Use of Wing Geometric Morphometric Analysis and mtDNA to Identify Africanization of Apis mellifera in the Central Highlands of Ecuador
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Geometric and Morphometric Qualities
2.3. Mitochondrial DNA
2.4. DNA Amplification
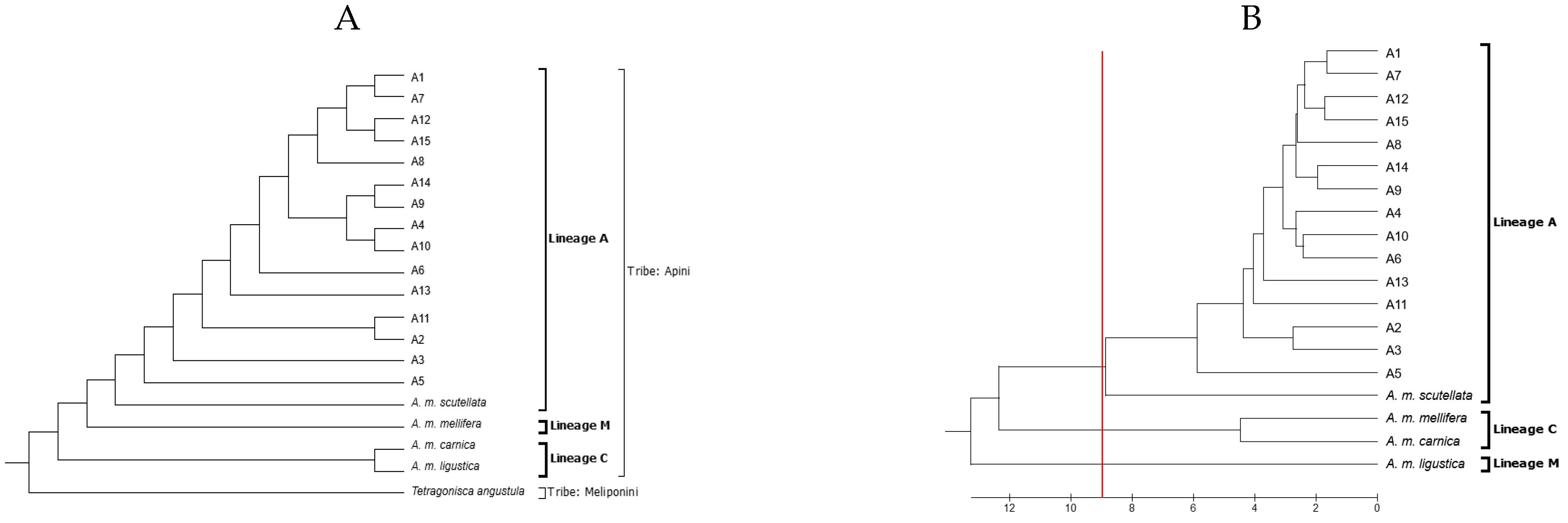
2.5. Sequencing and Phylogenetic Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- FAO. The Second Report on the State of the World’s Animal Genetic Resources for Food and Agriculture; Commission on Genetic Resources for Food and Agriculture Assessments: Rome, Italy, 2015. [Google Scholar]
- de Figueiró Santos, J.; Coelho, F.C.; Bliman, P.-A. Behavioral modulation of the coexistence between Apis melifera and Varroa destructor: A defense against colony colapse disorder? PLoS ONE 2016. [Google Scholar] [CrossRef]
- Ruttner, F.; Tassencourt, L.; Louveaux, J. Biometrical-statistical analysis of the geographic variability of Apis mellifera L. Material and methods. Apidologie 1978, 9, 363–381. [Google Scholar] [CrossRef]
- Alpatov, W. Biometrical studies on variation and races of the honey bee (Apis mellifera L.). Q. Rev. Biol. 1929, 4, 1–58. [Google Scholar] [CrossRef]
- Ruttner, F. Taxonomy and Biogeography of Honeybees; Springer: Berlin, Germany, 1988; p. 284. [Google Scholar]
- Ivanova, E.; Bouga, M.; Staykova, T.; Mladenovic, M.; Rasic, S.; Charistos, L.; Hatjina, F.; Petrov, P. The genetic variability of honey bees from the Southern Balkan Peninsula, based on alloenzymic data. J. Apic. Res. 2012, 51, 329–335. [Google Scholar] [CrossRef]
- Nunamaker, R.; Wilson, W. Isozyme changes in the honeybee, Apis mellifera L., during larval morphogenesis. Insect Biochem. 1982, 12, 99–104. [Google Scholar] [CrossRef]
- Charistos, L.; Hatjina, F.; Bouga, M.; Mladenovic, M.; Maistros, A.D. Morphological discrimination of Greek honey bee populations based on geometric Morphometrics analysis of wing shape. J. Apic. Sci. 2014, 58, 75–84. [Google Scholar] [CrossRef]
- De la Rúa, P.; Galián, J.; Serrano, J. Variabilidad del ADN mitocondrial en poblaciones de abejas de la miel (Apis mellifera L.) de la Región de Murcia. Investig. Agrar. Prod. Y Sanid. Anim. 1999, 14, 41–49. [Google Scholar]
- Pinto, M.A.; Henriques, D.; Neto, M.; Guedes, H.; Muñoz, I.; Azevedo, J.C.; De la Rúa, P. Maternal diversity patterns of Ibero-Atlantic populations reveal further complexity of Iberian honeybees. Apidologie 2013, 44, 430–439. [Google Scholar] [CrossRef]
- Szalanski, A.; Tripodi, A. Assessing the Utility of a PCR Diagnostics Marker for the Identification of Africanized Honey Bee, Apis mellifera L., (Hymenoptera: Apidae) in the United States. Sociobiology 2015, 61, 234–236. [Google Scholar] [CrossRef]
- Garnery, L.; Franck, P.; Baudry, E.; Vautrin, D.; Cornuet, J.; Solignac, M. Genetic diversity of the west European honey bee (Apis mellifera and A. m. iberica). Genet. Sel. Evol. 1998, 30, 31–47. [Google Scholar] [CrossRef]
- Cánovas, F.; de la Rúa, P.; Serrano, J.; Galián, J. Microsatellite variability reveals beekeeping influences on Iberian honeybee populations. Apidologie 2011, 42, 235–251. [Google Scholar] [CrossRef]
- Rangel, J.; Giresi, M.; Pinto, M.; Baum, K.; Rubink, W.; Coulson, R.; Johnston, J. Africanization of a feral honey bee (Apis mellifera) population in South Texas: Does a decade make a difference? Ecol. Evol. 2016, 6, 2158–2169. [Google Scholar] [CrossRef]
- Oyerinde, A.; Salako, E.; Rabiu, M. Morphometric taxonomy of honeybee races of Apis mellifera L. in Kaduna state. J. Entomol. Zool. Stud. 2017, 5, 825–829. [Google Scholar]
- Chávez-Galarza, J.; Henriques, D.; Johnston, J.S.; Carneiro, M.; Rufino, J.; Patton, J.C.; Pinto, M.A. Revisiting the Iberian honey bee (Apis mellifera iberiensis) contact zone: Maternal and genome-wide nuclear variations provide support for secondary contact from historical refugia. Mol. Ecol. 2015, 24, 2973–2992. [Google Scholar] [CrossRef] [PubMed]
- Porrini, L.P.; Quintana, S.; Brasesco, C.; Porrini, M.P.; Garrido, P.M.; Eguaras, M.J.; Müller, F.; Iriarte, P.F. Southern limit of Africanized honey bees in Argentina inferred by mtDNA and wing geometric morphometric analysis. J. Apic. Res. 2019, 59, 648–657. [Google Scholar] [CrossRef]
- Kandemir, İ.; Özkan, A.; Fuchs, S. Reevaluation of honeybee (Apis mellifera) microtaxonomy: A geometric morphometric approach. Apidologie 2011, 42, 618–627. [Google Scholar] [CrossRef]
- García, C.A.Y.; Rodrigues, P.J.; Tofilski, A.; Elen, D.; McCormak, G.P.; Oleksa, A.; Henriques, D.; Ilyasov, R.; Kartashev, A.; Bargain, C. Using the software DeepWings© to classify honey bees across Europe through wing geometric morphometrics. Insects 2022, 13, 1132. [Google Scholar] [CrossRef]
- Oldroyd, B.; Cornuet, J.-M.; Rowe, D.; Rinderer, T.; Crozier, R. Racial admixture of Apis mellifera in Tasmania, Australia: Similarities and differences with natural hybrid zones in Europe. Heredity 1995, 74, 315–325. [Google Scholar] [CrossRef]
- Kelomey, A.; Paraiso, A.; Sina, H.; Legout, H.; Adjanohoun, A.; Garnery, L.; Baba-Moussa, L. Genetic Variability of the Mitochondrial DNA in Honeybees (Apis mellifera L.) from Benin. J. Agric. Sci. Technol. 2017, 7, 557–566. [Google Scholar]
- Chapman, N.C.; Harpur, B.A.; Lim, J.; Rinderer, T.E.; Allsopp, M.H.; Zayed, A.; Oldroyd, B.P. A SNP test to identify Africanized honeybees via proportion of ‘African’ancestry. Mol. Ecol. Resour. 2015, 15, 1346–1355. [Google Scholar] [CrossRef] [PubMed]
- Felius, M.; Theunissen, B.; Lenstra, J. On the conservation of cattle—The role of breeds. J. Agric. Sci. 2015, 152–162. [Google Scholar] [CrossRef]
- Yepes, W.; Pardo, E.; Vargas, C.; Alfonso, L. Diversidad Genética del caballo criollo (Equus caballus) mediante genes asociados al pelaje en Valencia, Colombia. Rev. De Investig. Vet. Del Perú 2017, 28, 562–570. [Google Scholar] [CrossRef][Green Version]
- Winston, M. Killer Bees. The Africanized honey bee in the Americas; Harvard University Press: Cambridge, MA, USA, 1992. [Google Scholar]
- Garcia, R.; Oliveira, N.; Camargo, S.; Pires, B.; Oliveira, C.; Teixeira, R.; Pickler, M. Honey and propolis production, hygiene and defense behaviors of two generations of Africanized honey bees. Sci. Agric. 2013, 70, 74–81. [Google Scholar] [CrossRef]
- Nunes, L.; Araújo, E.; Marchini, L.; Moreti, A. Variation morphogeometrics of Africanized honey bees (Apis mellifera) in Brazil. Iheringia. Série Zool. 2012, 102, 321–326. [Google Scholar] [CrossRef]
- Gonçalves, L. Introduction of the African bees (Apis mellifera adansonii) into Brazil and some comments on their spread in South America. Am. Bee J. 1974, 114, 414–419. [Google Scholar]
- Medina-Flores, C.; Guzmán-Novoa, E.; Hamiduzzaman, M.; Aguilera Soto, J.; Carlos, L.; Marco, A. Africanización de colonias de abejas melíferas (Apis mellifera) en tres regiones climáticas del norte de México. Vet. México OA 2015, 2, 1–9. [Google Scholar]
- Branchiccela, B.; Aguirre, C.; Parra, G.; Estay, P.; Zunino, P.; Antúnez, K. Genetic changes in Apis mellifera after 40 years of Africanization. Apidologie 2014, 45, 752–756. [Google Scholar] [CrossRef][Green Version]
- Graciano, L. Niveles de infestación de Varroa destructor (Mesostigmata: Varroidae) en abejas africanizadas (Apis mellifera scutellata). Tesis de Maestria, Universidad Nacional de Colombia-Medellín, Medellín, Colombia, 2018. [Google Scholar]
- Kono, Y.; Kohn, J. Range and frequency of africanized honey bees in California (USA). PLoS ONE 2015, 10, e0137407. [Google Scholar] [CrossRef]
- Amakpe, F.; De Smet, L.; Brunain, M.; Frans, J.; Sinsin, B.; de Graaf, D. Characterization of native honey bee subspecies in Republic of Benin using morphometric and genetic tools. J. Apic. Sci. 2018, 62, 47–59. [Google Scholar] [CrossRef]
- Miguel, I.; Garnery, L.; Iriondo, M.; Baylac, M.; Manzano, C.; Steve, W.; Estonba, A. Origin, evolution and conservation of the honey bees from La Palma Island (Canary Islands): Molecular and morphological data. J. Apic. Res. 2016, 54, 427–440. [Google Scholar] [CrossRef]
- Ron, S.; Merino-Viteri, A.; Ortiz, D. Anfibios del Ecuador. Pontificia Universidad Católica del Ecuador, Version 2019.0; Museo de Zoología: Quito, Ecuador, 2019.
- Masaquiza, D.; Llerena, G.; Díaz, B.; Curbelo, L.; Carrasco, R.; Guapi, R. Caracterización de sistemas apícolas en la zona centro del Ecuador. Agrisost 2017, 23, 118–127. [Google Scholar]
- Vaziritabar, S.; Aghamirkarimi, A.; Mehdi, S. Evaluation of the defensive behavior in two honeybee races Iranian honeybee (Apis mellifera meda) and Carniolan honeybee (Apis mellifera carnica) and grooming behavior of different bee races in controlling Varroa destructor mite in honey. J. Entomol. Zool. Stud. 2016, 4, 586–602. [Google Scholar]
- MAGAP. Programa Nacional Apícola. In Ministerio de Agricultura, Ganadería; Acuacultura y Pesca: Quito, Ecuador, 2017. [Google Scholar]
- Meixner, M.; Pinto, M.; Bouga, M.; Kryger, P.; Ivanova, E.; Fuchs, S. Standard methods for characterising subspecies and ecotypes of Apis mellifera. J. Apic. Res. 2013, 52, 1–28. [Google Scholar] [CrossRef]
- Francoy, T.; Wittmann, D.; Drauschke, M.; Müller, S.; Steinhage, V.; Bezerra-Laure, M.; De Jong, D.; Gonçalves, L. Identification of Africanized honey bees through wing morphometrics: Two fast and efficient procedures. Apidologie 2008, 39, 488–494. [Google Scholar] [CrossRef]
- Rohlf, F. TpsUtil, version 1.40; Ecology & Evolution (Program); Suny at Stony Brook: New York, NY, USA, 2008.
- Rohlf, F.J. Shape statistics: Procrustes superimpositions and tangent spaces. J. Classification 1999, 16, 197–223. [Google Scholar] [CrossRef]
- Klingenberg, C. MorphoJ: An integrated software package for geometric morphometrics. Mol. Ecol. Resour. 2011, 11, 353–357. [Google Scholar] [CrossRef] [PubMed]
- Rohlf, F. Numerical Taxonomy and Multivariate Analysis System, Version 2.2; Stony Brook: New York, NY, USA, 2004.
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Truett, G.; Heeger, P.; Mynatt, R.; Truett, A.; Walker, J.; Warman, M. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 2000, 29, 52–54. [Google Scholar] [CrossRef]
- Garnery, L.; Solignac, M.; Celebrano, G.; Cornuet, J. A simple test using restricted PCR-amplified mitochondrial DNA to study the genetic structure of Apis mellifera L. Experientia 1993, 49, 1016–1021. [Google Scholar] [CrossRef]
- Klok, C.; Harrison, J. Atmospheric hypoxia limits selection for large body size in insects. PLoS ONE 2009, 4, e3876. [Google Scholar] [CrossRef][Green Version]
- Hidalgo, M.; Mena, S. Proyecto de viabilidad de implementación de una granja apícola en la parroquia de Nanegalito. Tesis de Ingeniería, Pontificia Universidad Católica del Ecuador, Quito, Ecuador, 2003. No publicado. [Google Scholar]
- Wallberg, A.; Han, F.; Wellhagen, G.; Dahle, B.; Kawata, M.; Haddad, N.; Simões, Z.; Allsopp, M.; Kandemir, I.; De la Rúa, P. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nat. Genet. 2014, 46, 1081–1088. [Google Scholar] [CrossRef] [PubMed]
- Clarke, K.; Oldroyd, B.; Javier, J.; Quezada-Euán, G.; Rinderer, T. Origin of honeybees (Apis mellifera L.) from the Yucatan peninsula inferred from mitochondrial DNA analysis. Mol. Ecol. 2001, 10, 1347–1355. [Google Scholar] [CrossRef] [PubMed]
- Tibatá, V.M.; Arias, E.; Corona, M.; Ariza Botero, F.; Figueroa-Ramírez, J.; Junca, H. Determination of the Africanized mitotypes in populations of honey bees (Apis mellifera L.) of Colombia. J. Apic. Res. 2018, 57, 219–227. [Google Scholar] [CrossRef]
- Acosta, J. Caracterización Morfométrica y Molecular de Apis mellifera Provenientes de Colmenas Localizadas en las Provincias de Pichincha, Imbabura y Carchi-Ecuador. Tesis de Ingeniería, Universidad de las Fuerzas Armadas ESPE, Sangolquí: Ecuador, 2018. [Google Scholar]
- Collet, T.; Ferreira, K.; Arias, M.; Soares, A.; Del Lama, M. Genetic structure of Africanized honeybee populations (Apis mellifera L.) from Brazil and Uruguay viewed through mitochondrial DNA COI–COII patterns. Heredity 2006, 97, 329–335. [Google Scholar] [CrossRef][Green Version]
- Franck, P.; Garnery, L.; Loiseau, A.; Oldroyd, B.; Hepburn, H.; Solignac, M.; Cornuet, J. Genetic diversity of the honeybee in Africa: Microsatellite and mitochondrial data. Heredity 2001, 86, 420–430. [Google Scholar] [CrossRef]
- Guzmán-Novoa, E.; Correa, A.; Espinosa, L.; Guzmán-Novoa, G. Colonización, impacto y control de las abejas melíferas africanizadas en México. Vet. México 2011, 42, 149–178. [Google Scholar]

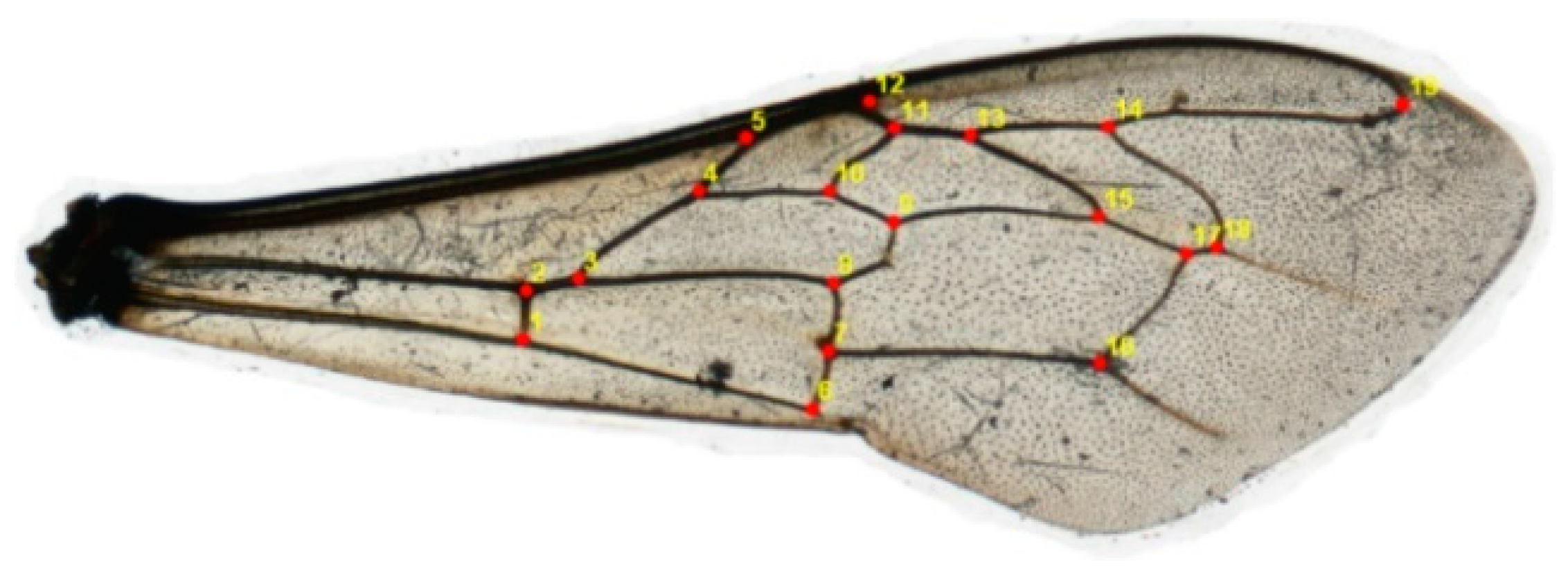


| Known Classification | N (Wings) | A. m. carnica (%) | A. m. ligustica (%) | A. m. mellifera (%) | A. m. scutellata (%) | AB* (%) |
|---|---|---|---|---|---|---|
| A. m. carnica | 50 | 90 | 10 | 0 | 0 | 0 |
| A. m. ligustica | 50 | 10 | 90 | 0 | 0 | 0 |
| A. m. mellifera | 50 | 0 | 2 | 98 | 0 | 0 |
| A. m. scutellata | 50 | 0 | 0 | 0 | 94 | 6 |
| AB* | 750 | 1.2 | 1.8 | 1.1 | 5.7 | 90.2 |
| Morphotype (%) | Altitudinal Floors (m.a.s.l.) | ||
|---|---|---|---|
| 2600–2800 | 2801–3000 | 3001–3274 | |
| n = 15 | n = 35 | n = 25 | |
| Africanized | 95.8 | 93 | 90 |
| European | 4.2 | 7 | 10 |
| Apiary | Bee | Identity (%) | Haplotype | Lineage |
|---|---|---|---|---|
| A1 | 1 | 99.87 | A26a | A |
| A2 | 2 | 99.87 | A26c | A |
| A3 | 3 | 99.6 | A30 | A |
| A4 | 4 | 100 | A30 | A |
| A5 | 5 | 99.49 | A26c | A |
| A6 | 6 | 99.70 | A26c | A |
| A7 | 7 | 96.83 | A1q | A |
| A8 | 8 | 99.87 | A26c | A |
| A9 | 9 | 99.83 | A1e | A |
| A10 | 10 | 100 | A1e | A |
| A11 | 11 | 96.14 | A26a | A |
| A12 | 12 | 99.66 | A1e | A |
| A13 | 13 | 100 | A26 | A |
| A14 | 14 | 100 | A1e | A |
| A15 | 15 | 94.14 | Seq1 | A |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Masaquiza, D.; Rodríguez, L.C.; Zapata, J.; Monar, J.; Vaca, M.; Porrini, L.; Eguaras, M.; Daniele, M.; Romero, D.; Arenal, A. Use of Wing Geometric Morphometric Analysis and mtDNA to Identify Africanization of Apis mellifera in the Central Highlands of Ecuador. Insects 2024, 15, 628. https://doi.org/10.3390/insects15080628
Masaquiza D, Rodríguez LC, Zapata J, Monar J, Vaca M, Porrini L, Eguaras M, Daniele M, Romero D, Arenal A. Use of Wing Geometric Morphometric Analysis and mtDNA to Identify Africanization of Apis mellifera in the Central Highlands of Ecuador. Insects. 2024; 15(8):628. https://doi.org/10.3390/insects15080628
Chicago/Turabian StyleMasaquiza, Diego, Lino Curbelo Rodríguez, José Zapata, Joffre Monar, Maritza Vaca, Leonardo Porrini, Martin Eguaras, Martin Daniele, Dora Romero, and Amilcar Arenal. 2024. "Use of Wing Geometric Morphometric Analysis and mtDNA to Identify Africanization of Apis mellifera in the Central Highlands of Ecuador" Insects 15, no. 8: 628. https://doi.org/10.3390/insects15080628
APA StyleMasaquiza, D., Rodríguez, L. C., Zapata, J., Monar, J., Vaca, M., Porrini, L., Eguaras, M., Daniele, M., Romero, D., & Arenal, A. (2024). Use of Wing Geometric Morphometric Analysis and mtDNA to Identify Africanization of Apis mellifera in the Central Highlands of Ecuador. Insects, 15(8), 628. https://doi.org/10.3390/insects15080628






