Detection of Ochyromera ligustri (Coleoptera: Curculionidae: Curculioninae) in Ligustrum spp. (Oleaceae) Using Newly Developed PCR Primers
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and Processing
2.2. Primer Design
2.3. DNA Extraction and Polymerase Chain Reaction Amplification
2.4. DNA Sequence Assembly
2.5. Primer Specificity
2.6. Primer Sensitivity
2.7. Application to Fruit Samples of Ligustrum spp.
2.8. Distribution Map
3. Results
3.1. Species Primer Design and Optimal PCR Conditions
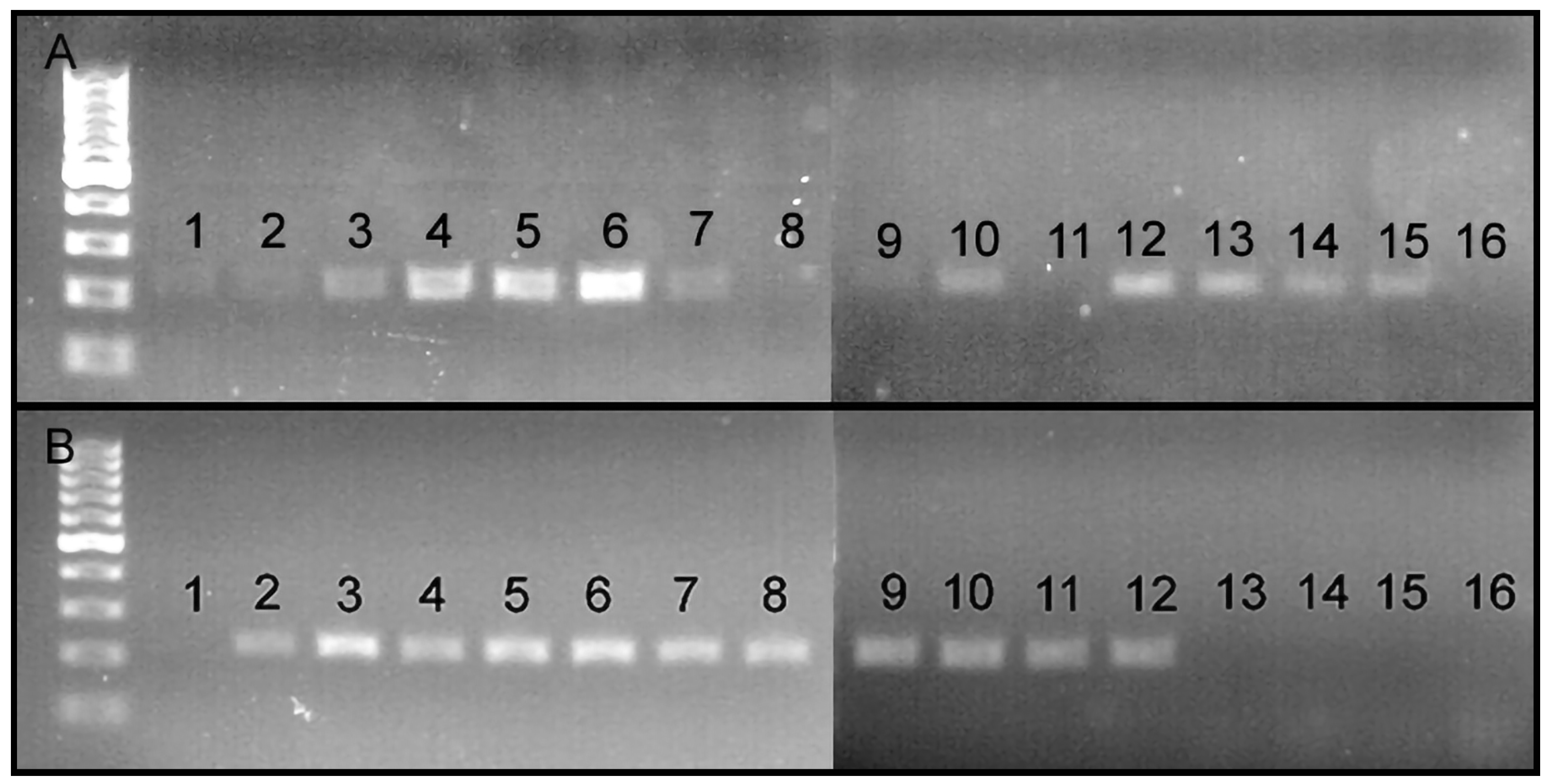
3.2. Primer Specificity
3.3. Primer Sensitivity
3.4. Application to the Fruit Samples of Ligustrum spp.
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dong, W.; Li, E.; Liu, Y.; Xu, C.; Wang, Y.; Liu, K.; Cui, X.; Sun, J.; Suo, Z.; Zhang, Z.; et al. Phylogenomic approaches untangle early divergences and complex diversifications of the olive plant family. BMC Biol. 2022, 20, 92. [Google Scholar] [CrossRef]
- Li, J.; Alexander, J.H.; Zhang, D. Paraphyletic Syringa (Oleaceae): Evidence from sequences of nuclear ribosomal DNA ITS and ETS regions. Syst. Bot. 2002, 27, 592–597. [Google Scholar] [CrossRef]
- Maddox, V.; Byrd, J.; Serviss, B. Identification and Control of Invasive Privets (Ligustrum spp.) in the Middle Southern United States. Invasive Plant Sci. Manag. 2010, 3, 482–488. [Google Scholar] [CrossRef]
- Dirr, M.A. Manual of Woody Landscape Plants: Their Identification, Ornamental Characteristics, Culture, Propagation, and Uses, 5th ed.; Stipes Publishing, LLC.: Champaign, IL, USA, 1998; pp. 1–563. [Google Scholar]
- Wilcox, J.; Beck, C.W. Effects of Ligustrum sinense Lour. (Chinese privet) on abundance and diversity of songbirds and native plants in a southeastern nature preserve. Southeast. Nat. 2007, 6, 535–550. [Google Scholar] [CrossRef]
- Greene, B.T.; Blossey, B. Lost in the weeds: Ligustrum sinense reduces native plant growth and survival. Biol. Invasions 2012, 14, 139–150. [Google Scholar] [CrossRef]
- Cash, J.S.; Anderson, C.J.; Gulsby, W.D. The ecological effects of Chinese privet (Ligustrum sinense) invasion: A synthesis. Invasive Plant Sci. Manag. 2020, 13, 3–13. [Google Scholar] [CrossRef]
- Foard, M.; Burnette, D.J.; Burge, D.R.; Marsico, T.D. Influence of river channelization and the invasive shrub, Ligustrum sinense, on oak (Quercus spp.) growth rates in bottomland hardwood forests. Appl. Veg. Sci. 2016, 19, 401–412. [Google Scholar] [CrossRef]
- Mowatt, J. Control of large-leaved privet (Ligustrum lucidum) and small-leaved privet (L. sinense) in urban bushland. In Proceedings of the Australian Weeds Conference, Gold Coast, QLD, Australia, 13 September 1981. [Google Scholar]
- Ahuja, G. Chemical Control of Chinese privet (Ligusrtum [sic] sinense). Ph.D. Dissertation, University of Georgia, Athens, GA, USA, December 2003. [Google Scholar]
- Faulkner, J.L.; Clebsch, E.E.; Sanders, W.L. Use of prescribed burning for managing natural and historic resources in Chickamauga and Chattanooga National Military Park, USA. Environ. Manag. 1989, 13, 603–612. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Hanula, J.L.; Horn, S.; Braman, S.K.; Sun, J.H. Biology of Leptoypha hospita (Hemiptera: Tingidae), a potential biological control agent of Chinese Privet. Ann. Entomol. Soc. Am. 2011, 104, 1327–1333. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Hanula, J.L.; Horn, S.; Jones, C.; Braman, S.K.; Sun, J.H. Fundamental host range of Leptoypha hospital (Hemiptera: Tingidae), a potential biological control agent of Chinese privet. Environ. Entomol. 2016, 45, 897–908. [Google Scholar] [CrossRef]
- Shaw, R.H.; Cock, M.J.; Evans, H.C. The natural enemies of privets (Ligustrum: Oleaceae): A literature review, with particular reference to biological control. CABI Rev. 2018, 13, 1–24. [Google Scholar] [CrossRef]
- Warner, R.E. The Genus Ochyromera New to the Western Hemisphere, with a New Species and Additions to the Junk-Schenkling Coleopterorum Catalogus. (Curculionidae: Prionomerinae, Endaeini). Coleopt. Bull. 1961, 15, 121–124. Available online: http://www.jstor.org/stable/3998995 (accessed on 27 December 2022).
- Wray, D.L. Biology and life history of the ligustrum weevil (Curculionidae). Coleopt. Bull. 1961, 15, 119–120. [Google Scholar]
- Kojima, H.; Morimoto, K.; Horikawa, M. Two new species of the genus Ochyromera (Coleoptera: Curculionidae) from Japan. ESAKIA 1998, 38, 113–122. [Google Scholar] [CrossRef]
- O‘Brien, C.W.; Wibmer, G.J. Annotated checklist of the weevils (Curculionidae sensu lato) of North America, Central America, and the West Indies (Coleoptera: Curculionoidea). Mem. Am. Entomol. Soc. 1982, 34, 1–382. [Google Scholar]
- Johnson, W.T.; Lyon, H.H. Insects That Feed on Trees and Shrubs, 2nd ed.; Cornell University Press: Ithaca, NY, USA, 1988; pp. 1–556. [Google Scholar]
- Cuda, J.P.; Zeller, M.C. First Record of Ochyromera ligustri (Coleoptera: Curculionidae) from Chinese Privet in Florida. Fla. Entomol. 1998, 81, 582–584. [Google Scholar] [CrossRef]
- Parra, J.R.P.; Coelho, A., Jr. Insect Rearing Techniques for Biological Control Programs, a Component of Sustainable Agriculture in Brazil. Insects 2022, 13, 105. [Google Scholar] [CrossRef] [PubMed]
- Van Klinken, R.D. Host specificity testing: Why do we do it and how we can do it better. In Host Specificity Testing of Exotic Arthropod Biological Control Agents: The Biological Basis for Improvement in Safety. Proceedings of the International Symposium on Biological Control of Weeds, Bozeman, MT, USA, 4–14 July 1999; Van Driesche, R.G., Heard, T.A., McClay, A.S., Reardon, R.C., Eds.; USDA Forest Service Forest Health Technology Enterprise Team: Morgantown, WV, USA, 1999. [Google Scholar]
- Atkins, S.D.; Clark, I.M.; Pande, S.; Hirsch, P.R.; Kerry, B.R. The use of real-time PCR and species-specific primers for the identification and monitoring of Paecilomyces lilacinus. FEMS Microbiol. Ecol. 2005, 51, 257–264. [Google Scholar] [CrossRef]
- Franck, P.; Maalouly-Matar, M.; Olivares, J. Molecular Tools for the Detection and the Identification of Hymenoptera Parasitoids in Tortricid Fruit Pests. Int. J. Mol. Sci. 2017, 18, 2031. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.N.; Santander, R.D.; Talamas, E.J.; Jentsch, P.J.; Bon, M.-C.; Aćimović, S.G. Molecular Identification of Trissolcus japonicus, Parasitoid of the Brown Marmorated Stink Bug, by Species-Specific PCR. Insects 2021, 12, 467. [Google Scholar] [CrossRef]
- Gariepy, T.D.; Haye, T.; Zhang, J. A molecular diagnostic tool for the preliminary assessment of host–parasitoid associations in biological control programmes for a new invasive pest. Mol. Ecol. 2014, 23, 3912–3924. [Google Scholar] [CrossRef] [PubMed]
- Hrcek, J.; Miller, S.E.; Quicke, D.L.J.; Smith, M.A. Molecular detection of trophic links in a complex insect host–parasitoid food web. Mol. Ecol. Resour. 2011, 11, 786–794. [Google Scholar] [CrossRef]
- Jenkins, C.; Chapman, T.A.; Micallef, J.L.; Reynolds, O.L. Molecular Techniques for the Detection and Differentiation of Host and Parasitoid Species and the Implications for Fruit Fly Management. Insects 2012, 3, 763–788. [Google Scholar] [CrossRef]
- Pook, V.G.; Athey, K.J.; Chapman, E.G.; Clutts-Stoelb, S.A.; Sharkey, M.J. New PCR primers enhance investigation of host-parasitoid food webs. Entomol. Exp. Appl. 2017, 162, 309–314. [Google Scholar] [CrossRef]
- Lantero, E.; Matallanas, B.; Pascual, S.; Callejas, C. PCR Species-Specific Primers for Molecular Gut Content Analysis to Determine the Contribution of Generalist Predators to the Biological Control of the Vector of Xylella fastidiosa. Sustainability 2018, 10, 2207. [Google Scholar] [CrossRef]
- Avanesyan, A.; Lamp, W.O. Use of Molecular Gut Content Analysis to Decipher the Range of Food Plants of the Invasive Spotted Lanternfly, Lycorma delicatula. Insects 2020, 11, 215. [Google Scholar] [CrossRef] [PubMed]
- Day, W.H. Estimating Mortality Caused by Parasites and Diseases of Insects: Comparisons of the Dissection and Rearing Methods. Environ. Entomol. 1994, 23, 543–550. [Google Scholar] [CrossRef]
- Kelley, J.T.; Danforth, B.; Day, W.; Hoffmann, M.P. Determining Parasitoid Species Composition in a Host Population: A Molecular Approach. Ann. Entomol. Soc. Am. 2000, 93, 640–647. [Google Scholar] [CrossRef]
- Agustí, N.; Bourguet, D.; Spataro, T.; Delos, M.; Eychenne, N.; Folcher, L.; Arditi, R. Detection, identification and geographical distribution of European corn borer larval parasitoids using molecular markers. Mol. Ecol. 2005, 14, 3267–3274. [Google Scholar] [CrossRef] [PubMed]
- Morgulis, A.; Coulouris, G.; Raytselis, Y.; Madden, T.L.; Agarwala, R.; Schäffer, A.A. Database indexing for production MegaBLAST searches. Bioinformatics 2008, 24, 1757–1764. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Eurofins Oligo Analysis Tool. Available online: https://eurofinsgenomics.com/en/resources/tools/oligo-analysis/ (accessed on 19 October 2022).
- Hebert, P.D.; Penton, E.H.; Burns, J.M.; Janzen, D.H.; Hallwachs, W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl. Acad. Sci. USA 2004, 101, 14812–14817. [Google Scholar] [CrossRef]
- Geneious Prime 2022.0.1. Available online: https://www.geneious.com (accessed on 2 September 2022).
- ArcGIS Pro, version 3.0.0; Software. Desktop; Esri Inc.: Redlands, CA, USA, 2022.
- Champlot, S.; Berthelot, C.; Pruvost, M.; Bennett, E.A.; Grange, T.; Geigl, E.M. An efficient multistrategy DNA decontamination procedure of PCR reagents for hypersensitive PCR applications. PLoS ONE 2010, 5, e13042. [Google Scholar] [CrossRef]
- Rychlik, W.; Spencer, W.J.; Rhoads, R.E. Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res. 1990, 18, 6409–6412. [Google Scholar] [CrossRef] [PubMed]
- Bell, D.A.; DeMarini, D.M. Excessive cycling converts PCR products to randomlength higher molecular weight fragments. Nucleic Acids Res. 1991, 19, 5079. [Google Scholar] [CrossRef] [PubMed]
- Bulman, L.S.; Kimberley, M.O.; Gadgil, P.D. Estimation of the efficiency of pest detection surveys. N. Z. J. For. Sci. 1999, 29, 102–115. [Google Scholar]
- Bakuza, J.S.; Denwood, M.J.; Nkwengulila, G.; Mable, B.K. Estimating the prevalence and intensity of Schistosoma mansoni infection among rural communities in Western Tanzania: The influence of sampling strategy and statistical approach. PLoS Negl. Trop. Dis. 2017, 11, e0005937. [Google Scholar] [CrossRef] [PubMed]
- McCravy, K.W. A Review of Sampling and Monitoring Methods for Beneficial Arthropods in Agroecosystems. Insects 2018, 9, 170. [Google Scholar] [CrossRef]
- Brynildsrud, O. COVID-19 prevalence estimation by random sampling in population—Optimal sample pooling under varying assumptions about true prevalence. BMC Med. Res. Methodol. 2020, 20, 196. [Google Scholar] [CrossRef]
- Abd-Elgawad, M.M.M. Optimizing Sampling and Extraction Methods for Plant-Parasitic and Entomopathogenic Nematodes. Plants 2021, 10, 629. [Google Scholar] [CrossRef]
- Sawicki, R.; Korona-Glowniak, I.; Boguszewska, A.; Stec, A.; Polz-Dacewicz, M. Sample pooling as a strategy for community monitoring for SARS-CoV-2. Sci. Rep. 2021, 11, 3122. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, A.; Vinci, M.; L’Episcopo, F.; Ragalmuto, A.; Neri, V.; Roccella, M.; Quatrosi, G.; Vetri, L.; Calì, F. Implementation of Sample Pooling Procedure Using a Rapid SARS-CoV-2 Diagnostic Real-Time PCR Test Performed Prior to Hospital Admission of People with Intellectual Disabilities. Int. J. Environ. Res. Public Health 2021, 18, 9317. [Google Scholar] [CrossRef] [PubMed]
- Karunakaran, C.; Jayas, D.S.; White, N.D.G. X-ray image analysis to detect infestations caused by insects in grain. Cereal Chem. 2003, 80, 553–557. [Google Scholar] [CrossRef]





| Gene | Primer Name and Direction | Sequence (5′ → 3′) | Source |
|---|---|---|---|
| COI | LepF1 (F) | ATTCAACCAATCATAAAGATATTGG | [38] |
| COI | LepR1 (R) | TAAACTTCTGGATGTCCAAAAAATCA | [38] |
| COI | COI_O_ligustri_F1 (F) | TTACTACCTCCTTCACTAATTTTACTTC | Newly developed |
| COI | COI_O_ligustri_R1 (R) | CCGCTCTAGTGTCATTCCTAT | Newly developed |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, I.; Roda, A.; Misiaszek, B.; Sparks, T.; Diaz, R. Detection of Ochyromera ligustri (Coleoptera: Curculionidae: Curculioninae) in Ligustrum spp. (Oleaceae) Using Newly Developed PCR Primers. Insects 2024, 15, 320. https://doi.org/10.3390/insects15050320
Kang I, Roda A, Misiaszek B, Sparks T, Diaz R. Detection of Ochyromera ligustri (Coleoptera: Curculionidae: Curculioninae) in Ligustrum spp. (Oleaceae) Using Newly Developed PCR Primers. Insects. 2024; 15(5):320. https://doi.org/10.3390/insects15050320
Chicago/Turabian StyleKang, Ilgoo, Amy Roda, Brandi Misiaszek, Tanner Sparks, and Rodrigo Diaz. 2024. "Detection of Ochyromera ligustri (Coleoptera: Curculionidae: Curculioninae) in Ligustrum spp. (Oleaceae) Using Newly Developed PCR Primers" Insects 15, no. 5: 320. https://doi.org/10.3390/insects15050320
APA StyleKang, I., Roda, A., Misiaszek, B., Sparks, T., & Diaz, R. (2024). Detection of Ochyromera ligustri (Coleoptera: Curculionidae: Curculioninae) in Ligustrum spp. (Oleaceae) Using Newly Developed PCR Primers. Insects, 15(5), 320. https://doi.org/10.3390/insects15050320






