Identification and Functional Analysis of the fruitless Gene in a Hemimetabolous Insect, Nilaparvata lugens
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Husbandry
2.2. Cloning of N. lugens Fruitless (Nlfru)
2.3. Semi-Quantitative RT-PCR
2.4. Real-Time Quantitative PCR (qPCR) Analysis
2.5. RNA Interference (RNAi)
2.6. Recording Courtship Behavior and Measurement of Courtship Signals
2.7. Lethality Statistics
3. Results
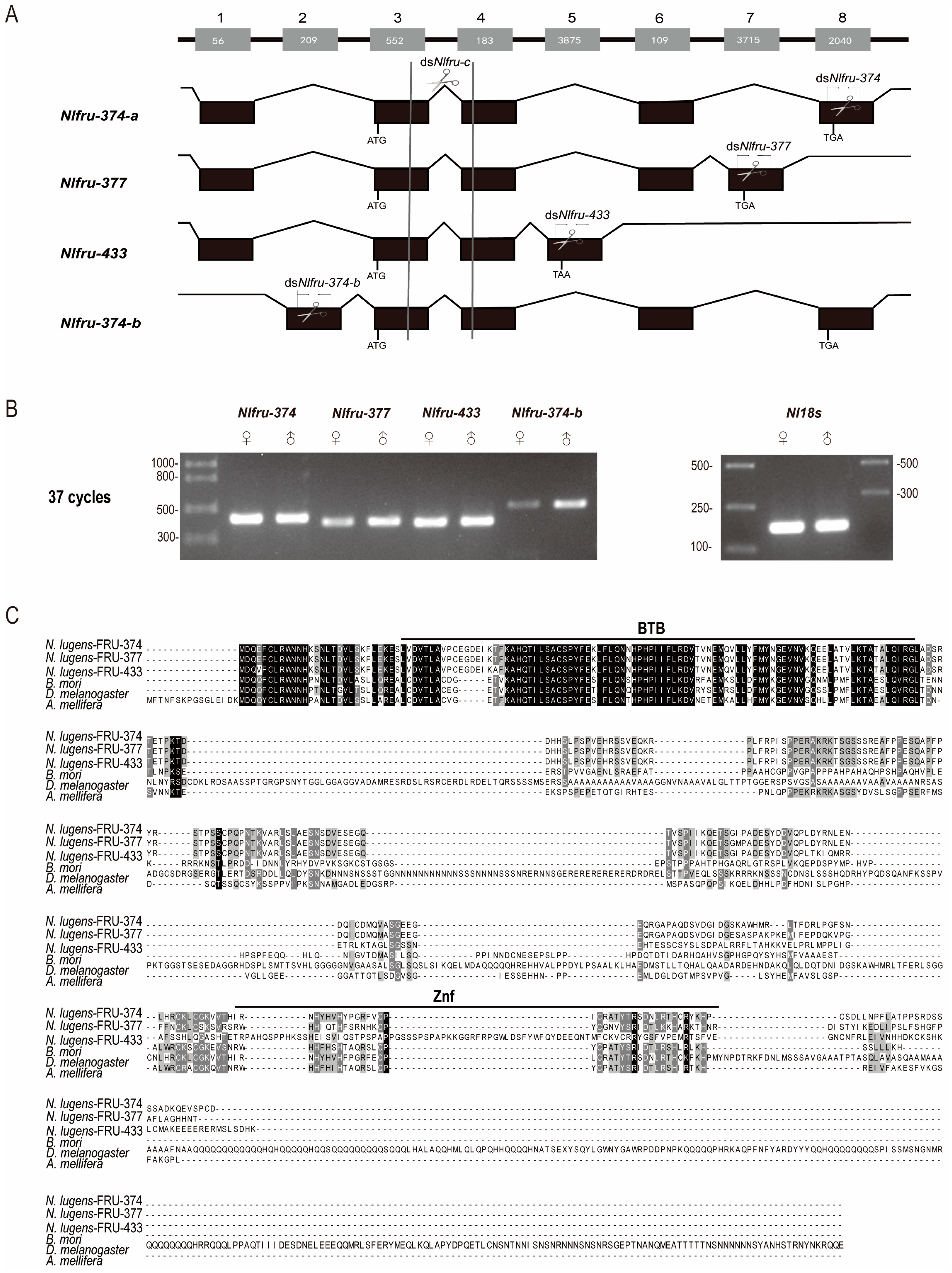
3.1. The Homologs of Fruitless in N. lugens
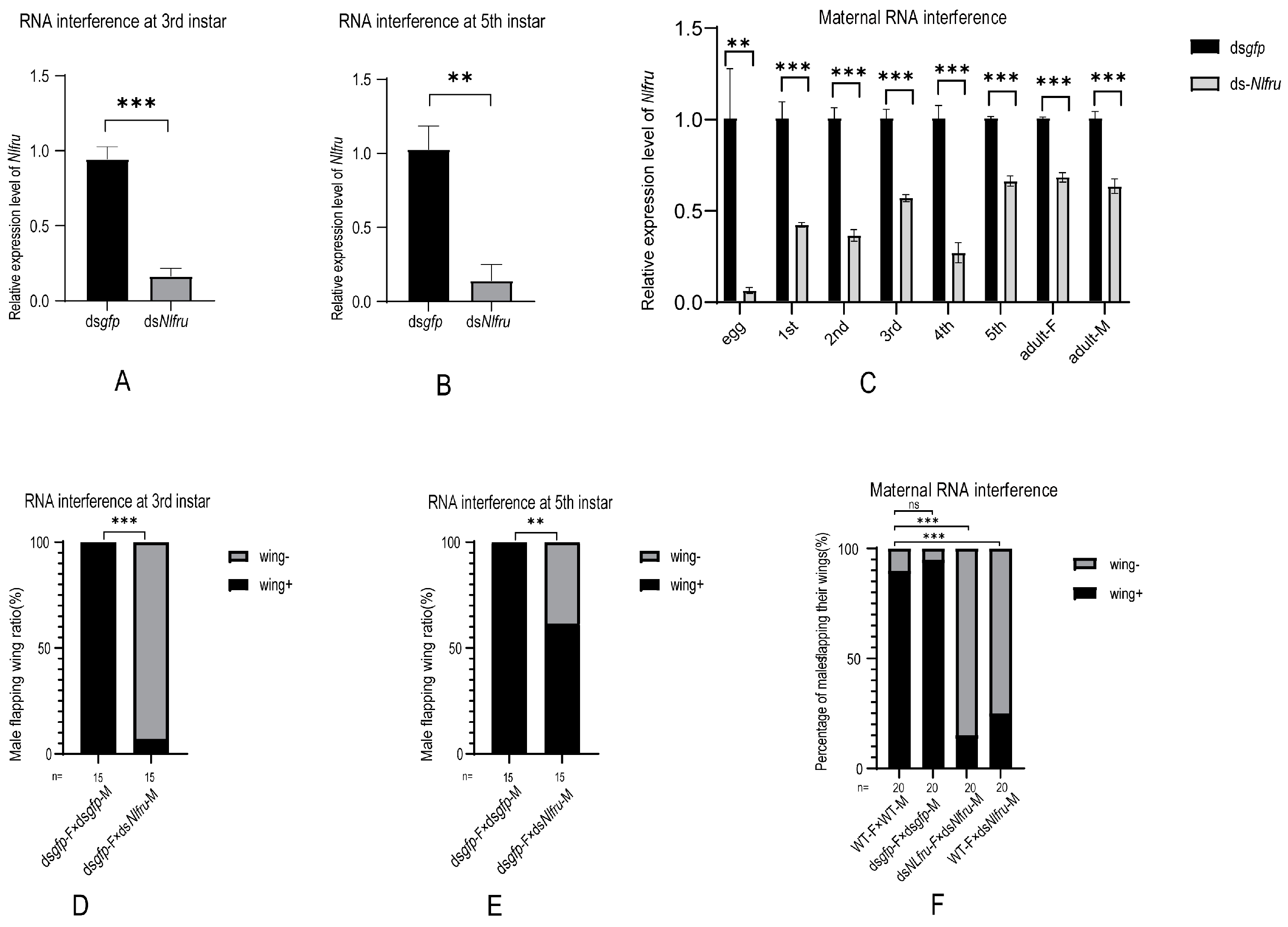
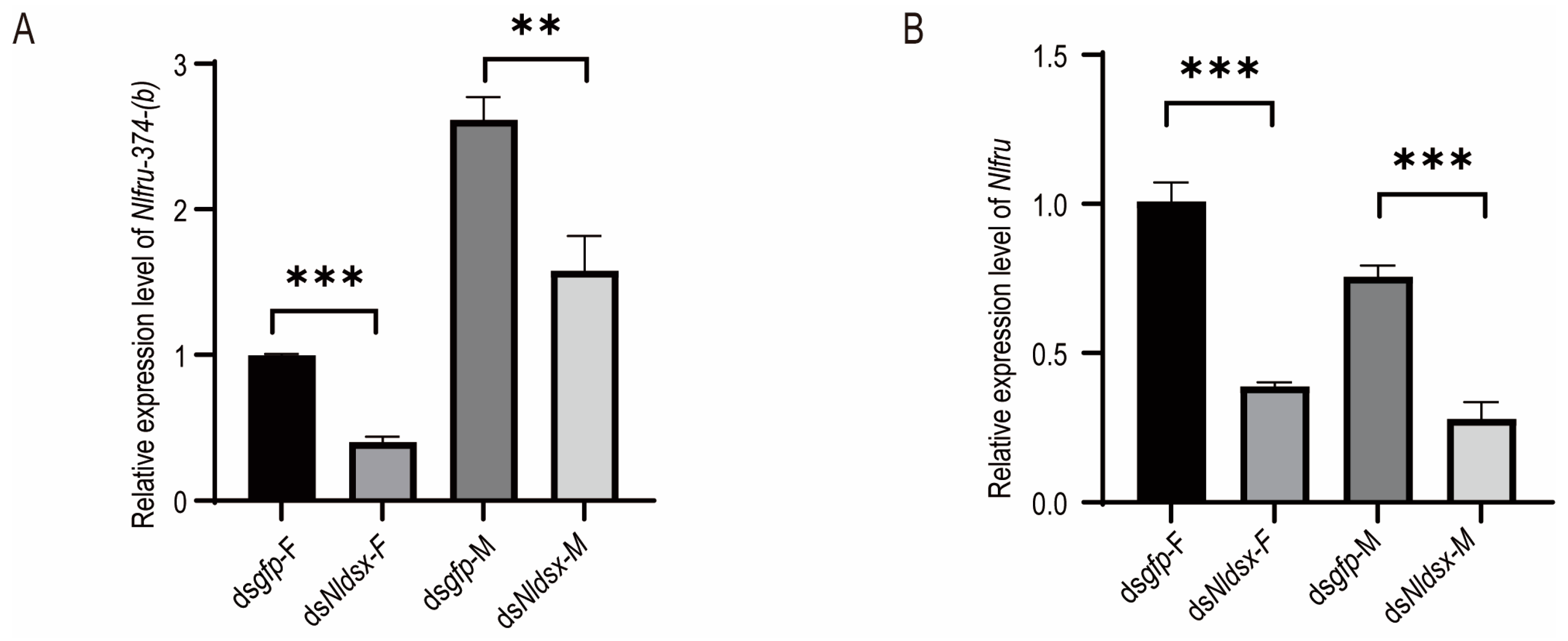
3.2. Nlfru Controls the Wing-Flapping Behavior of Males and the Receptivity of Females
3.3. Nlfru Does Not Affect the Courtship Songs
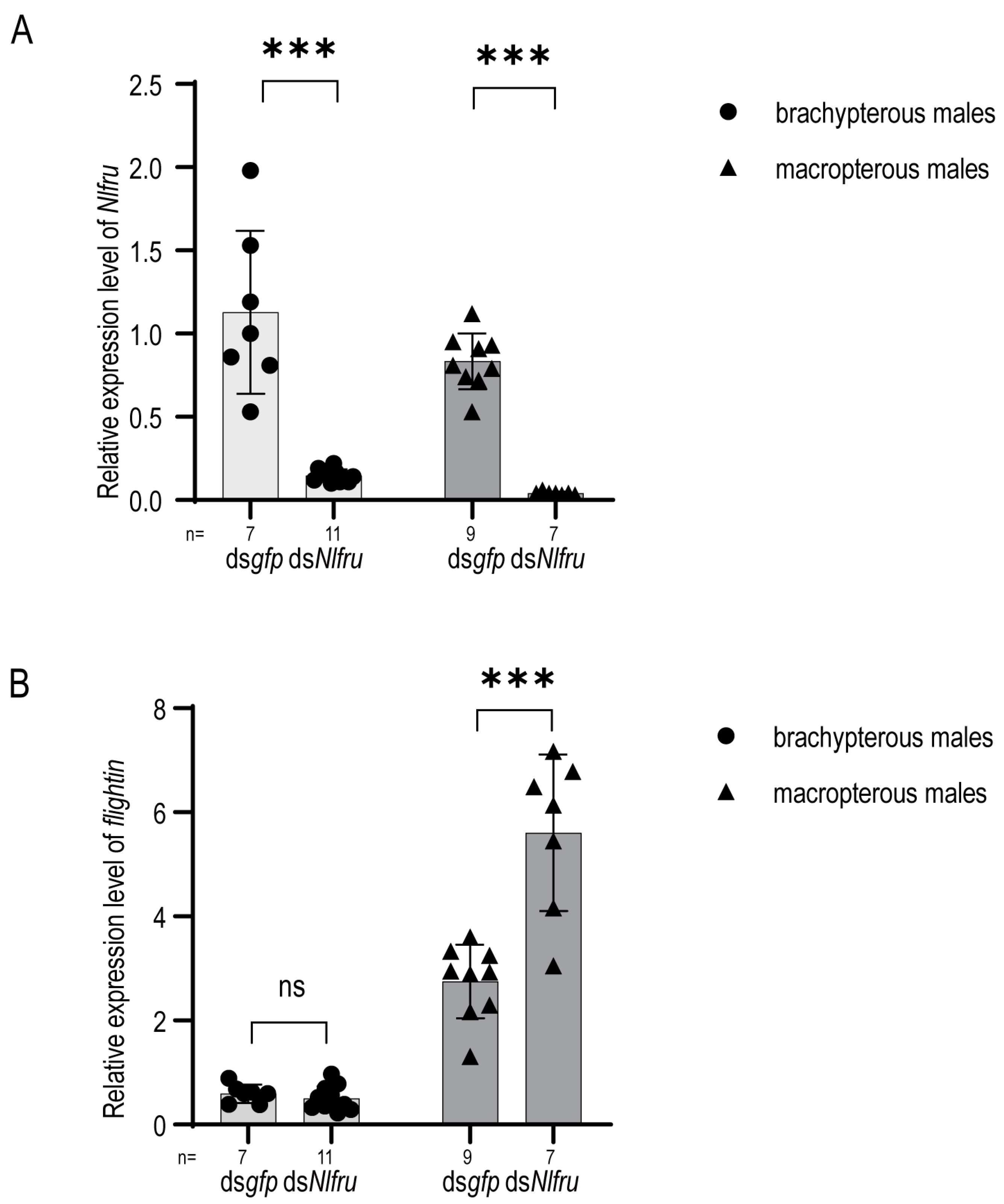
3.4. Nlfru Regulates the Expression of Flightin in Males
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lloyd, J.E. Bioluminescent communication in insects. Annu. Rev. Entomol. 1971, 16, 97–122. [Google Scholar] [CrossRef]
- Mukai, H.; Takanashi, T.; Yamawo, A. Elaborate mating dances: Multimodal courtship displays in jewel bugs. Ecology 2022, 103, e3632. [Google Scholar] [CrossRef]
- Funk, D.H.; Tallamy, D.W. Courtship role reversal and deceptive signals in the long-tailed dance fly, Rhamphomyia longicauda. Anim. Behav. 2000, 59, 411–421. [Google Scholar] [CrossRef]
- Eberhard, W.G. Copulatory courtship and cryptic female choice in insects. Biol. Rev. 1991, 66, 1–31. [Google Scholar] [CrossRef]
- Hall, J.C. The mating of a fly. Science 1994, 264, 1702–1714. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, D.A.; Fields, W.L.; Hall, J.C. Spectral analysis of Drosophila courtship songs: D. melanogaster, D. simulans, and their interspecific hybrid. Behav. Genet. 1988, 18, 675–703. [Google Scholar] [CrossRef] [PubMed]
- Villella, A.; Hall, J.C. Neurogenetics of courtship and mating in Drosophila. Adv. Genet. 2008, 62, 67–184. [Google Scholar] [PubMed]
- Salvemini, M.; Polito, C.; Saccone, G. Fruitless alternative splicing and sex behaviour in insects: An ancient and unforgettable love story? J. Genet. 2010, 89, 287–299. [Google Scholar] [CrossRef]
- Usui-Aoki, K.; Ito, H.; Ui-Tei, K.; Takahashi, K.; Lukacsovich, T.; Awano, W.; Nakata, H.; Piao, Z.F.; Nilsson, E.E.; Tomida, J.-Y. Formation of the male-specific muscle in female Drosophila by ectopic fruitless expression. Nat. Cell Biol. 2000, 2, 500–506. [Google Scholar] [CrossRef] [PubMed]
- Ryner, L.C.; Goodwin, S.F.; Castrillon, D.H.; Anand, A.; Villella, A.; Baker, B.S.; Hall, J.C.; Taylor, B.J.; Wasserman, S.A. Control of male sexual behavior and sexual orientation in Drosophila by the fruitless gene. Cell 1996, 87, 1079–1089. [Google Scholar] [CrossRef]
- Anand, A.; Villella, A.; Ryner, L.C.; Carlo, T.; Goodwin, S.F.; Song, H.-J.; Gailey, D.A.; Morales, A.; Hall, J.C.; Baker, B.S. Molecular genetic dissection of the sex-specific and vital functions of the Drosophila melanogaster sex determination gene fruitless. Genetics 2001, 158, 1569–1595. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, S.F.; Taylor, B.J.; Villella, A.; Foss, M.; Ryner, L.C.; Baker, B.S.; Hall, J.C. Aberrant splicing and altered spatial expression patterns in fruitless mutants of Drosophila melanogaster. Genetics 2000, 154, 725–745. [Google Scholar] [CrossRef]
- Zollman, S.; Godt, D.; Privé, G.G.; Couderc, J.-L.; Laski, F.A. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc. Natl. Acad. Sci. USA 1994, 91, 10717–10721. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Fujitani, K.; Usui, K.; Shimizu-Nishikawa, K.; Tanaka, S.; Yamamoto, D. Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc. Natl. Acad. Sci. USA 1996, 93, 9687–9692. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Foss, M.; Goodwin, S.F.; Carlo, T.; Taylor, B.J.; Hall, J.C. Spatial, temporal, and sexually dimorphic expression patterns of the fruitless gene in the Drosophila central nervous system. J. Neurobiol. 2000, 43, 404–426. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Chen, J.; Pan, Y. From fruitless to sex: On the generation and diversification of an innate behavior. Genes. Brain Behav. 2021, 20, e12772. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Baker, B.S. Genetic identification and separation of innate and experience-dependent courtship behaviors in Drosophila. Cell 2014, 156, 236–248. [Google Scholar] [CrossRef] [PubMed]
- Salvemini, M.; Robertson, M.; Aronson, B.; Atkinson, P.; Polito, L.C.; Saccone, G. Ceratitis capitata transformer-2 gene is required to establish and maintain the autoregulation of Cctra, the master gene for female sex determination. Int. J. Dev. Biol. 2009, 53, 109. [Google Scholar] [CrossRef]
- Basrur, N.S.; De Obaldia, M.E.; Morita, T.; Herre, M.; von Heynitz, R.K.; Tsitohay, Y.N.; Vosshall, L.B. Fruitless mutant male mosquitoes gain attraction to human odor. Elife 2020, 9, e63982. [Google Scholar] [CrossRef] [PubMed]
- Ohbayashi, F. Structural and Functional Analyses on the Bombyx mori Genes Homologous to Drosophila Doublesex and Fruitless; University of Tokyo: Tokyo, Japan, 2001; p. 71. [Google Scholar]
- Ueno, M.; Nakata, M.; Kaneko, Y.; Iwami, M.; Takayanagi-Kiya, S.; Kiya, T. Fruitless is sex-differentially spliced and is important for the courtship behavior and development of silkmoth Bombyx mori. Insect Biochem. Mol. Biol. 2023, 159, 103989. [Google Scholar] [CrossRef]
- Saccone, G. A history of the genetic and molecular identification of genes and their functions controlling insect sex determination. Insect Biochem. Mol. Biol. 2022, 151, 103873. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.-I.; Hachiya, T.; Koganezawa, M.; Tazawa, T.; Yamamoto, D. Fruitless and doublesex coordinate to generate male-specific neurons that can initiate courtship. Neuron 2008, 59, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Dauwalder, B. The Roles of fruitless and doublesex in the Control of Male Courtship. Int. Rev. Neurobiol. 2011, 99, 87–105. [Google Scholar] [CrossRef] [PubMed]
- Rideout, E.J.; Billeter, J.C.; Goodwin, S.F. The sex-determination genes and specify a neural substrate required for courtship song. Curr. Biol. 2007, 17, 1473–1478. [Google Scholar] [CrossRef] [PubMed]
- Siwicki, K.K.; Kravitz, E.A. Fruitless, doublesex and the genetics of social behavior in Drosophila melanogaster. Curr. Opin. Neurobiol. 2009, 19, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Butlin, R. The variability of mating signals and preferences in the brown planthopper, Nilaparvata lugens (Homoptera: Delphacidae). J. Insect Behav. 1993, 6, 125–140. [Google Scholar] [CrossRef]
- Zhuo, J.-C.; Hu, Q.-L.; Zhang, H.-H.; Zhang, M.-Q.; Jo, S.B.; Zhang, C.-X. Identification and functional analysis of the doublesex gene in the sexual development of a hemimetabolous insect, the brown planthopper. Insect Biochem. Mol. Biol. 2018, 102, 31–42. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.-J.; Zhang, C.-X.; Xue, J.; Ye, Y.-X.; Jiang, Y.-Q.; Zhuo, J.-C.; Huang, H.-J.; Cheng, R.-L. Efficient RNAi of Rice Planthoppers Using Microinjection. Protoc. Exch. 2015, 12, 1–8. [Google Scholar]
- Zhuo, J.-C.; Xue, J.; Lu, J.-B.; Huang, H.-J.; Xu, H.-J.; Zhang, C.-X. Effect of RNAi-mediated knockdown of NlTOR gene on fertility of male Nilaparvata lugens. J. Insect Physiol. 2017, 98, 149–159. [Google Scholar] [CrossRef]
- Pan, W.; Kong, X.; Xu, J.; Pan, W. Measurement and analysis system of vibration for the detection of insect acoustic signals. In Proceedings of the 2016 Asia-Pacific International Symposium on Electromagnetic Compatibility (APEMC), Shenzhen, China, 17–21 May 2016; pp. 1090–1092. [Google Scholar]
- Xue, J.; Zhang, X.-Q.; Xu, H.-J.; Fan, H.-W.; Huang, H.-J.; Ma, X.-F.; Wang, C.-Y.; Chen, J.-G.; Cheng, J.-A.; Zhang, C.-X. Molecular characterization of the flightin gene in the wing-dimorphic planthopper, Nilaparvata lugens, and its evolution in Pancrustacea. Insect Biochem. Mol. Biol. 2013, 43, 433–443. [Google Scholar] [CrossRef] [PubMed]
- Ustinova, J.; Mayer, F. Alternative starts of transcription, several paralogues, and almost-fixed interspecific differences of the gene in a hemimetabolous insect. J. Mol. Evol. 2006, 63, 788–800. [Google Scholar] [CrossRef]
- Liu, Y.T.; Xie, J.X.; Wang, W.L.; Lei, Y.Y.; Zhou, X.G.; Zhang, Y.J.; Xie, W. Splicing and Expression Regulation of fruitless Gene in Bemisia tabaci (Hemiptera: Aleyrodidae). Horticulturae 2023, 9, 962. [Google Scholar] [CrossRef]
- Just, J.; Laslo, M.; Lee, Y.J.; Yarnell, M.; Zhang, Z.; Angelini, D.R. Distinct developmental mechanisms influence sexual dimorphisms in the milkweed bug Oncopeltus fasciatus. Proc. R. Soc. B 2023, 290, 20222083. [Google Scholar] [CrossRef] [PubMed]
- Greenspan, R.J.; Ferveur, J.-F. Courtship in drosophila. Annu. Rev. Genet. 2000, 34, 205–232. [Google Scholar] [CrossRef] [PubMed]
- Shirangi, T.R.; McKeown, M. Sex in flies: What ‘body–mind’ dichotomy? Dev. Biol. 2007, 306, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Heinrichs, V.; Ryner, L.C.; Baker, B.S. Regulation of sex-specific selection of fruitless 5′ splice sites by transformer and transformer-2. Mol. Cell. Biol. 1998, 18, 450–458. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Whitworth, C.; Pozmanter, C.; Neville, M.C.; Van Doren, M. Doublesex regulates fruitless expression to promote sexual dimorphism of the gonad stem cell niche. PLoS Genet. 2021, 17, e1009468. [Google Scholar] [CrossRef]
- Claridge, M.; Hollander, J.D.; Morgan, J. Variation in courtship signals and hybridization between geographically definable populations of the rice brown planthopper, Nilaparvata lugens (Stål). Biol. J. Linn. Soc. 1985, 24, 35–49. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, B.; Mao, Z.; Chen, Y.; Ying, J.; Wang, H.; Sun, Z.; Li, J.; Zhang, C.; Zhuo, J. Identification and Functional Analysis of the fruitless Gene in a Hemimetabolous Insect, Nilaparvata lugens. Insects 2024, 15, 262. https://doi.org/10.3390/insects15040262
Wang B, Mao Z, Chen Y, Ying J, Wang H, Sun Z, Li J, Zhang C, Zhuo J. Identification and Functional Analysis of the fruitless Gene in a Hemimetabolous Insect, Nilaparvata lugens. Insects. 2024; 15(4):262. https://doi.org/10.3390/insects15040262
Chicago/Turabian StyleWang, Biyun, Zeping Mao, Youyuan Chen, Jinjun Ying, Haiqiang Wang, Zongtao Sun, Junmin Li, Chuanxi Zhang, and Jichong Zhuo. 2024. "Identification and Functional Analysis of the fruitless Gene in a Hemimetabolous Insect, Nilaparvata lugens" Insects 15, no. 4: 262. https://doi.org/10.3390/insects15040262




