Wing Plasticity Is Associated with Growth and Energy Metabolism in Two Color Morphs of the Pea Aphid
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plants and Insects
2.2. Aphid Wing Induction
2.3. Aphid Growth, Fecundity, and Starvation Tolerance
2.4. Metabolite Extraction and Analysis
2.5. Gene Expression Analysis
2.6. Statistical Analysis
3. Results
3.1. Wing Production, Growth, Reproduction, and Starvation Tolerance
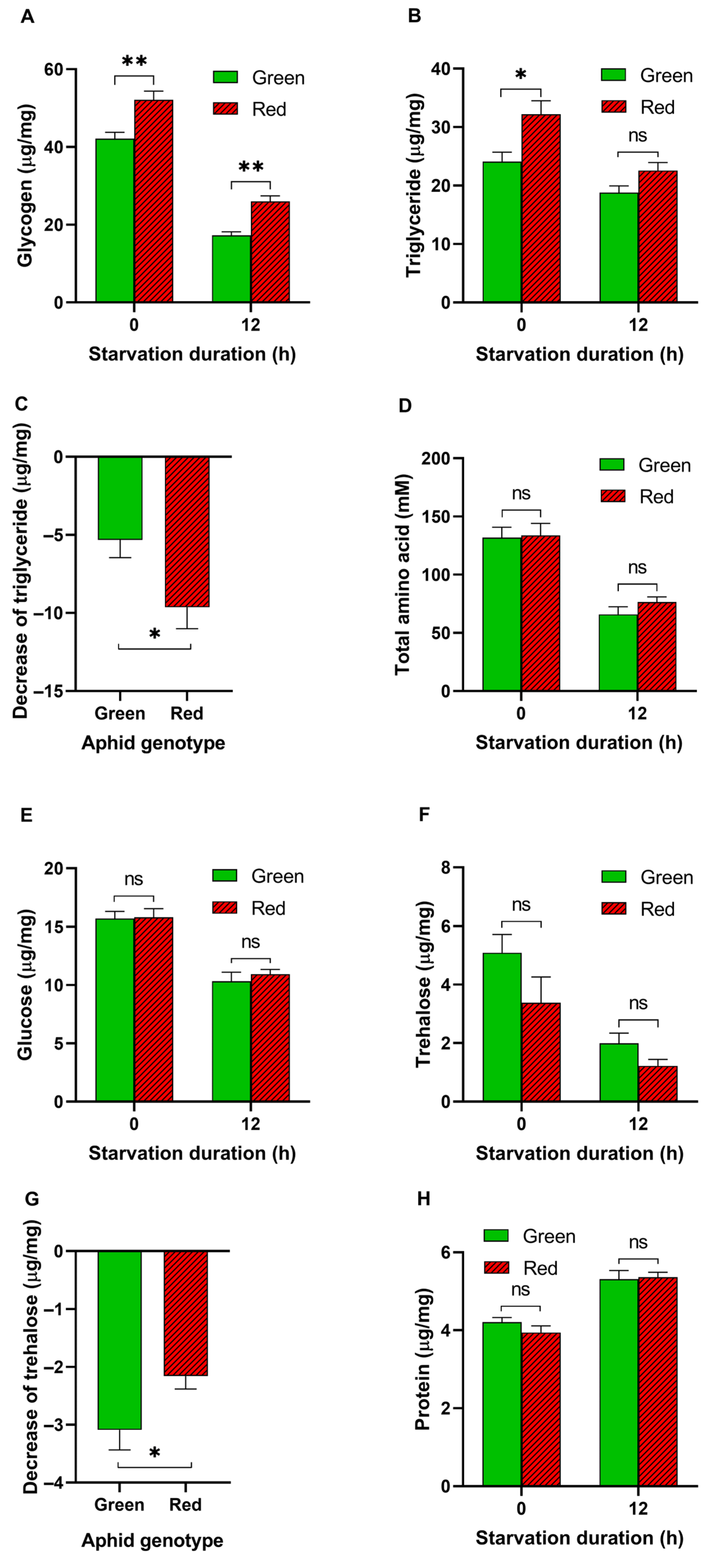
3.2. Energy Metabolites in Aphids before and after Starvation
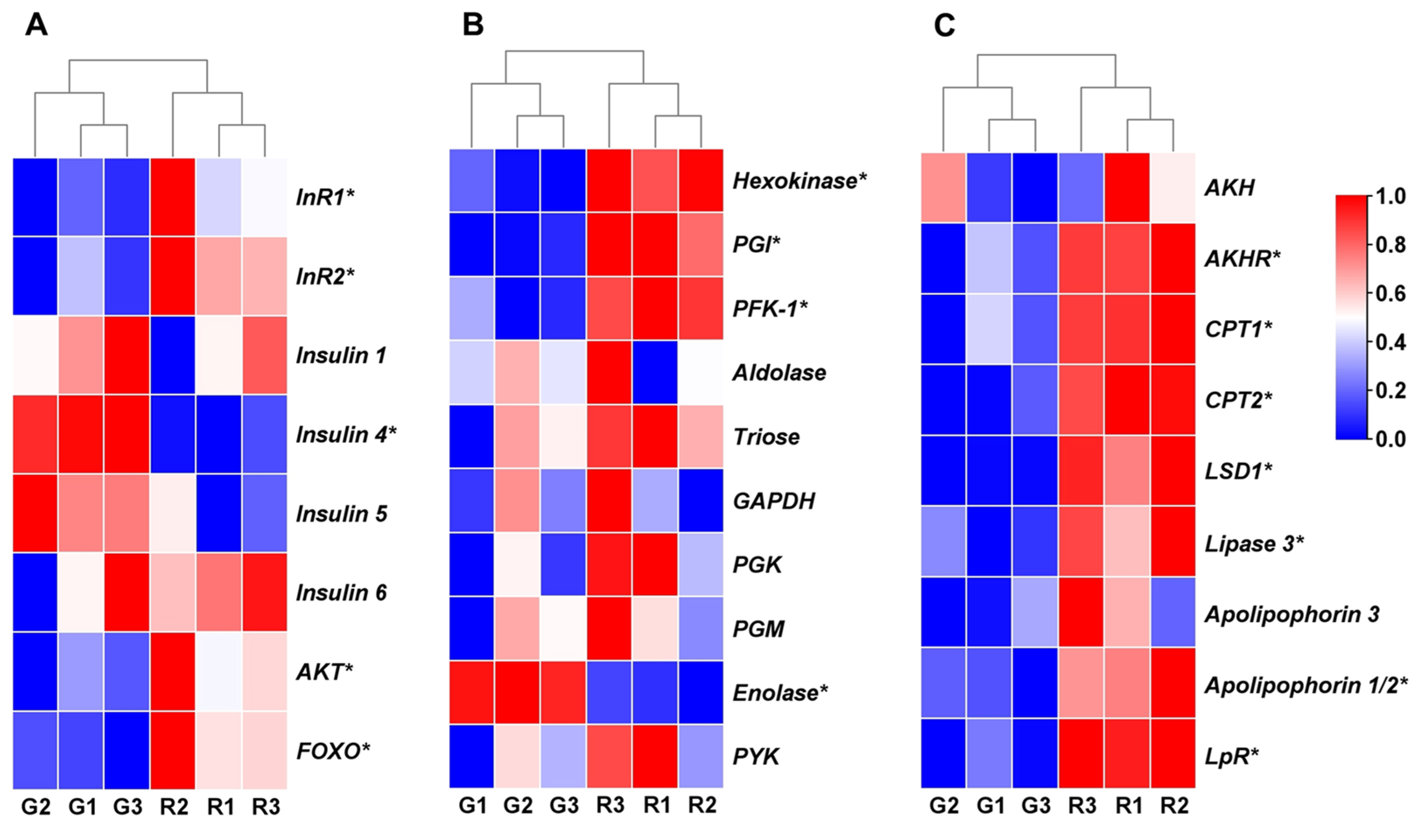
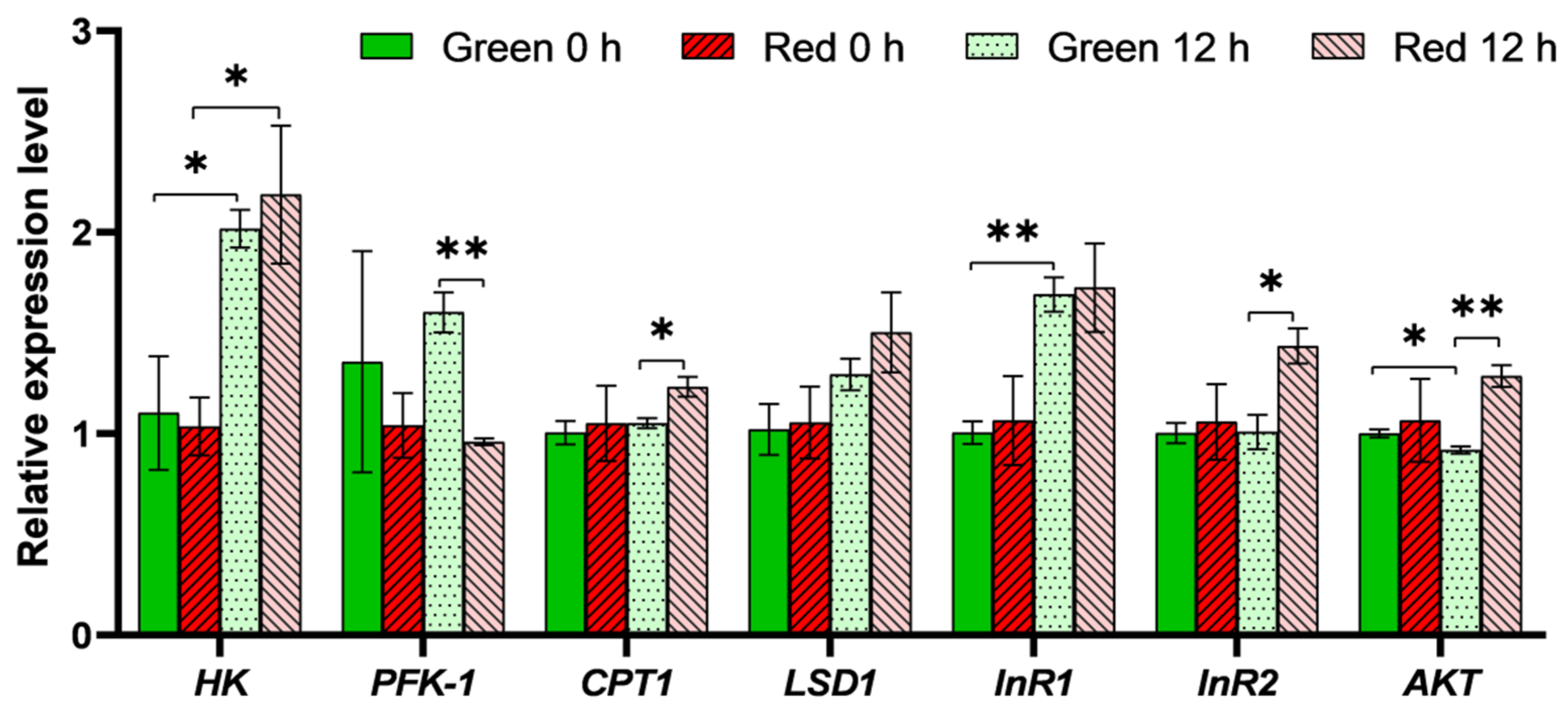
3.3. Expression Analysis of Genes Involved in Insulin Pathway and Energy Metabolism
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hayes, A.M.; Lavine, M.D.; Gotoh, H.; Lin, X.; Lavine, L.C. Mechanisms regulating phenotypic plasticity in wing polyphenic insects. Adv. Insect Physiol. 2019, 56, 43–72. [Google Scholar]
- Weisser, W.W.; Braendle, C. Body colour and genetic variation in winged morph production in the pea aphid. Entomol. Exp. Appl. 2001, 99, 217–223. [Google Scholar] [CrossRef]
- Zera, A.J.; Denno, R.F. Physiology and ecology of dispersal polymorphism in insects. Annu. Rev. Entomol. 1997, 42, 207–230. [Google Scholar] [CrossRef]
- Zhang, C.X.; Brisson, J.A.; Xu, H.J. Molecular mechanisms of wing polymorphism in insects. Annu. Rev. Entomol. 2019, 64, 297–314. [Google Scholar] [CrossRef]
- Vellichirammal, N.N.; Gupta, P.; Hall, T.A.; Brisson, J.A. Ecdysone signaling underlies the pea aphid transgenerational wing polyphenism. Proc. Natl. Acad. Sci. USA 2017, 114, 1419–1423. [Google Scholar] [CrossRef]
- Yuan, Y.; Wang, Y.; Ye, W.; Yuan, E.; Di, J.; Chen, X.; Xing, Y.; Sun, Y.; Ge, F. Functional evaluation of the insulin/insulin-like growth factor signaling pathway in determination of wing polyphenism in pea aphid. Insect Sci. 2023, 30, 816–828. [Google Scholar] [CrossRef]
- Shang, F.; Niu, J.; Ding, B.Y.; Zhang, W.; Wei, D.D.; Wei, D.; Jiang, H.B.; Wang, J.J. The miR-9b microRNA mediates dimorphism and development of wing in aphids. Proc. Natl. Acad. Sci. USA 2020, 117, 8404–8409. [Google Scholar] [CrossRef]
- Yuan, E.; Guo, H.; Chen, W.; Du, B.; Mi, Y.; Qi, Z.; Yuan, Y.; Zhu-Salzman, K.; Ge, F.; Sun, Y. A novel gene REPTOR2 activates the autophagic degradation of wing disc in pea aphid. Elife 2023, 12, e83023. [Google Scholar] [CrossRef]
- Parker, B.J.; Brisson, J.A. A laterally transferred viral gene modifies aphid wing plasticity. Curr. Biol. 2019, 29, 2098–2103. [Google Scholar] [CrossRef]
- Ahsaei, S.M.; Tabadkani, S.M.; Hosseininaveh, V.; Allahyari, H.; Bigham, M. Differential accumulation of energy by the color morphs of the pea aphid Acyrthosiphon pisum (Hemiptera: Aphididae) mirrors their ecological adaptations. Eur. J. Entomol. 2013, 110, 241–245. [Google Scholar] [CrossRef]
- Markkula, M. Studies on the pea aphid, Acyrthosiphon pisum Harris (Hom., Aphididae) with special reference to the difference in biology of the green and red forms. Ann. Agric. Fenn. Suppl. 1963, 2, 1–30. [Google Scholar]
- Sutherland, O.R.W. The role of crowding in the production of winged forms by two strains of the pea aphid, Acyrthosiphon pisum. J. Insect Physiol. 1969, 15, 1385–1410. [Google Scholar] [CrossRef]
- Cao, H.-H.; Zhang, Z.-F.; Wang, X.-F.; Liu, T.-X. Nutrition versus defense: Why Myzus persicae (green peach aphid) prefers and performs better on young leaves of cabbage. PLoS ONE 2018, 13, e0196219. [Google Scholar] [CrossRef]
- Ahmed, M.A.; Ban, N.; Hussain, S.; Batool, R.; Zhang, Y.-J.; Liu, T.-X.; Cao, H.-H. Preference and performance of the green peach aphid, Myzus persicae on three Brassicaceae vegetable plants and its association with amino acids and glucosinolates. PLoS ONE 2022, 17, e0269736. [Google Scholar] [CrossRef]
- Seifter, S.; Dayton, S.; Novic, B.; Muntwyler, E. The estimation of glycogen with the anthrone reagent. Arch. Biochem. 1950, 25, 191–200. [Google Scholar]
- Wu, J.; Lan, H.; Zhang, Z.-F.; Cao, H.-H.; Liu, T.-X. Performance and transcriptional response of the green peach aphid Myzus persicae to the restriction of dietary amino acids. Front. Physiol. 2020, 11, 487. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “One for All, All for One” Bioinformatics Platform for Biological Bigdata Mining. Mol. Plant. 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Yang, C.; Pan, H.; Liu, Y.; Zhou, X. Selection of reference genes for expression analysis using quantitative real-time PCR in the pea aphid, Acyrthosiphon pisum (Harris) (Hemiptera, Aphidiae). PLoS ONE 2014, 9, e110454. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Kammer, A.E.; Heinrich, B. Insect flight metabolism. Adv. Insect Physiol. 1978, 13, 133–228. [Google Scholar]
- Zhang, D.W.; Xiao, Z.-J.; Zeng, B.P.; Li, K.; Tang, Y.L. Insect behavior and physiological adaptation mechanisms under starvation stress. Front. Physiol. 2019, 10, 163. [Google Scholar] [CrossRef] [PubMed]
- Artacho, P.; Figueroa, C.C.; Cortes, P.A.; Simon, J.C.; Nespolo, R.F. Short-term consequences of reproductive mode variation on the genetic architecture of energy metabolism and life-history traits in the pea aphid. J. Insect Physiol. 2011, 57, 986–994. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Brown, M.R. Signaling and function of insulin-like peptides in insects. Annu. Rev. Entomol. 2006, 51, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.J.; Xue, J.; Lu, B.; Zhang, X.C.; Zhuo, J.C.; He, S.F.; Ma, X.F.; Jiang, Y.Q.; Fan, H.W.; Xu, J.Y.; et al. Two insulin receptors determine alternative wing morphs in planthoppers. Nature 2015, 519, 464–467. [Google Scholar] [CrossRef]
- Vellichirammal, N.N.; Madayiputhiya, N.; Brisson, J.A. The genome wide transcriptional response underlying the pea aphid wing polyphenism. Mol. Ecol. 2016, 25, 4146–4160. [Google Scholar] [CrossRef]

| Primer Name | Primer Sequences (5′ to 3′) | GenBank Accession Number |
|---|---|---|
| NADH-Forward | CGAGGAGAACATGCTCTTAGAC GATAGCTTGGGCTGGACATATAG | NM_001162323.2 |
| NADH-Reverse | ||
| HK-Forward | TGTACATGGGCGAGATCGTG | XM_003242192.4 |
| HK-Reverse | GGATACGAATGACGTGCCGA | |
| PFK-1-Forward | ACCGTGATCCGTGACCTCTA | XM_029487259.1 |
| PFK-1-Reverse | TCATGTTCAGCCCTGGCAAT | |
| CPT1-Forward | CTGAGCATTCGTGGGCTGAT | XM_029488698.1 |
| CPT1-Reverse | GGAGGTTCCAATTCAGGAGCA | |
| LSD1-Forward | GCCGACAAACCGAGTACAGA | XM_003240065.4 |
| LSD1-Reverse | CACGAAGCACTGGACGGTAT | |
| InR1-Forward | ATCTGTCCACCGGAATGTGG | MK510961.1 |
| InR1-Reverse | TCGGCTGGACATTCCTTCAC | |
| Insulin R2-Forward | CGAGCCTCCTGAAATCACGTA | XM_008184754.3 |
| Insulin R2-Reverse | TGGCATTTTTGTCGTCCTTGTG | |
| AKT-Forward | TGGCTGTTCAAACGAGGAGA | XM_008185246.3 |
| AKT-Reverse | CGGTCATTGGACCTTCTGGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, H.; Wang, X.; Wang, J.; Lu, Z.; Liu, T. Wing Plasticity Is Associated with Growth and Energy Metabolism in Two Color Morphs of the Pea Aphid. Insects 2024, 15, 279. https://doi.org/10.3390/insects15040279
Cao H, Wang X, Wang J, Lu Z, Liu T. Wing Plasticity Is Associated with Growth and Energy Metabolism in Two Color Morphs of the Pea Aphid. Insects. 2024; 15(4):279. https://doi.org/10.3390/insects15040279
Chicago/Turabian StyleCao, Hehe, Xi Wang, Jiawei Wang, Zhaozhi Lu, and Tongxian Liu. 2024. "Wing Plasticity Is Associated with Growth and Energy Metabolism in Two Color Morphs of the Pea Aphid" Insects 15, no. 4: 279. https://doi.org/10.3390/insects15040279
APA StyleCao, H., Wang, X., Wang, J., Lu, Z., & Liu, T. (2024). Wing Plasticity Is Associated with Growth and Energy Metabolism in Two Color Morphs of the Pea Aphid. Insects, 15(4), 279. https://doi.org/10.3390/insects15040279






