACTGGAGTAACTAAAGGATT | 30 | |||
| CM-RPA-2 | F: TACCATTTATTGTTCTAGCTGCACGTCGT | 29 | 254 | |
| R: TTGAATATGAACTGGAGTAACTAAAGGATT | 30 | |||
| CM-RPA-3 | F: TTTTATCCCAGCCAATCCTTTAGTTACTCC | 30 | 199 | |
| R: ATTAATAGGGTAGAATTGAATACCTCGGAAC | 31 | |||
| RPA-LFD | CM-RPA-2 | mF: [FAM]-TACCATTTATTGTTCTAGCTGCACGTCGT | 29 | 254 |
| mR: [Biotin]-TTGAATATGAACTGGAGTAACTAAAGGATT | 30 | |||
| Probe | [FAM]-ATTTTATTAGTATTAATTAATCCTTACTTAC [TNF]TTGGTGACCCTGATAAT-[C3 spacer] | 48 |
| Repeat | Dilution Factor | ||||||
|---|---|---|---|---|---|---|---|
| 20 (1.25 × 10−2 ng) | 21 (6.25 × 10−3 ng) | 22 (3.13 × 10−3 ng) | 23 (1.56 × 10−3 ng) | 24 (7.8 × 10−4 ng) | 25 (3.9 × 10−4 ng) | 26 (1.95 × 10−4 ng) | |
| I | +++ | +++ | ++ | ++ | + | − | − |
| II | +++ | +++ | ++ | ++ | + | − | − |
| Case | Sample | Species Identified by Sequencing |
|---|---|---|
| S2018-18 | Chrysomya pinguis (Walker) | |
| S2018-168 | Chrysomya megacephala | |
| S2018-219 | 1 | Synthesiomyia nudiseta (van der Wulp) |
| 2 | Chrysomya rufifacies | |
| S2020-107 | Sarcophaga peregrina | |
| S2022-18 | 1–3 | Sarcophaga peregrina |
| S2022-34 | 1, 2 | Aldrichina grahami (Aldrich) |
| S2022-108 | 1–3 | Chrysomya megacephala |
| S2022-110 | 1 | Lucilia sericata |
| 2, 3 | Chrysomya megacephala | |
| S2022-113 | 1 | Creophilus maxillosus (Linnaeus) |
| 2, 7 | Chrysomya megacephala | |
| 3 | Megaselia scalaris | |
| 4 | Dohrniphora cornuta (Bigot) | |
| 5, 6 | Muscina stabulans (Fallén) | |
| S2022-139 | 1–3 | Chrysomya megacephala |
| S2022-150 | 1–4 | Chrysomya megacephala |
| S2023-35 | 1 | Chrysomya rufifacies |
| S2023-107 | 1, 2, 4–9 | Chrysomya pinguis |
| 3 | Chrysomya rufifacies | |
| S2023-159 | Chrysomya megacephala | |
| S2023-161 | Sarcophaga peregrina | |
| S2023-202 | 1, 4 | Sarcophaga peregrina |
| 2 | Chrysomya megacephala | |
| 3 | Musca domestica (Linnaeus) | |
| N1 | 1–5 | Sarcophaga crassipalpis |
| N2 | Lucilia sericata | |
| N3 | Sarcophaga crassipalpis | |
| N4 | Sarcophaga portschinskyi (Rohdendorf) | |
| Sarcophaga peregrina | ||
| N5 | Sarcophaga crassipalpis | |
| N6 | Megaselia scalaris | |
| N7 | Sarcophaga crassipalpis | |
| Chrysomya rufifacies | ||
| N8 | Sarcophaga crassipalpis | |
| Lucilia sericata | ||
| N9 | Sarcophaga crassipalpis | |
| N10 | 1–3 | Calliphora vicina |
| N11 | 1, 2 | Calliphora vicina |
| 3 | Lucilia sericata | |
| N12 | 1–3 | Calliphora vicina |
| N13 | Calliphora vicina | |
| N14 | 1, 2 | Calliphora vomitoria (Linnaeus) |
| 3, 4 | Triceratopyga calliphoroides (Rohdendorf) | |
| 6 | Dermestes coarctatus (Harold) | |
| N15 | 2 | Lucilia sericata |
| 3 | Sarcophaga portschinskyi | |
| N16 | 1–3 | Lucilia ampullacea (Villeneuve) |
| N17 | 1 | Lucilia sericata |
| 2, 3 | Chrysomya megacephala | |
| N18 | 1, 2 | Lucilia sericata |
| 3 | Sarcophaga dux (Thomson) | |
| N19 | 1, 2 | Megaselia scalaris |
| 3 | Blattella germanica (Linnaeus) | |
| N20 | 1, 2 | Sarcophaga crassipalpis |
| 3 | Lucilia sericata | |
| N21 | 1–3 | Lucilia sericata |
| N22 | 1–3 | Megaselia scalaris |
| N23 | 1, 2 | Chrysomya megacephala |
| 3 | Chrysomya rufifacies | |
| N24 | 1 | Chrysomya megacephala |
| 2 | Sarcophaga dux | |
| 3, 4 | Chrysomya rufifacies | |
| N25 | 1 | Megaselia scalaris |
| 2 | Synthesiomyia nudiseta | |
| 3 | Sarcophaga crassipalpis | |
| 4 | Sarcophaga nathani (Lopes) | |
| N26 | 1 | Blattella germanica |
| 3–4 | Sarcophaga portschinskyi | |
| N27 | 1–4 | Lucilia sericata |
| N28 | 1–3 | Calliphora vicina |
| N29 | 1, 4 | Calliphora nigribarbis (Vollenhoven) |
| 3 | Lucilia sericata | |
| 5, 6 | Protophormia terraenovae (Robineau-Desvoidy) |
| Species Name | Total Cases | Cases from South China | Cases from North China |
|---|---|---|---|
| Chrysomya megacephala | 11 | 8 | 3 |
| Lucilia sericata | 11 | 1 | 10 |
| Sarcophaga crassipalpis | 8 | 8 | |
| Chrysomya rufifacies | 6 | 2 | 4 |
| Calliphora vicina | 5 | 5 | |
| Megaselia scalaris | 5 | 1 | 4 |
| Sarcophaga peregrina | 5 | 4 | 1 |
| Sarcophaga portschinskyi | 3 | 3 | |
| Chrysomya pinguis | 2 | 2 | |
| Blattella germanica | 2 | 2 | |
| Sarcophaga dux | 2 | 2 | |
| Sarcophaga nathani | 2 | 2 | |
| Calliphora nigribarbis | 1 | 1 | |
| Protophormia terraenovae | 1 | 1 | |
| Muscina stabulans | 1 | 1 | |
| Triceratopyga calliphoroides | 1 | 1 | |
| Creophilus maxillosus | 1 | 1 | |
| Calliphora vomitoria | 1 | 1 | |
| Lucilia ampullaceal | 1 | 1 | |
| Musca domestica | 1 | 1 | |
| Dohrniphora cornuta | 1 | 1 | |
| Aldrichina grahami | 1 | 1 | |
| Synthesiomyia nudiseta | 1 | 1 | |
| Dermestes coarctatus | 1 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, C.; Tang, X.; Yang, F.; Zhang, X.; Shang, Y.; Xia, Y.; Wang, Y.; Guo, S.; Zha, L.; Guo, Y.; et al. Rapid and Accurate Detection of Chrysomya megacephala (Diptera: Calliphoridae) Using Recombinase Polymerase Amplification Combined with Lateral Flow Dipstick. Insects 2024, 15, 1008. https://doi.org/10.3390/insects15121008
Ye C, Tang X, Yang F, Zhang X, Shang Y, Xia Y, Wang Y, Guo S, Zha L, Guo Y, et al. Rapid and Accurate Detection of Chrysomya megacephala (Diptera: Calliphoridae) Using Recombinase Polymerase Amplification Combined with Lateral Flow Dipstick. Insects. 2024; 15(12):1008. https://doi.org/10.3390/insects15121008
Chicago/Turabian StyleYe, Chengxin, Xuan Tang, Fengqin Yang, Xiangyan Zhang, Yanjie Shang, Yang Xia, Yuanxing Wang, Shaojiang Guo, Lagabaiyila Zha, Yadong Guo, and et al. 2024. "Rapid and Accurate Detection of Chrysomya megacephala (Diptera: Calliphoridae) Using Recombinase Polymerase Amplification Combined with Lateral Flow Dipstick" Insects 15, no. 12: 1008. https://doi.org/10.3390/insects15121008
APA StyleYe, C., Tang, X., Yang, F., Zhang, X., Shang, Y., Xia, Y., Wang, Y., Guo, S., Zha, L., Guo, Y., & Wen, D. (2024). Rapid and Accurate Detection of Chrysomya megacephala (Diptera: Calliphoridae) Using Recombinase Polymerase Amplification Combined with Lateral Flow Dipstick. Insects, 15(12), 1008. https://doi.org/10.3390/insects15121008








