A Mysterious Asian Firefly Genus, Oculogryphus Jeng, Engel & Yang (Coleoptera, Lampyridae): The First Complete Mitochondrial Genome and Its Phylogenetic Implications
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
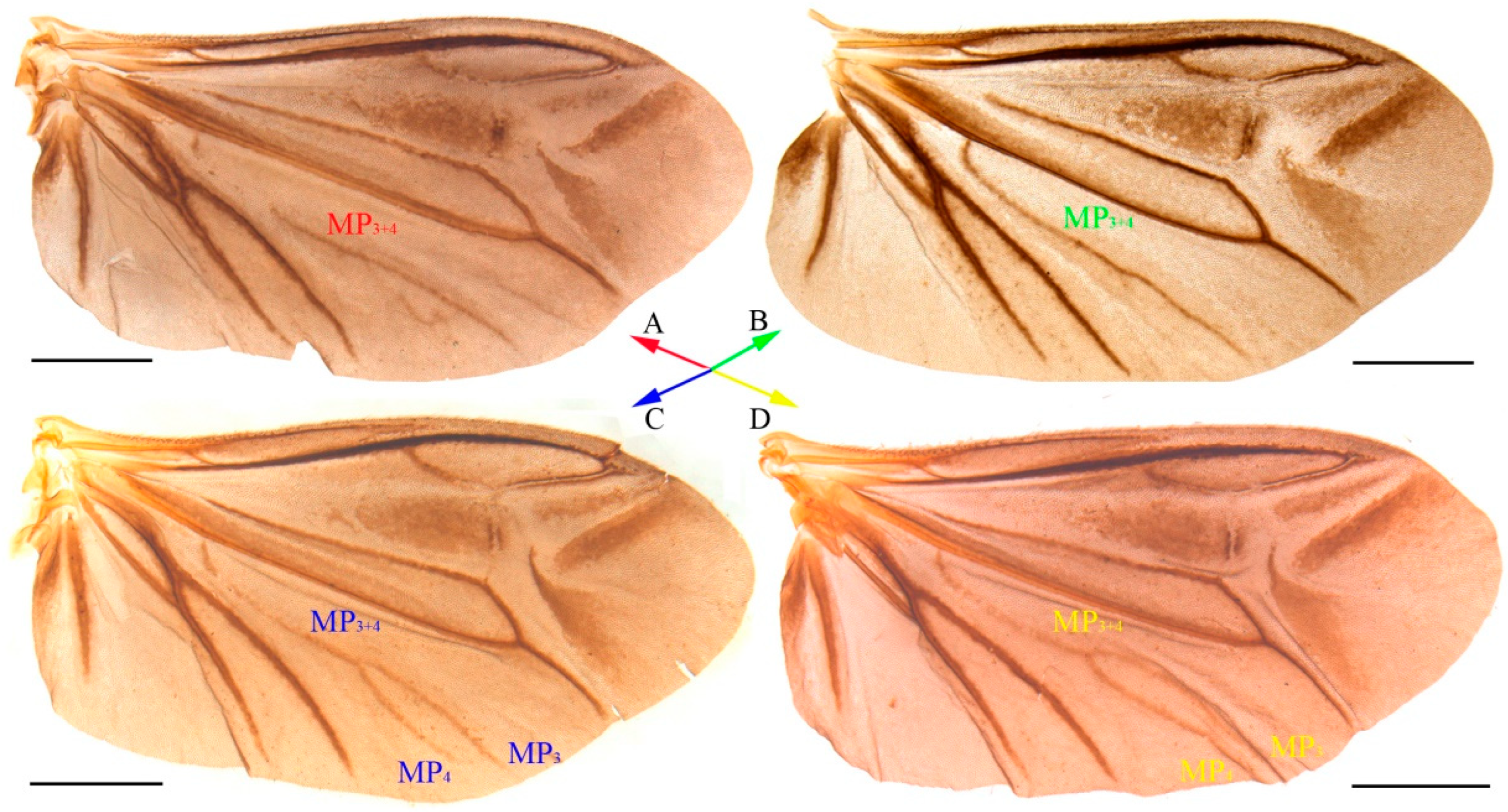
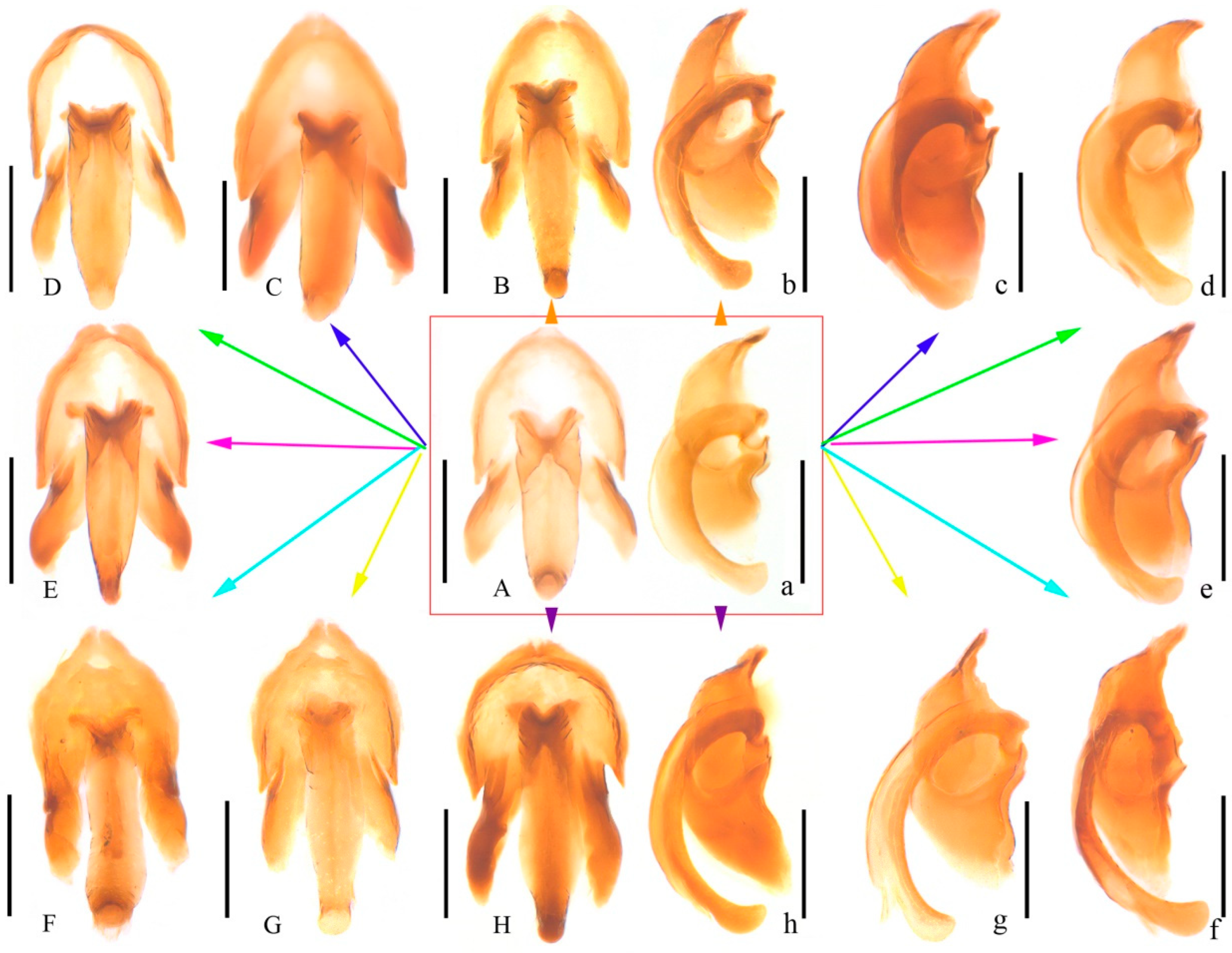
2.2. Morphological Technique
2.3. DNA Extraction, Mitochondrial Genome Sequencing, and Assembly
2.4. Genome Annotation and Sequence Analyses
2.5. Dataset Assembly and Phylogenetic Analyses
3. Results
3.1. Taxonomy
- Class Insecta Linnaeus, 1758
- Order Coleoptera Linnaeus, 1758
- Family Lampyridae Rafinesque, 1815
- Genus Oculogryphus Jeng, Engel & Yang, 2007
- Oculogryphus chenghoiyanae Yiu & Jeng, 2018
- Oculogryphus chenghoiyanae Yiu & Jeng, 2018 [21]: 67.
3.2. General Features of the Mitochondrial Genome
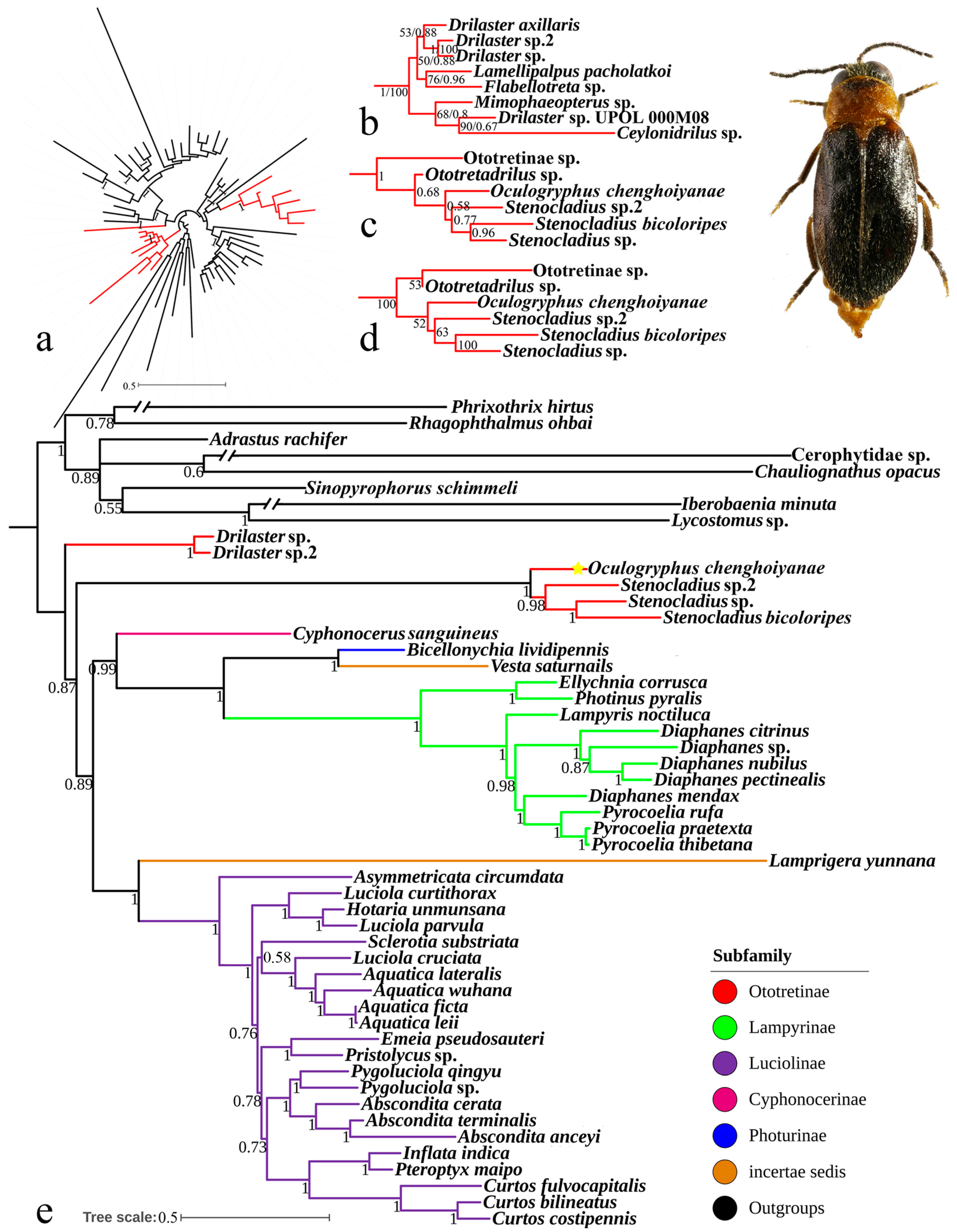
3.3. Phylogenetic Analyses
4. Discussion
4.1. Species Identification
4.2. Comparisons of the Mitogenomes between Oculogryphus and Stenocladius
4.3. Phylogenetic Position of Oculogryphus
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lawrence, J.F. Rhinorphipidae, a new beetle family from Australia, with comments on the phylogeny of the Elateriformia. Invertebr. Syst. 1988, 2, 1–53. [Google Scholar] [CrossRef]
- Lawrence, J.F.; Newton, A.F. Families and subfamilies of Coleoptera (with selected genera, notes, references and data on family-group names). In Biology, Phylogeny, and the Classification of Coleoptera: Papers Celebrating the 80th Birthday of Roy, A. Crowson; Pakaluk, J., Slipinski, S.A., Eds.; Muzeum I InstytutZoologii PAN: Warszawa, Hungary, 1995; pp. 779–1092. [Google Scholar]
- Lawrence, J.F.; Ślipiski, A.; Seago, A.E.; Thayer, M.K.; Newton, A.F.; Marvaldi, A.E. Phylogeny of the Coleoptera based on morphological characters of adults and larvae. Ann. Zool. 2011, 61, 1–217. [Google Scholar] [CrossRef]
- Bouchard, P.; Bousquet, Y.; Davies, A.E.; Alonso-Zarazaga, M.A.; Lawrence, J.F.; Lyal, C.H.C.; Newton, A.F.; Reid, C.A.M.; Schmitt, M.; Ślipiński, A.; et al. Family-group names in Coleoptera (Insecta). ZooKeys 2011, 88, 1–972. [Google Scholar] [CrossRef]
- Branham, M.A. Lampyridae Latreille, 1817. In Coleoptera, Beetles: Morphology and Systematics (Elateroidea, Bostrichiformia, Cucujiformia partim); Leschen, R.A.B., Beutel, R.G., Lawrence, J.F., Eds.; Walter de Gruyter: Berlin, Germany, 2010; Volume 2, pp. 141–149. [Google Scholar]
- Olivier, E. Coleoptera. Fam. Lampyridae. In Genera Insectorum, Fasc. 53; Wytsman, P., Ed.; Verteneuil, V. and Desmet, L.: Brussels, Belgium, 1907; pp. 74–79. [Google Scholar]
- Olivier, E. Lampyridae. In Coleopterorum Catalogus, Pars. 9; Schenkling, S., Ed.; W. Junk: Berlin, Germany, 1910; pp. 1–68. [Google Scholar]
- McDermott, F.A. The taxonomy of the Lampyridae (Coleoptera). Trans. Am. Entomol. Soc. 1964, 90, 1–72. Available online: https://www.jstor.org/stable/25077867 (accessed on 10 January 2024).
- Crowson, R.A. A review of the classification of Cantharoidea (Coleoptera), with the definition of two new families, Cneoglossidae and Omethidae. Rev. Univ. Madrid 1972, 21, 35–77. [Google Scholar]
- Nakane, T. Lampyrid insects of the world. In The Reconstruction of Firefly Environments, Reconquista, Special No. 1; The Association of Natural Restoration of Japan (editorial commission); Saiteku: Tokyo, Japan, 1991; pp. 3–11, (In Japanese with English summary). [Google Scholar]
- Branham, M.A.; Wenzel, J.W. The evolution of bioluminescence in cantharoids (Coleoptera: Elateroidea). Fla. Entomol. 2001, 84, 565–586. [Google Scholar] [CrossRef]
- Branham, M.A.; Wenzel, J.W. The origin of photic behavior and the evolution of sexual communication in fireflies (Coleoptera: Lampyridae). Cladistics 2003, 19, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Jeng, M.L. Comprehensive Phylogenetics, Systematics, and Evolution of Neoteny of Lampyridae (Insecta: Coleoptera). Ph.D. Thesis, University of Kansas, Lawrence, Kansas, 2008. [Google Scholar]
- Stanger-Hall, K.F.; Lloyd, J.E.; Hillis, D.M. Phylogeny of north American fireflies (Coleoptera: Lampyridae): Implications for the evolution of light signals. Mol. Phylogenetics Evol. 2007, 45, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Dong, Z.W.; Liu, G.C.; He, J.; Zhao, R.P.; Wang, W.; Peng, Y.Q.; Li, X.Y. Phylogenetic analysis provides insights into the evolution of Asian fireflies and adult bioluminescence. Mol. Phylogenetics Evol. 2019, 140, 106600. [Google Scholar] [CrossRef]
- Martin, G.J.; Stanger-Hall, K.F.; Branham, M.A.; Silveira, L.F.L.; Lower, S.E.; Hall, D.W.; Li, X.Y.; Lemmon, A.R.; Lemmon, E.M.; Bybee, S.M. Higher-level phylogeny and reclassification of Lampyridae (Coleoptera: Elateroidea). Insect Syst. Diver. 2019, 3, 11. [Google Scholar] [CrossRef]
- Martin, G.J.; Branham, M.A.; Whiting, M.F.; Bybee, S.M. Total evidence phylogeny and the evolution of adult bioluminescencein fireflies (Coleoptera: Lampyridae). Mol. Phylogenetics Evol. 2017, 107, 564–575. [Google Scholar] [CrossRef] [PubMed]
- Jeng, M.L.; Engel, M.S.; Yang, P.S. Oculogryphus, a remarkable new genus of fireflies from Asia (Coleoptera: Lampyridae). Am. Mus. Novit. 2007, 3600, 1–19. [Google Scholar] [CrossRef]
- Jeng, M.L.; Branham, M.A.; Engel, M.S. A second species of Oculogryphus (Coleoptera, Lampyridae), with notes on the phylogenetic affinities of the genus. ZooKeys 2011, 97, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Jeng, M.L.; Engel, M.S. Description of Oculogryphus shuensis sp. n. (Coleoptera, Lampyridae), the first species of the genus in the Sino-Japanese realm, with a modified key to the subfamily Ototretinae. ZooKeys 2014, 378, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Yiu, V.; Jeng, M.L. Oculogryphus chenghoiyanae sp. n. (Coleoptera, Lampyridae): A new ototretine firefly from Hong Kong with descriptions of its bioluminescent behavior and ultraviolet-induced fluorescence in females. ZooKeys 2018, 739, 65–78. [Google Scholar] [CrossRef] [PubMed]
- Janisova, K.; Bocakova, M. Revision of the subfamily Ototretinae (Coleoptera: Lampyridae). Zool. Anz. 2013, 252, 1–19. [Google Scholar] [CrossRef]
- Resch, M.C.; Shrubovych, J.; Bartel, D.; Szucsich, N.U.; Timelthaler, G.; Bu, Y.; Manfred, W.G.; Pass, G. Where taxonomy based on subtle morphological differences is perfectly mirrored by huge genetic distances: DNA barcoding in Protura (Hexapoda). PLoS ONE 2014, 9, e90653. [Google Scholar] [CrossRef] [PubMed]
- Derocles, S.A.; Plantegenest, M.; Rasplus, J.Y.; Marie, A.; Evans, D.M.; Lunt, D.H.; Le-Ralec, A. Are generalist Aphidiinae (Hym. Braconidae) mostly cryptic species complexes? Syst. Entomol. 2016, 41, 379–391. [Google Scholar] [CrossRef]
- Kundrata, R.; Blank, S.M.; Prosvirov, A.S.; Sormove, E.; Gimmel, M.L.; Vondráček, D.; Kramp, K. One less mystery in Coleoptera systematics: The position of Cydistinae (Elateroformia incertae sedis) resolved by multigene phylogenetic analysis. Zool. J. Linn. Soc. 2019, 187, 1259–1277. [Google Scholar] [CrossRef]
- Evangelista, D.; Thouzé, F.; Kohli, M.K.; Lopez, P.; Legendre, F. Topological support and data quality can only be assessed through multiple tests in reviewing Blattodea phylogeny. Mol. Phylogenetics Evol. 2018, 128, 112–122. [Google Scholar] [CrossRef]
- Yoshizawa, K.; Johnson, K.P.; Sweet, A.D.; Yao, I.; Ferreira, R.L.; Cameron, S.L. Mitochondrial phylogenomics and genome rearrangements in the barklice (Insecta: Psocodea). Mol. Phylogenetics Evol. 2018, 119, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Plese, B.; Kenny, N.J.; Rossi, M.E.; Cárdenas, P.; Schuster, A.; Taboada, S.; Koutsouveli, V.; Riesgo, A. Mitochondrial evolution in Demospongiae (Porifera): Phylogeny, divergence time, and genome biology. Mol. Phylogenetics Evol. 2021, 155, 107011. [Google Scholar] [CrossRef] [PubMed]
- Dowton, M.; Belshaw, R.; Austin, A.D.; Quicke, D.L.J. Simultaneous molecular and morphological analysis of Braconid relationships (Insecta: Hymenoptera: Braconidae) indicates independent mt-tRNA gene inversions within a single wasp family. J. Mol. Evol. 2002, 54, 210–226. [Google Scholar] [CrossRef] [PubMed]
- Cameron, S.L. Insect mitochondrial genomics: Implications for evolution and phylogeny. Annu. Rev. Entomol. 2014, 59, 95–117. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L.; Brown, W.M. Big trees from little genomes: Mitochondrial gene order as a phylogenetic tool. Curr. Opin. Genet. Dev. 1998, 8, 668–674. [Google Scholar] [CrossRef]
- Shao, R.; Campbell, N.J.H.; Schmidt, E.R.; Barker, S.C. Increased rate of gene rearrangement in the mitochondrial genomes of three orders of hemipteroid insects. Mol. Biol. Evol. 2001, 18, 1828–1832. [Google Scholar] [CrossRef]
- Mao, M.; Austin, A.D.; Johnson, N.F.; Dowton, M. Coexistence of minicircular and a highly rearranged mtDNA molecule suggests that recombination shapes mitochondrial genome organization. Mol. Biol. Evol. 2014, 31, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wei, S.J.; Tang, P.; Wu, Q.; Shi, M.; Sharkey, M.J.; Chen, X.X. Multiple lines of evidence from mitochondrial genomes resolve phylogenetic relationships of parasitic wasps in Braconidae. Genome Biol. Evol. 2016, 8, 2651–2662. [Google Scholar] [CrossRef]
- Chen, L.; Chen, P.Y.; Xue, X.F.; Hua, H.Q.; Li, Y.X.; Zhang, F.; Wei, S.J. Extensive gene rearrangements in the mitochondrial genomes of two egg parasitoids, Trichogramma japonicum and Trichogramma ostriniae (Hymenoptera: Chalcidoidea: Trichogrammatidae). Sci. Rep. 2018, 8, 7034. [Google Scholar] [CrossRef]
- Sankoff, D.; Leduc, G.; Antoine, N.; Paquin, B.; Lang, B.F.; Cedergren, R. Gene order comparisons for phylogenetic inference: Evolution of the mitochondrial genome. Proc. Natl. Acad. Sci. USA 1992, 89, 6575–6579. [Google Scholar] [CrossRef]
- Guo, E.; Yang, Y.; Kong, L.F.; Yu, H.; Liu, S.K.; Liu, Z.J.; Li, Q. Mitogenomic phylogeny of Trochoidea (Gastropoda: Vetigastropoda): New insights from increased complete genomes. Zool. Scr. 2020, 50, 43–57. [Google Scholar] [CrossRef]
- Ge, X.Y.; Liu, T.; Kang, Y.; Liu, H.Y.; Yang, Y.X. First complete mitochondrial genomes of Ototretinae (Coleoptera, Lampyridae) with evolutionary insights into the gene rearrangement. Genomics 2022, 114, 110305. [Google Scholar] [CrossRef]
- Zheng, Y.; Roberts, R.J.; Kasif, S. Identification of genes with fast-evolving regions in microbial genomes. Nucleic Acids Res. 2004, 32, 6347–6357. [Google Scholar] [CrossRef] [PubMed]
- Kundrata, R.; Bocakova, M.; Bocak, L. The comprehensive phylogeny of the superfamily Elateroidea (Coleoptera: Elateriformia). Mol. Phylogenetics Evol. 2014, 76, 162–171. [Google Scholar] [CrossRef]
- Kundrata, R.; Bocak, L. The phylogeny and limits of Elateridae (Insecta, Coleoptera): Is there a common tendency of click beetles to soft-bodiedness and neoteny? Zool. Scr. 2011, 40, 364–378. [Google Scholar] [CrossRef]
- Sagegami-Oba, R.; Takahashi, N.; Oba, Y. The evolutionary process of bioluminescence and aposematism in cantharoid beetles (Coleoptera: Elateroidea) inferred by the analysis of 18S ribosomal DNA. Gene 2007, 400, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. Unraveling heteroplasmy patterns with NOVOPlasty. Genom. Bioinf. 2020, 2, lqz011. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Ogoh, K.; Ohba, N.; Liang, X.; Ohmiya, Y. Mitochondrial genomes of two luminous beetles, Rhagophthalmus lufengensis and R. ohbai (Arthropoda, Insecta, Coleoptera). Gene 2007, 392, 196–205. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Jühling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Pütz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo Metazoan Mitochondrial Genome Annotation. Mol. Phylogenetics Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lowe, T.M. tRNAscan-SE: Searching for tRNA genes in genomic sequences. In Methods in Molecular Biology; Humana: New York, NY, USA, 2019; Volume 1962, pp. 1–14. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Julio, R.; Sánchez-DelBarri, J.C.; Messeguer, X.; Rozas, R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 2004, 19, 2496–2497. [Google Scholar]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Agarwala, R.; Barrett, T.; Beck, J.; Benson, D.A.; Bollin, C.; Bolton, E.; Bourexis, D.; Brister, J.R.; Bryant, S.H.; Lanese, K.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2016, 44, D7–D19. [Google Scholar] [CrossRef]
- Katoh, K.; Kuma, K.; Toh, H.; Miyata, T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005, 33, 511–518. [Google Scholar] [CrossRef]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; Von-Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Lartillot, N.; Rodrigue, N.; Stubbs, D.; Richer, J. PhyloBayes MPI: Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 2013, 62, 611–615. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.S.; Kim, I.; Sohn, H.D.; Jin, B.R. The mitochondrial genome of the firefly, Pyrocoelia rufa: Complete DNA sequence, genome organization, and phylogenetic analysis with other insects. Mol. Phylogenetics Evol. 2004, 32, 978–985. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Fu, X. The complete mitochondrial genome of the firefly, Abscondita anceyi (Olivier) (Coleoptera: Lampyridae). Mitochondrial DNA Part B 2018, 3, 442–443. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.Y.; Yuan, L.L.; Kang, Y.; Liu, T.; Liu, H.Y.; Yang, Y.X. Characterization of the first complete mitochondrial genome of Cyphonocerinae (Coleoptera: Lampyridae) with implications for phylogeny and evolution of fireflies. Insects 2021, 12, 570. [Google Scholar] [CrossRef]
- Tuxen, S.L. Taxonomists Glossary of Genitalia in Insects; Scandinavian University Press: Copenhagen, Denmark, 1970. [Google Scholar]
- Vrana, P.; Wheeler, Q.D. Individual organisms as terminal entities laying the species problem to rest. Cladistics 1992, 8, 67–72. [Google Scholar] [CrossRef]
- Usener, J.L.; Cognato, A.I. Phylogenetic review of desert firefly taxonomic characters (Coleoptera: Lampyridae: Microphotus). Insect Syst. Evol. 2006, 37, 71–80. [Google Scholar]
- Babbucci, M.; Basso, A.; Scupola, A.; Patarnello, T.; Negrisolo, E. Is it an ant or a butterfly? Convergent evolution in the mitochondrial gene order of Hymenoptera and Lepidoptera. Genome Biol. Evol. 2014, 12, 3326–3343. [Google Scholar] [CrossRef] [PubMed]
- Basso, A.; Babbucci, M.; Pauletto, M.; Riginella, E.; Patarnello, T.; Negrisolo, E. The highly rearranged mitochondrial genomes of the crabs Maja crispata and Maja squinado (Majidae) and gene order evolution in Brachyura. Sci. Rep. 2017, 7, 4096. [Google Scholar] [CrossRef] [PubMed]
- Moritz, C.; Dowling, T.E.; Brown, W.M. Evolution of animal mitochondrial DNA: Relevance for population biology and systematic. Annu. Rev. Ecol. Syst. 1987, 18, 269–292. [Google Scholar] [CrossRef]
- Boore, J.L. The duplication/random loss model for gene rearrangement exemplified by mitochondrial genomes of deuterostome animals. Comp. Genom. 2000, 1, 133–147. [Google Scholar]
- Bernt, M.; Middendorf, M. A method for computing an inventory of metazoan mitochondrial gene order rearrangements. BMC Bioinform. 2011, 12, S6. [Google Scholar] [CrossRef] [PubMed]
- Shao, R.; Dowton, M.; Murrell, A.; Barker, S.C. Rates of gene rearrangement and nucleotide substitution are correlated in the mitochondrial genomes of insects. Mol. Biol. Evol. 2003, 20, 1612–1619. [Google Scholar] [CrossRef]
- Liu, Q.Q.; He, J.; Song, F.; Tian, L.; Cai, W.Z.; Li, H. Positive correlation of the gene rearrangements and evolutionary rates in the mitochondrial genomes of Thrips (Insecta: Thysanoptera). Insects 2022, 13, 585. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.-X.; Kang, Y.; Ge, X.-Y.; Yuan, S.-L.; Li, X.-Y.; Liu, H.-Y. A Mysterious Asian Firefly Genus, Oculogryphus Jeng, Engel & Yang (Coleoptera, Lampyridae): The First Complete Mitochondrial Genome and Its Phylogenetic Implications. Insects 2024, 15, 464. https://doi.org/10.3390/insects15070464
Yang Y-X, Kang Y, Ge X-Y, Yuan S-L, Li X-Y, Liu H-Y. A Mysterious Asian Firefly Genus, Oculogryphus Jeng, Engel & Yang (Coleoptera, Lampyridae): The First Complete Mitochondrial Genome and Its Phylogenetic Implications. Insects. 2024; 15(7):464. https://doi.org/10.3390/insects15070464
Chicago/Turabian StyleYang, Yu-Xia, Ya Kang, Xue-Ying Ge, Shuai-Long Yuan, Xue-Yan Li, and Hao-Yu Liu. 2024. "A Mysterious Asian Firefly Genus, Oculogryphus Jeng, Engel & Yang (Coleoptera, Lampyridae): The First Complete Mitochondrial Genome and Its Phylogenetic Implications" Insects 15, no. 7: 464. https://doi.org/10.3390/insects15070464
APA StyleYang, Y.-X., Kang, Y., Ge, X.-Y., Yuan, S.-L., Li, X.-Y., & Liu, H.-Y. (2024). A Mysterious Asian Firefly Genus, Oculogryphus Jeng, Engel & Yang (Coleoptera, Lampyridae): The First Complete Mitochondrial Genome and Its Phylogenetic Implications. Insects, 15(7), 464. https://doi.org/10.3390/insects15070464





