Antennal Transcriptome Evaluation and Analysis for Odorant-Binding Proteins, Chemosensory Proteins, and Suitable Reference Genes in the Leaf Beetle Pest Diorhabda rybakowi Weise (Coleoptera: Chrysomelidae)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing
2.2. Sample Collection
2.3. RNA Extraction and cDNA Synthesis
2.4. cDNA Library Construction, Assembly, and Gene Annotation
2.5. Identification of Candidate Reference Genes and Olfactory Genes
2.6. Sequence Analysis and Phylogenetic Tree Construction
2.7. Reverse-Transcription Quantitative PCR (RT-qPCR)
2.8. Analysis of the Stability of Candidate Reference Genes
2.9. Tissue Expression Profiles of OBPs and CSPs
3. Results
3.1. Antennal Transcriptome and Functional Annotation
3.2. Identification of Candidate Reference Genes, OBPs, and CSPs
3.2.1. Candidate Reference Genes
3.2.2. Identification of OBPs
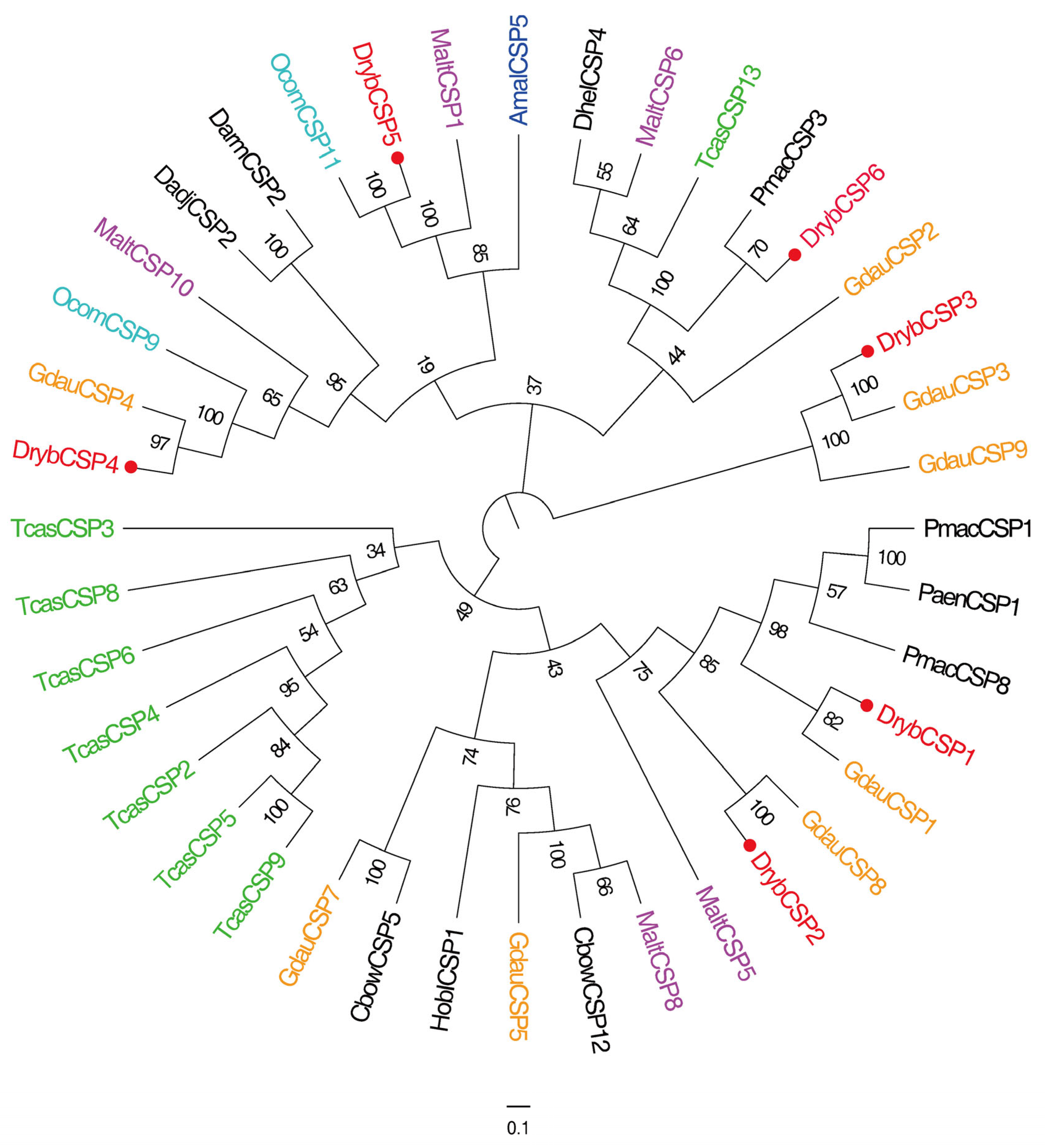
3.2.3. Identification of CSPs
3.3. Validation and Design of RT-qPCR Primers
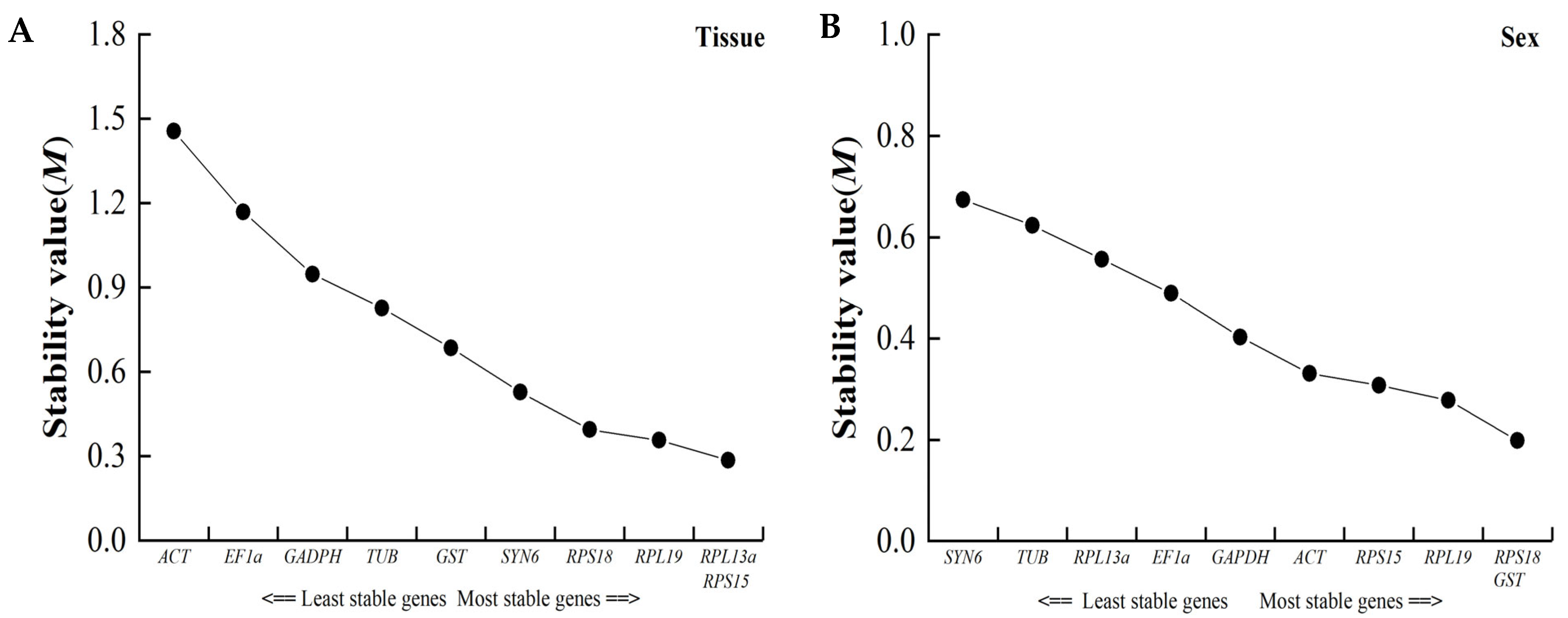
3.4. Stability of Candidate Reference Genes under Different Conditions
3.4.1. Delta Ct Analysis
3.4.2. GeNorm Analysis
3.4.3. NormFinder Analysis
3.4.4. BestKeeper Analysis
3.4.5. RefFinder Analysis
3.5. Relative Gene Expression of OBPs and CSPs
4. Discussion
4.1. Antennal Transcriptome Analysis
4.2. Evaluation of Reference Genes Stability
4.3. OBP Gene Identification and Expression Profiling
4.4. CSP Gene Identification and Expression Profiling
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, S.K.; Bao, P.; Yang, H.M. Damage of certain prominent insect pests on deserted grassland in Alshan and their control. Pratacult. Sci. 2000, 17, 44–46, 50. [Google Scholar]
- Xi, B.X.; Shang, S.Q.; Hu, G.X.; Chen, J.G.; Liu, Y.J.; Wang, Y.; Cui, X.N. Ultrastructural morphology on antennal sensilla of the adult Diorhabda rybakowi on grassland. Pratacult. Sci. 2023, 40, 1920–1931. [Google Scholar] [CrossRef]
- Liu, N.Y.; Gou, W.S.; Ma, W.X.; Tang, L.; Hu, G.X.; Sun, Y.D. Effect of temperature on the growth, development and reproduction of Diorhabda rybakowi (Coleoptera: Chrysomelidae). Plant Prot. 2023, 49, 220–226. [Google Scholar] [CrossRef]
- Li, D.H.; Liu, N.Y.; Gou, W.S.; Ma, W.X.; Hu, G.X. Effects of different host plants on development and reproduction of Diorhabda rybakowi (Coleoptera: Chrysomelidae). Chin. J. Biol. Control 2023, 39, 91–98. [Google Scholar] [CrossRef]
- Conchou, L.; Lucas, P.; Meslin, C.; Proffit, M.; Staudt, M.; Ronou, M. Insect odorscapes: From plant volatiles to natural olfactory scenes. Front. Physiol. 2019, 10, 972. [Google Scholar] [CrossRef]
- Hansson, B.S.; Stensmyr, M.C. Evolution of insect olfaction. Neuron 2011, 72, 698–711. [Google Scholar] [CrossRef] [PubMed]
- Loreto, F.; DAuria, S. The 40-Year Mystery of Insect Odorant-Binding Proteins? Trends Plant Sci. 2021, 27, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Taszakowski, A.; Maslowski, A.; Daane, K.M.; Brozek, J. Closer view of antennal sensory organs of two Leptoglossus species (Insecta, Hemiptera, Coreidae). Sci. Rep. 2023, 13, 617. [Google Scholar] [CrossRef] [PubMed]
- Steinbrecht, R.A. Odorant-binding proteins: Expression and function. Ann. N. Y. Acad. Sci. 1998, 855, 323–332. [Google Scholar] [CrossRef]
- Calvello, M.; Guerra, N.; Brandazza, A.; Dambrosio, C.; Scaloni, A.; Dani, F.R.; Turillazzi, S.; Pelosi, P. Soluble proteins of chemical communication in the social wasp Polistes dominulus. Cell. Mol. Life Sci. 2003, 60, 1933–1943. [Google Scholar] [CrossRef]
- Rondoni, G.; Roman, A.; Meslin, C.; Montagne, N.; Conti, E.; Jacquin-Joly, E. Antennal transcriptome analysis and identification of candidate chemosensory genes of the harlequin ladybird beetle, Harmonia axyridis (Pallas) (Coleoptera: Coccinellidae). Insects 2021, 12, 209. [Google Scholar] [CrossRef] [PubMed]
- Li, X.M.; Zhu, X.Y.; Wang, Z.Q.; Wang, Y.; He, P.; Chen, G.; Sun, L.; Deng, D.G.; Zhang, Y.N. Candidate chemosensory genes identified in Colaphellus bowringi by antennal transcriptome analysis. BMC Genom. 2015, 16, 1028. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Shimomura, K.; Hosoi, A.; Sato, Y.; Oikawa, Y.; Seino, Y.; Kuribara, T.; Yajima, S.; Tomizawa, M. Antennal transcriptome analysis of chemosensory genes in the cowpea beetle, Callosobruchus maculatus (F.). PLoS ONE 2006, 17, e0262817. [Google Scholar] [CrossRef]
- Lechuga-Paredes, P.; Segura-Leon, O.L.; Cibrian-Tovar, J.; Torres-Huerta, B.; Velazquez-Gonzalez, J.C.; Cruz-Jaramillo, J.L. Odorant-binding and chemosensory proteins in Anthonomus eugenii (Coleoptera: Curculionidae) and their tissue expression. Int. J. Mol. Sci. 2023, 24, 3406. [Google Scholar] [CrossRef] [PubMed]
- Yue, Y.; Ma, C.; Zhang, Y.; Chen, H.S.; Guo, J.Y.; Liu, T.H.; Zhou, Z.S. Characterization and functional analysis of OcomOBP7 in Ophraella communa Lesage. Insects 2023, 14, 190. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhou, Y.T.; Tan, Y.; Zhou, X.R.; Pang, B.P. Identification of odorant-binding protein genes in Galeruca daurica (Coleoptera: Chrysomelidae) and analysis of their expression profiles. Bull. Entomol. Res. 2017, 107, 550–561. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.S.; Qin, J.H.; Cao, Y.Z.; Li, K.B.; Yin, J. Two classic OBPs modulate the responses of female Holotrichia oblita to three major ester host plant volatiles. Insect Mol. Biol. 2021, 30, 390–399. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Pentzold, S.; Kunert, M.; Groth, M.; Brandt, W.; Pasteels, J.M.; Boland, W.; Burse, A. A subset of chemosensory genes differs between two populations of a specialized leaf beetle after host plant shift. Ecol. Evol. 2018, 8, 8055–8075. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.F.; Liu, X.Y.; Zhao, X.; Qin, J.H.; Cao, Y.Z.; Li, K.B.; Zhou, J.J.; Wang, S.S.; Yin, J. Evidence of the involvement of a Plus-C odorant-binding protein HparOBP14 in host plant selection and oviposition of the scarab beetle Holotrichia parallela. Insects 2021, 12, 430. [Google Scholar] [CrossRef]
- Oliveira, D.S.; Brito, N.F.; Franco, T.A.; Moreira, M.F.; Leal, W.S.; Melo, A.C.A. Functional characterization of odorant binding protein 27 (RproOBP27) from Rhodnius prolixus antennae. Front. Physiol. 2018, 9, 1175. [Google Scholar] [CrossRef]
- Wang, Y.L.; Jin, Y.C.; Chen, Q.; Wen, M.; Zhao, H.B.; Duan, H.X.; Ren, B.Z. Selectivity and ligand-based molecular modeling of an odorant-binding protein from the leaf beetle Ambrostoma quadriimpressum (Coleoptera: Chrysomelidae) in relation to habitat-related volatiles. Sci. Rep. 2017, 7, 15374. [Google Scholar] [CrossRef] [PubMed]
- Li, J.Q.; Zhu, R.; Yao, W.C.; Yu, H.P.; Huang, J.R.; Wang, Z.; Sun, X.Y.; Yuan, D.H.; Sun, Y.Y.; Emam, S.S.; et al. Chemosensory protein 2 of male Athetis lepigone is involved in the perception of sex pheromones and maize volatiles. J. Agric. Food Chem. 2023, 71, 6277–6287. [Google Scholar] [CrossRef]
- Ma, C.; Cui, S.W.; Tian, Z.Y.; Zhang, Y.; Chen, G.M.; Gao, X.Y.; Tian, Z.Q.; Chen, H.S.; Guo, J.Y.; Zhou, Z.S. OcomCSP12, a chemosensory protein expressed specifically by ovary, mediates reproduction in Ophraella communa (Coleoptera: Chrysomelidae). Front. Physiol. 2019, 10, 1290. [Google Scholar] [CrossRef]
- Gao, P.; Tan, J.J.; Su, S.; Wang, S.J.; Peng, X.; Chen, M.H. Overexpression of the chemosensory protein CSP7 gene contributed to lambda-cyhalothrin resistance in the bird cherry-oat aphid Rhopalosiphum padi. J. Agric. Food Chem. 2023, 71, 17005–17013. [Google Scholar] [CrossRef]
- Gao, P.; Zhang, S.Q.; Tan, J.J.; Li, X.H.; Chen, M.H. Chemosensory proteins are associated with thiamethoxam tolerance in bird cherry-oat aphid Rhopalosiphum padi Pestic. Biochem. Physiol. 2023, 192, 105393. [Google Scholar] [CrossRef]
- Shakeel, M.; Rodriguez, A.; Bin Tahir, U.; Jin, F.L. Gene expression studies of reference genes for quantitative real-time PCR: An overview in insects. Biotechnol. Lett. 2018, 40, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Chapman, J.R.; Waldenstrom, J. With reference to reference genes: A systematic review of endogenous controls in gene expression studies. PLoS ONE 2015, 10, e0141853. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Yang, C.X.; Zhang, Y.J.; Pan, H.P. Selection of reference genes for the normalization of RT-qPCR data in gene expression studies in insects: A systematic review. Front. Physiol. 2018, 9, 1560. [Google Scholar] [CrossRef]
- Sellamuthu, G.; Bily, J.; Joga, M.R.; Synek, J.; Roy, A. Identifying optimal reference genes for gene expression studies in Eurasian spruce bark beetle, Ips typographus (Coleoptera: Curculionidae: Scolytinae). Sci. Rep. 2022, 12, 4671. [Google Scholar] [CrossRef]
- Brah, G.S.; Kaur, G.; Singh, S.; Shukla, J.N.; Pandher, S. Identification and validation of stage-specific reference genes for gene expression analysis in Callosobruchus maculatus (Coleoptera: Bruchidae). Gene Expr. Patterns 2022, 43, 119233. [Google Scholar] [CrossRef]
- Ma, L.; Jiang, T.; Liu, X.; Xiao, H.J.; Peng, Y.C.; Zhang, W.N. Evaluation of candidate reference genes for gene expression analysis in the brassica leaf beetle, Phaedon brassicae (Coleoptera: Chrysomelidae). PLoS ONE 2021, 16, e0251920. [Google Scholar] [CrossRef] [PubMed]
- Lord, J.C.; Hartzer, K.; Toutges, M.; Oppert, B. Evaluation of quantitative PCR reference genes for gene expression studies in Tribolium castaneum after fungal challenge. J. Microbiol. Methods 2010, 80, 219–221. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.Q.; Yang, Y.C.; Chai, Y.P.; Gao, L.L.; Ma, R.Y. Identification and evaluation of reference genes for quantitative PCR normalization in Alligator Weed Flea Beetle (Coleoptera: Chrysomelidae). J. Insect Sci. 2021, 21, 9. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Fu, L.M.; Niu, B.F.; Zhu, Z.W.; Wu, S.T.; Li, W.Z. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Manni, M.; Berkeley, M.R.; Seppey, M.; Simao, F.A.; Zdobnov, E.M. BUSCO update: Novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 2021, 38, 4647–4654. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011, 12, 323. [Google Scholar] [CrossRef]
- Cui, X.N.; Liu, D.G.; Sun, K.K.; He, Y.; Shi, X.Q. Expression profiles and functional characterization of two odorant-binding proteins from the apple buprestid beetle Agrilus mali (Coleoptera: Buprestidae). J. Econ. Entomol. 2018, 111, 1420–1432. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for saequence lignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef]
- Gascuel, O. BIONJ: An improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997, 14, 685–695. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, 45. [Google Scholar] [CrossRef]
- Silver, N.; Best, S.; Jiang, J.; Thein, S.L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 2006, 7, 33. [Google Scholar] [CrossRef]
- Vandesompele, J.; De-Preter, K.; Pattyn, F.; Poppe, B.; Van-Roy, N.; De-Paepe, A.; Spememan, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Orntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.L.; Wang, J.Y.; Zhang, B.H. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genom. 2023, 23, 125. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Method 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Ma, C.; Zhao, C.C.; Cui, S.W.; Zhang, Y.; Chen, G.M.; Chen, H.S.; Wan, F.H.; Zhou, Z.S. Identification of candidate chemosensory genes of Ophraella communa LeSage (Coleoptera: Chrysomelidae) based on antennal transcriptome analysis. Sci. Rep. 2019, 9, 15551. [Google Scholar] [CrossRef]
- Ha, T.S.; Smith, D.P. Recent insights into insect olfactory receptors and odorant-binding proteins. Insects 2022, 13, 926. [Google Scholar] [CrossRef] [PubMed]
- Lizana, P.; Mutis, A.; Quiroz, A.; Venthur, H. Insights into chemosensory proteins from non-model insects: Advances and perspectives in the context of pest management. Front. Physiol. 2022, 13, 924750. [Google Scholar] [CrossRef] [PubMed]
- Renou, M.; Anton, S. Insect olfactory communication in a complex and changing world. Curr. Opin. Insect Sci. 2020, 42, 1–7. [Google Scholar] [CrossRef]
- Wang, X.X.; Liu, H.Y.; Xie, G.L.; Wang, W.K.; Yang, Y.X. Identification and expression analyses of the olfactory-related genes in different tissues’ transcriptome of a predacious soldier beetle, Podabrus annulatus (Coleoptera, Cantharidae). Arch. Insect Biochem. Physiol. 2023, 112, e21997. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Gonzalez, A.; Rubio-Melendez, M.E.; Ballesteros, G.I.; Ramirez, C.C.; Palma-Millanao, R. Sex-and tissue-specific expression of odorant-binding proteins and chemosensory proteins in adults of the scarab beetle Hylamorpha elegans (Burmeister) (Coleoptera: Scarabaeidae). PeerJ 2019, 7, e7054. [Google Scholar] [CrossRef]
- Sanchez-Gracia, A.; Vieira, F.G.; Rozas, J. Molecular evolution of the major chemosensory gene families in insects. Heredity 2009, 103, 208–216. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, M.R. A maturing understanding of the composition of the insect gene repertoire. Curr. Opin. Insect Sci. 2015, 7, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Johnson, P.K. Putting the genome in insect phylogenomics. Curr. Opin. Insect Sci. 2019, 36, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhao, X.X.; Li, M.Z.; He, K.; Huang, C.; Zhou, Y.N.; Li, Z.; Walters, J.R. Insect genomes: Progress and challenges. Insect Mol. Biol. 2019, 28, 739–758. [Google Scholar] [CrossRef]
- Romain, F.; Waterhouse, M.R. Exploring new genomic territories with emerging model insects. Curr. Opin. Insect Sci. 2022, 51, 100902. [Google Scholar] [CrossRef]
- Huggett, J.F.; Whale, A.S.; De-Spiegelaere, W.; Nour, A.A.; Bae, Y.K.; Benes, V.; Burke, D.; Cleveland, M.; Corbisier, P.; Devonshire, A.S.; et al. The digital MIQE guidelines update: Minimum information for publication of quantitative digital PCR experiments for 2020. Clin. Chem. 2020, 66, 1012–1029. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J.; Zheng, H.L.; Liu, Y.Y.; Li, H.W.; Jiang, Y.H.; Lin, L.B.; Deng, X.Y.; Zhang, Q.L. Selection of reference genes for quantitative real-time PCR in Aquatica leii (Coleoptera: Lampyridae) under five different experimental conditions. Front. Physiol. 2020, 11, 555233. [Google Scholar] [CrossRef] [PubMed]
- Marabita, F.; De Candia, P.; Torri, A.; Tegner, J.; Abrignani, S.; Rossi, R.L. Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Brief. Bioinform. 2016, 17, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.Q.; Zhu, L.; Li, Y.; Liu, W.; Ma, W.H.; Lei, C.L.; Wang, X.P. A de novo transcriptome and valid reference genes for quantitative real-time PCR in Colaphellus bowringi. PLoS ONE 2015, 10, e0118693. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, J.Q.; Chen, G.M.; Ma, C.; Chen, H.S.; Gao, X.Y.; Tian, Z.Q.; Cui, S.W.; Tian, Z.Y.; Guo, J.Y.; et al. Identification and validation of reference genes for quantitative gene expression analysis in Ophraella communa. Front. Physiol. 2020, 11, 355. [Google Scholar] [CrossRef]
- Leal, W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 337–391. [Google Scholar] [CrossRef] [PubMed]
- Sims, C.; Birkett, M.A.; Withall, D.M. Enantiomeric discrimination in insects: The role of OBPs and ORs. Insects 2022, 13, 368. [Google Scholar] [CrossRef] [PubMed]
- He, P.; Zhang, J.; Liu, N.Y.; Zhang, Y.N.; Yang, K.; Dong, S.L. Distinct expression profiles and different functions of odorant binding proteins in Nilaparvata lugens Stal. PLoS ONE 2011, 6, e28921. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, J.W.; Sun, W.X.; Li, W.; Gao, H.Y.; Liu, T.X.; Qu, M.J. Candidate chemosensory genes identified in the adult antennae of Sympiezomias velatus and binding property of odorant-binding protein 15. Front. Physiol. 2022, 13, 907667. [Google Scholar] [CrossRef]
- Bai, P.H.; Wang, H.M.; Liu, B.S.; Li, M.; Liu, B.M.; Gu, X.S.; Tang, R. Botanical volatiles selection in mediating electrophysiological responses and reproductive behaviors for the fall webworm moth Hyphantria cunea. Front. Physiol. 2020, 11, 486. [Google Scholar] [CrossRef]
- Tomaselli, S.; Crescenzi, O.; Sanfelice, D.; Ab, E.; Wechselberger, R.; Angeli, S.; Scaloni, A.; Boelens, R.; Tancredi, T.; Pelosi, P.; et al. Solution structure of a chemosensory protein from the desert locust Schistocerca gregaria. Biochemistry 2006, 45, 10606–10613. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.X.; Xuan, N.; Rajashekar, B.; Arnaud, P.; Offmann, B.; Picimbon, J.F. Comprehensive history of CSP genes: Evolution, phylogenetic distribution and functions. Genes 2020, 11, 413. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Ahmed, M.Z.; Li, N.; Ali, S.A.I.; Wang, M.Q. Functional characteristics of chemosensory proteins in the sawyer beetle Monochamus alternatus Hope. Bull. Entomol. Res. 2019, 109, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Li, C.B.; Sun, K.K.; Li, D.X.; Liu, D.G. Functional characterization of chemosensory protein AmalCSP5 from apple buprestid beetle, Agrilus mali (Coleoptera: Buprestidae). J. Econ. Entomol. 2021, 114, 348–359. [Google Scholar] [CrossRef] [PubMed]




| Gene_ID | Gene Abbr | Genbank Number | ORF (aa) | Blastx Annotation | Acc. Number | E-Value | Identity (%) |
|---|---|---|---|---|---|---|---|
| TRINITY_DN13598_c2_g3 | ACT | OR797776 | 376 | actin, muscle [Agrilus planipennis] | XP_018335426.1 | 0 | 99.73 |
| TRINITY_DN13848_c0_g3 | GAPDH | OR797777 | 332 | glyceraldehyde-3-phosphate dehydrogenase 2-like [Diabrotica virgifera virgifera] | XP_028133871.1 | 0 | 93.66 |
| TRINITY_DN13241_c1_g2 | EF1a | OR797778 | 462 | elongation factor A [Diabrotica undecimpunctata howardi] | APQ43052.1 | 0 | 97.77 |
| TRINITY_DN8410_c0_g1 | TUB | OR797779 | 447 | tubulin beta-1 chain [Anoplophora glabripennis] | XP_018568298.1 | 0 | 100.00 |
| TRINITY_DN10540_c0_g1 | RPL13a | OR797780 | 204 | 60S ribosomal protein L13a [Sitophilus oryzae] | XP_030748989.1 | 3.00 × 10−126 | 94.61 |
| TRINITY_DN14590_c0_g1 | RPS18 | OR797781 | 152 | ribosomal protein S18 [Phaedon cochleariae] | AFQ22730.1 | 5.00 × 10−89 | 99.23 |
| TRINITY_DN10601_c0_g1 | RPL19 | OR797782 | 199 | ribosomal protein L19 [Chrysomela tremula] | ACY71295.1 | 3.00 × 10−136 | 97.49 |
| TRINITY_DN14218_c0_g1 | SYN6 | OR797783 | 153 | syntaxin-6 [Anoplophora glabripennis] | XP_018565804.1 | 5.00 × 10−84 | 92.54 |
| TRINITY_DN1008_c0_g1 | GST | OR797784 | 220 | glutathione S-transferase 1 [Anoplophora glabripennis] | XP_018568551.1 | 4.00 × 10−118 | 75.45 |
| TRINITY_DN11144_c0_g1 | RPS15 | OR797785 | 148 | ribosomal protein S15 [Stegobium paniceum] | ALG76024.1 | 2.00 × 10−76 | 94.59 |
| Gene_ID | Gene Name | Genebank Number | Complete ORF (aa) | Signal Peptide | Group | BLAST Annotation | Acc. Number | E-Value | Identity (%) |
|---|---|---|---|---|---|---|---|---|---|
| Odorant-binding proteins, OBPs | |||||||||
| TRINITY_DN10570_c0_g1 | DrybOBP1 | OR797799 | 126 | 1-16 | Minus-C OBP | odorant-binding protein 26 [Pyrrhalta maculicollis] | APC94196.1 | 2.00 × 10−25 | 51.92 |
| TRINITY_DN10570_c0_g2 | DrybOBP2 | OR797786 | 130 | 1-16 | Minus-C OBP | odorant-binding protein 26 [Pyrrhalta maculicollis] | APC94196.1 | 4.00 × 10−38 | 51.64 |
| TRINITY_DN10673_c0_g1 | DrybOBP3 | OR797787 | 143 | 1-17 | Minus-C OBP | odorant-binding protein [Galeruca daurica] | AQY18990.1 | 7.00 × 10−58 | 65.6 |
| TRINITY_DN12287_c4_g1 | DrybOBP4 | OR797800 | 137 | 1-16 | Minus-C OBP | odorant-binding protein [Galeruca daurica] | AQY18989.1 | 1.00 × 10−86 | 89.78 |
| TRINITY_DN12797_c0_g5 | DrybOBP5 | OR797788 | 135 | 1-18 | Minus-C OBP | odorant-binding protein [Galeruca daurica] | AQY18986.1 | 1.00 × 10−66 | 77.1 |
| TRINITY_DN12797_c0_g7 | DrybOBP6 | OR797789 | 134 | 1-18 | Minus-C OBP | odorant-binding protein 5 [Pyrrhalta maculicollis] | APC94193.1 | 9.00 × 10−35 | 46.32 |
| TRINITY_DN14706_c2_g1 | DrybOBP7 | OR797790 | 167 | 1-17 | Minus-C OBP | odorant-binding protein [Galeruca daurica] | AQY18987.1 | 1.00 × 10−58 | 63.64 |
| TRINITY_DN14781_c6_g3 | DrybOBP8 | OR797791 | 130 | 1-17 | Minus-C OBP | odorant-binding protein 29 [Pyrrhalta maculicollis] | APC94190.1 | 4.00 × 10−51 | 64.12 |
| TRINITY_DN29911_c0_g1 | DrybOBP9 | OR797792 | 136 | 1-16 | Minus-C OBP | odorant-binding protein [Galeruca daurica] | AQY18985.1 | 1.00 × 10−11 | 28.89 |
| TRINITY_DN14470_c4_g1 | DrybOBP10 | OR797801 | 120 | 1-23 | Classic-OBP | odorant-binding protein [Galeruca daurica] | AQY18968.1 | 4.00 × 10−61 | 77.5 |
| TRINITY_DN11053_c0_g1 | DrybOBP11 | OR797793 | 220 | 0 | Plus-c OBP | odorant-binding protein 25 [Colaphellus bowringi] | ALR72513.1 | 2.00 × 10−53 | 45.81 |
| Chemosensory proteins, CSPs | |||||||||
| TRINITY_DN11935_c0_g1 | DrybCSP1 | OR797794 | 129 | 1-18 | / | chemosensory protein | ARM20137.1 | 2.00 × 10−51 | 79.84 |
| TRINITY_DN13680_c2_g1 | DrybCSP2 | OR797795 | 131 | 1-19 | / | ejaculatory bulb-specific protein 3-like [Diorhabda carinulata] | XP_057656967.1 | 3.00 × 10−57 | 91.89 |
| TRINITY_DN14709_c1_g3 | DrybCSP3 | OR797796 | 136 | 1-21 | / | chemosensory protein | ARM20139.1 | 4.00 × 10−76 | 93.38 |
| [Galeruca daurica] | |||||||||
| TRINITY_DN14908_c1_g2 | DrybCSP4 | OR797797 | 124 | 1-22 | / | ejaculatory bulb-specific protein 3-like | XP_056639215.1 | 3.00 × 10−79 | 91.87 |
| TRINITY_DN8389_c0_g1 | DrybCSP5 | OR797802 | 117 | 1-22 | / | chemosensory protein 11 | UMT69263.1 | 5.00 × 10−64 | 83.9 |
| [Ophraella communa] | |||||||||
| TRINITY_DN9992_c0_g1 | DrybCSP6 | OR797798 | 232 | 1-18 | / | chemosensory protein | ARM20146.1 | 2.00 × 10−82 | 65.9 |
| [Galeruca daurica] | |||||||||
| TRINITY_DN11935_c0_g1 | DrybCSP1 | OR797794 | 129 | 1-18 | / | chemosensory protein | ARM20137.1 | 2.00 × 10−51 | 79.84 |
| [Galeruca daurica] | |||||||||
| TRINITY_DN13680_c2_g1 | DrybCSP2 | OR797795 | 131 | 1-19 | / | ejaculatory bulb-specific protein 3-like [Diorhabda carinulata] | XP_057656967.1 | 3.00 × 10−57 | 91.89 |
| TRINITY_DN14709_c1_g3 | DrybCSP3 | OR797796 | 136 | 1-21 | / | chemosensory protein | ARM20139.1 | 4.00 × 10−76 | 93.38 |
| [Galeruca daurica] | |||||||||
| TRINITY_DN14908_c1_g2 | DrybCSP4 | OR797797 | 124 | 1-22 | / | ejaculatory bulb-specific protein 3-like | XP_056639215.1 | 3.00 × 10−79 | 91.87 |
| [Diorhabda carinulata] | |||||||||
| TRINITY_DN8389_c0_g1 | DrybCSP5 | OR797802 | 117 | 1-22 | / | chemosensory protein 11 | UMT69263.1 | 5.00 × 10−64 | 83.9 |
| [Ophraella communa] | |||||||||
| TRINITY_DN9992_c0_g1 | DrybCSP6 | OR797798 | 232 | 1-18 | / | chemosensory protein | ARM20146.1 | 2.00 × 10−82 | 65.9 |
| [Galeruca daurica] | |||||||||
| Rank | Tissue | Sex | ||
|---|---|---|---|---|
| Gene | SV | Gene | SV | |
| 1 | SYN6 | 0.047 | RPL19 | 0.169 |
| 2 | RPS18 | 0.125 | ACT | 0.249 |
| 3 | RPL19 | 0.583 | RPS15 | 0.322 |
| 4 | RPL13a | 0.659 | EF1a | 0.346 |
| 5 | TUB | 0.755 | RPS18 | 0.445 |
| 6 | RPS15 | 0.815 | GST | 0.495 |
| 7 | GAPDH | 0.834 | RPL13a | 0.507 |
| 8 | GST | 1.347 | TUB | 0.71 |
| 9 | EF1a | 1.816 | GADPH | 0.775 |
| 10 | ACT | 2.457 | SYN6 | 0.802 |
| Rank | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| CV ± SD | CV ± SD | CV ± SD | CV ± SD | CV ± SD | CV ± SD | CV ± SD | CV ± SD | CV ± SD | CV ± SD | |
| Tissue | RPL13a | GST | RPL19 | RPS18 | RPS15 | SYN6 | GAPDH | TUB | EF1a | ACT |
| 0.88 ± 0.18 | 1.68 ± 0.42 | 1.72 ± 0.38 | 1.76 ± 0.39 | 1.76 ± 0.41 | 2.41 ± 0.73 | 3.44 ± 0.88 | 4.27 ± 1.07 | 7.36 ± 1.78 | 9.22 ± 2.14 | |
| Sex | GAPDH | GST | RPS18 | RPS15 | ACT | RPL19 | SYN6 | EF1a | TUB | RPL13a |
| 2.25 ± 0.54 | 3.48 ± 0.85 | 3.82 ± 0.8 | 4.11 ± 0.91 | 4.46 ± 0.93 | 5.07 ± 1.08 | 5.91 ± 1.67 | 6.44 ± 1.38 | 7.55 ± 1.7 | 7.80 ± 1.54 |
| Conditions | Reference Gene | RefFinder | ΔCt | GeNorm | NormFinder | BestKeeper | Recommendation | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Stability | Rank | Stability | Rank | Stability | Rank | Stability | Rank | Stability | Rank | |||
| Tissue | ||||||||||||
| ACT | 10.000 | 10 | 2.600 | 10 | 1.455 | 9 | 2.457 | 10 | 2.140 | 10 | RPL13a, RPS18 | |
| TUB | 6.400 | 6 | 1.320 | 6 | 0.827 | 6 | 0.755 | 5 | 1.074 | 8 | ||
| RPS18 | 2.210 | 2 | 1.030 | 1 | 0.394 | 3 | 0.125 | 2 | 0.388 | 3 | ||
| GST | 6.620 | 7 | 1.600 | 8 | 0.685 | 5 | 1.347 | 8 | 0.422 | 5 | ||
| SYN6 | 2.780 | 3 | 1.090 | 2 | 0.527 | 4 | 0.047 | 1 | 0.732 | 6 | ||
| GAPDH | 7.240 | 8 | 1.440 | 7 | 0.947 | 7 | 0.834 | 7 | 0.879 | 7 | ||
| EF1a | 9.000 | 9 | 2.040 | 9 | 1.168 | 8 | 1.816 | 9 | 1.783 | 9 | ||
| RPL13a | 1.860 | 1 | 1.120 | 3 | 0.285 | 1 | 0.659 | 4 | 0.163 | 1 | ||
| RPL19 | 2.910 | 4 | 1.130 | 4 | 0.356 | 2 | 0.583 | 3 | 0.381 | 2 | ||
| RPL15 | 3.310 | 5 | 1.200 | 5 | 0.285 | 1 | 0.815 | 6 | 0.412 | 4 | ||
| Sex | ||||||||||||
| ACT | 3.160 | 3 | 0.560 | 2 | 0.331 | 4 | 0.249 | 2 | 0.926 | 5 | RPL19, GST | |
| TUB | 8.710 | 9 | 0.790 | 8 | 0.623 | 8 | 0.710 | 8 | 1.696 | 9 | ||
| RPS18 | 2.510 | 2 | 0.610 | 4 | 0.199 | 1 | 0.445 | 5 | 0.797 | 2 | ||
| GST | 3.220 | 4 | 0.650 | 6 | 0.199 | 1 | 0.495 | 6 | 0.849 | 3 | ||
| SYN6 | 9.740 | 10 | 0.880 | 10 | 0.674 | 9 | 0.802 | 10 | 1.670 | 9 | ||
| GAPDH | 4.700 | 6 | 0.840 | 9 | 0.403 | 5 | 0.775 | 9 | 0.539 | 1 | ||
| EF1a | 5.600 | 7 | 0.620 | 5 | 0.489 | 6 | 0.346 | 4 | 1.376 | 7 | ||
| RPL13a | 7.480 | 8 | 0.680 | 7 | 0.557 | 7 | 0.507 | 7 | 1.540 | 8 | ||
| RPL19 | 2.060 | 1 | 0.540 | 1 | 0.279 | 2 | 0.169 | 1 | 1.079 | 6 | ||
| RPL15 | 3.460 | 5 | 0.570 | 3 | 0.308 | 3 | 0.322 | 3 | 0.905 | 4 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xi, B.-X.; Cui, X.-N.; Shang, S.-Q.; Li, G.-W.; Dewer, Y.; Li, C.-N.; Hu, G.-X.; Wang, Y. Antennal Transcriptome Evaluation and Analysis for Odorant-Binding Proteins, Chemosensory Proteins, and Suitable Reference Genes in the Leaf Beetle Pest Diorhabda rybakowi Weise (Coleoptera: Chrysomelidae). Insects 2024, 15, 251. https://doi.org/10.3390/insects15040251
Xi B-X, Cui X-N, Shang S-Q, Li G-W, Dewer Y, Li C-N, Hu G-X, Wang Y. Antennal Transcriptome Evaluation and Analysis for Odorant-Binding Proteins, Chemosensory Proteins, and Suitable Reference Genes in the Leaf Beetle Pest Diorhabda rybakowi Weise (Coleoptera: Chrysomelidae). Insects. 2024; 15(4):251. https://doi.org/10.3390/insects15040251
Chicago/Turabian StyleXi, Bo-Xin, Xiao-Ning Cui, Su-Qin Shang, Guang-Wei Li, Youssef Dewer, Chang-Ning Li, Gui-Xin Hu, and Yan Wang. 2024. "Antennal Transcriptome Evaluation and Analysis for Odorant-Binding Proteins, Chemosensory Proteins, and Suitable Reference Genes in the Leaf Beetle Pest Diorhabda rybakowi Weise (Coleoptera: Chrysomelidae)" Insects 15, no. 4: 251. https://doi.org/10.3390/insects15040251
APA StyleXi, B.-X., Cui, X.-N., Shang, S.-Q., Li, G.-W., Dewer, Y., Li, C.-N., Hu, G.-X., & Wang, Y. (2024). Antennal Transcriptome Evaluation and Analysis for Odorant-Binding Proteins, Chemosensory Proteins, and Suitable Reference Genes in the Leaf Beetle Pest Diorhabda rybakowi Weise (Coleoptera: Chrysomelidae). Insects, 15(4), 251. https://doi.org/10.3390/insects15040251








