BmToll9-1 Is a Positive Regulator of the Immune Response in the Silkworm Bombyx mori
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing and Tissue Dissection
2.2. RNA Extraction, cDNA Synthesis, and qPCR
2.3. RNAi Protocol
2.4. Antibacterial Activity Assays
2.5. Data Analysis
3. Results
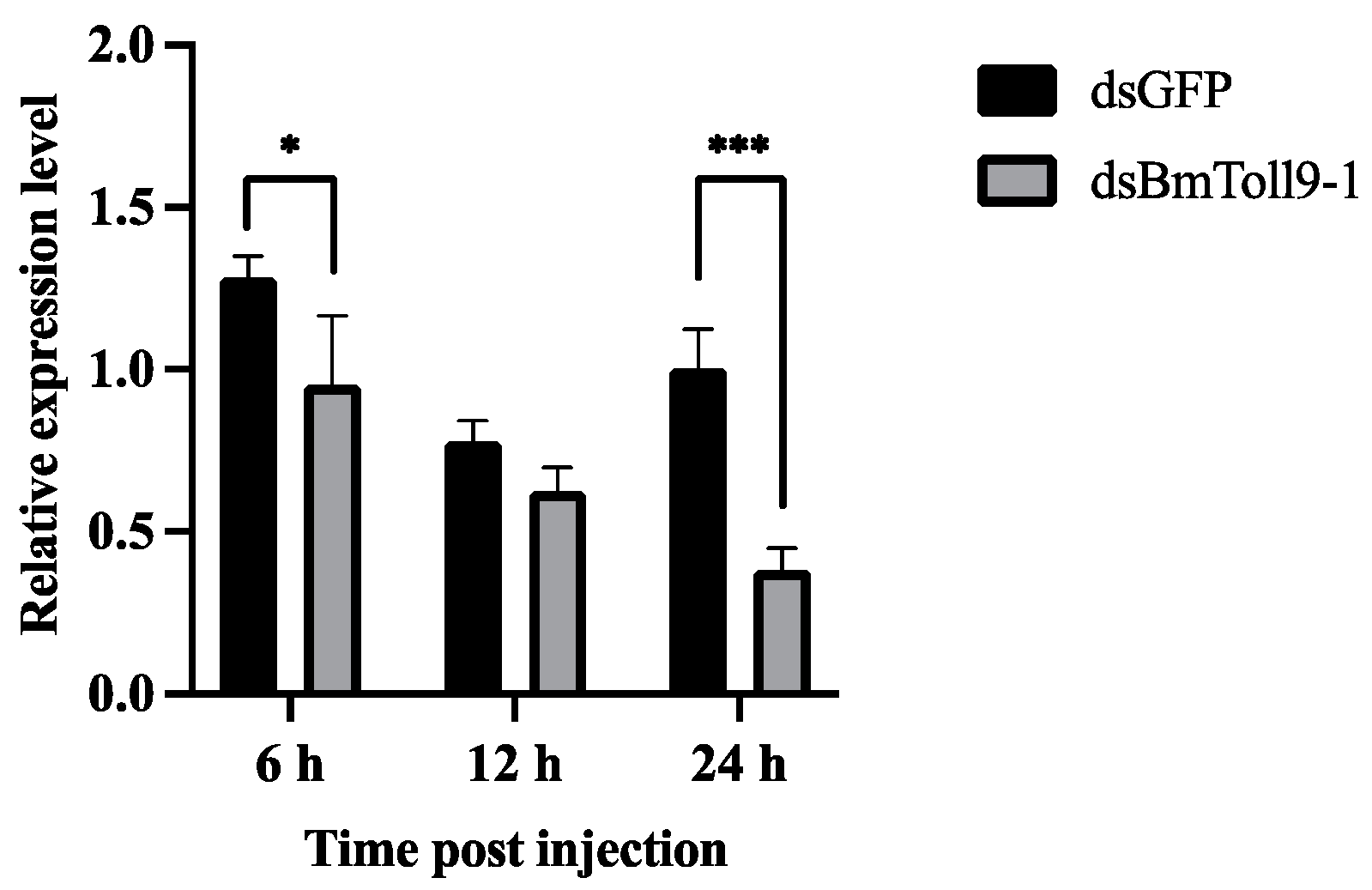
3.1. The BmToll9-1 Gene Was Effectively Silenced in the Midgut
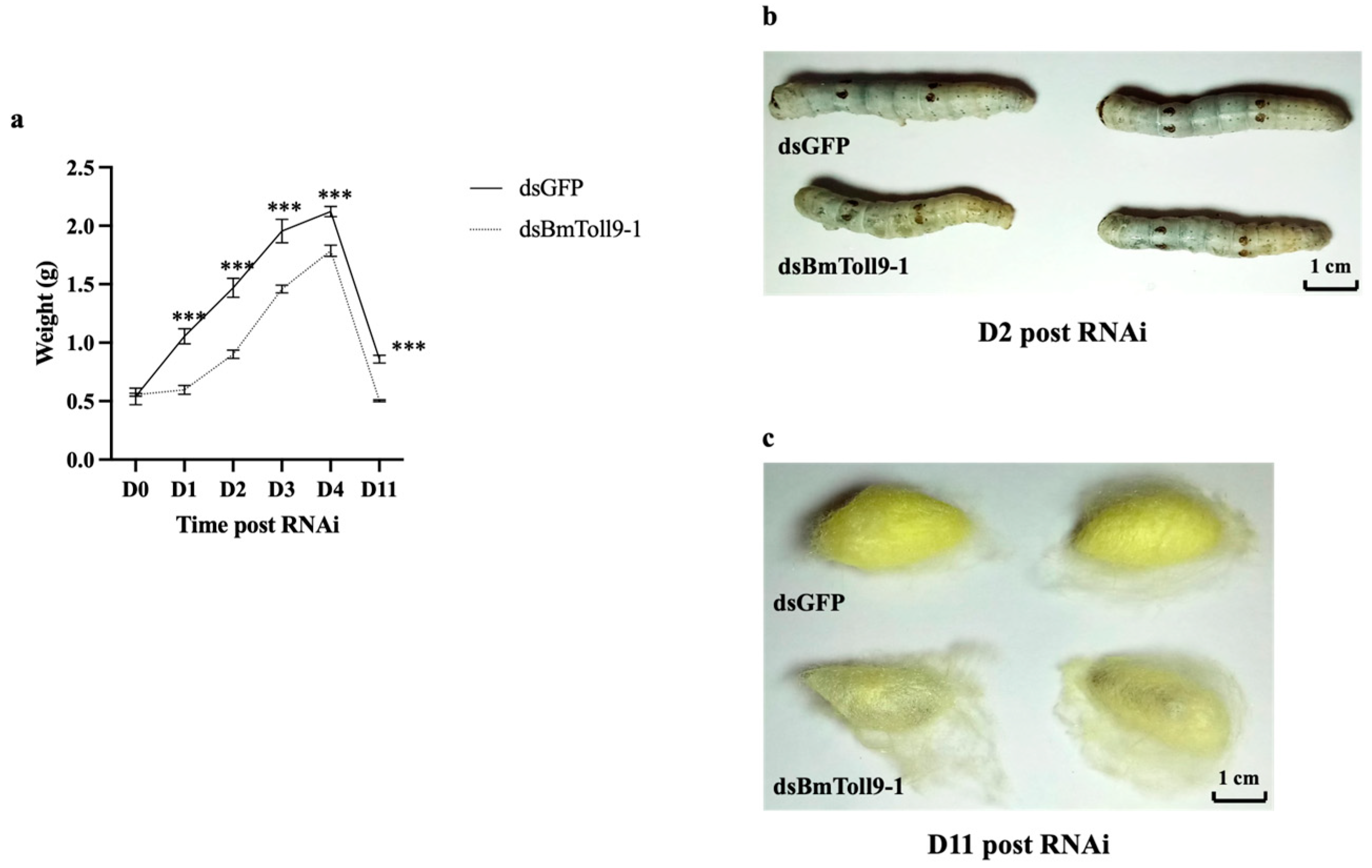
3.2. RNAi of BmToll9-1 Gene Affected the Growth of the Silkworm Larvae and Cocoons
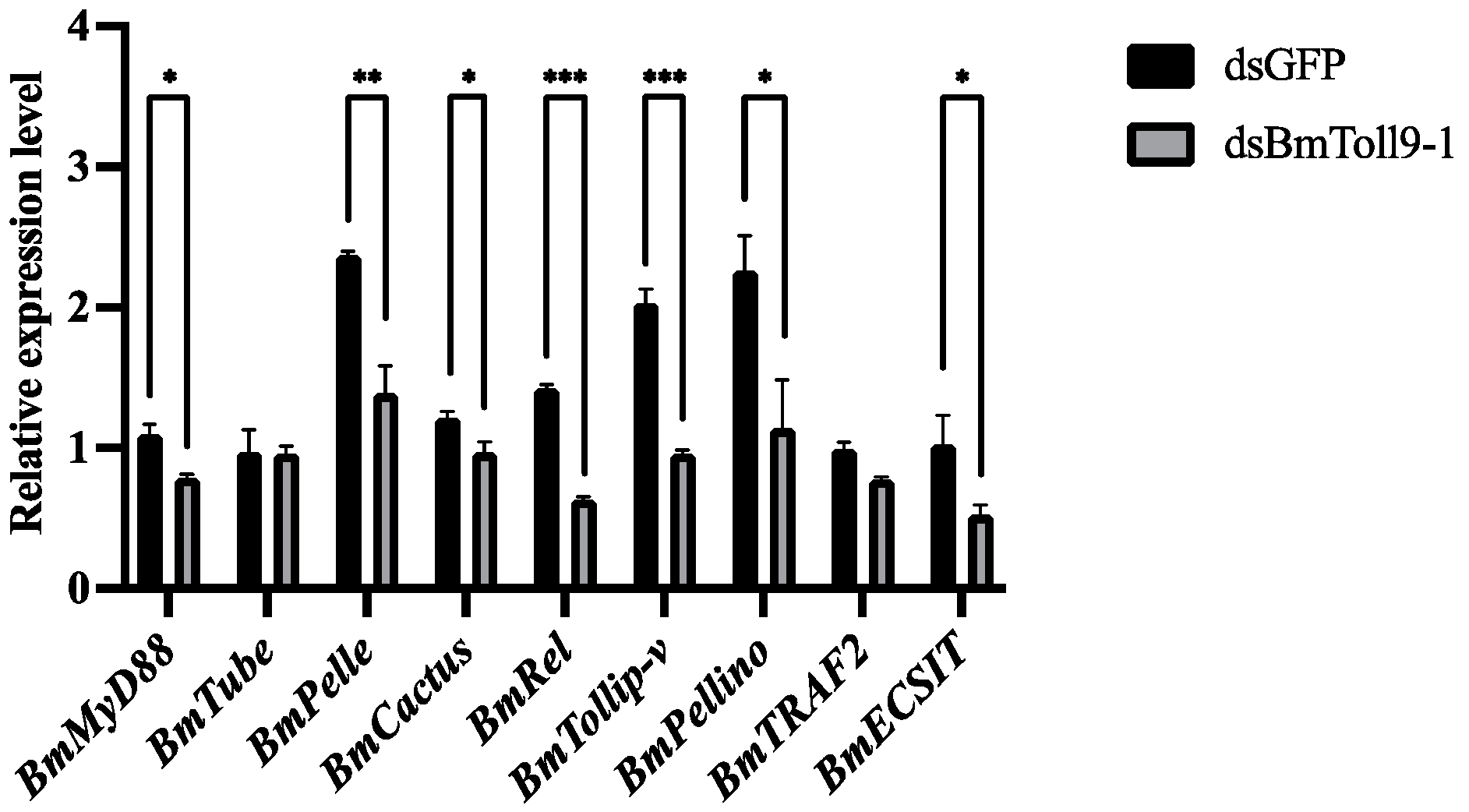
3.3. RNAi of the BmToll9-1 Gene Reduced the Expression of Signaling GENES in the Toll Pathway
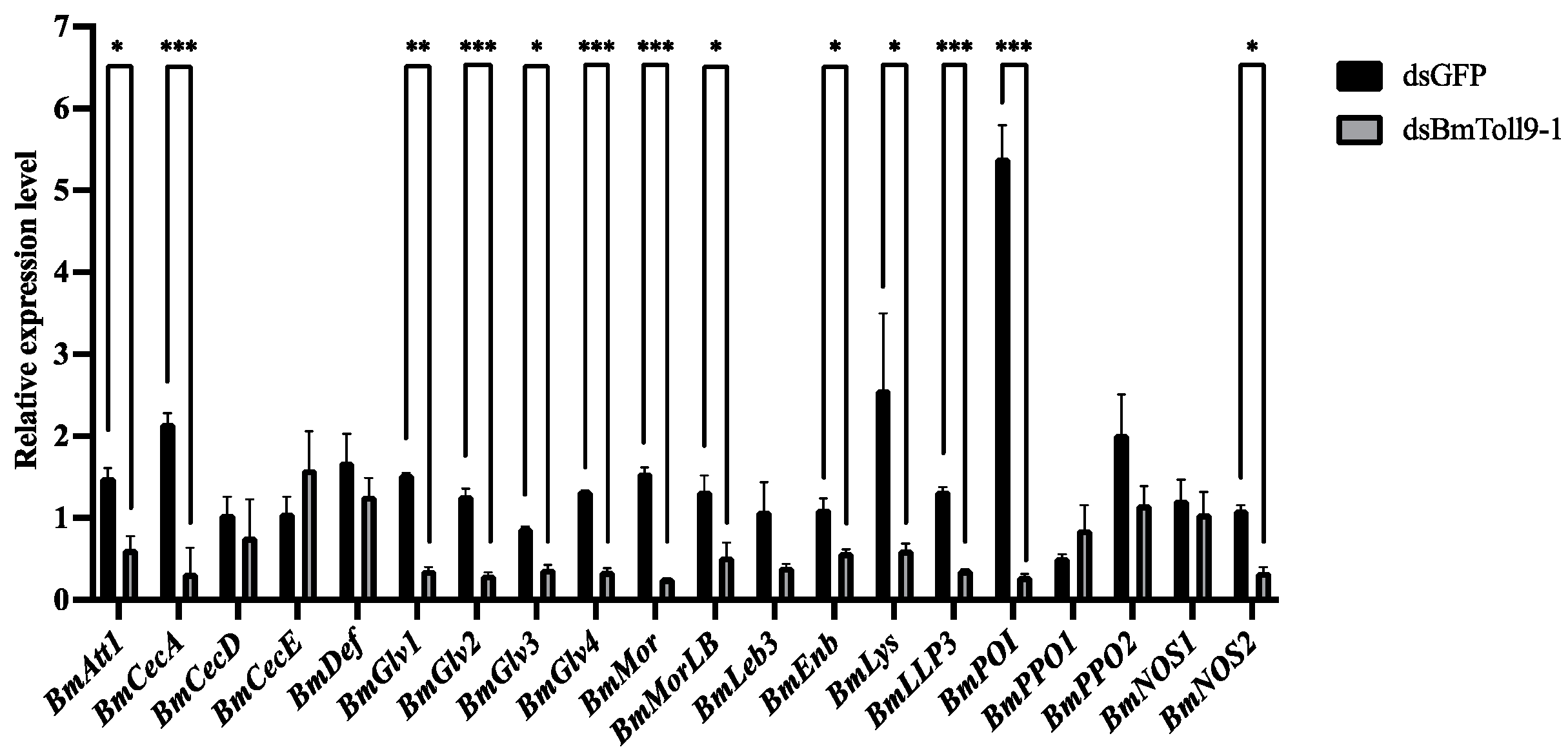
3.4. RNAi of the BmToll9-1 Gene Reduced the Expressions of the Downstream Effector Genes
3.5. RNAi of BmToll9-1 Decreased Antibacterial Activity
4. Discussion
4.1. BmToll9-1 May Modulate Gut Homeostasis
4.2. BmToll9-1 Is Positively Involved in the Toll Pathway
4.3. BmToll9-1 Positively Regulates the Immune Effectors
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cheng, T.; Zhang, Y.; Liu, C.; Xu, P.; Gao, Z.; Xia, Q.; Xiang, Z. Identification and Analysis of Toll-Related Genes in the Domesticated Silkworm, Bombyx mori. Dev. Comp. Immunol. 2008, 32, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Merkling, S.H.; van Rij, R.P. Beyond RNAi: Antiviral Defense Strategies in Drosophila and Mosquito. J. Insect Physiol. 2013, 59, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Chintapalli, R.T.V.; Hillyer, J.F. Hemolymph Circulation in Insect Flight Appendages: Physiology of the Wing Heart and Circulatory Flow in the Wings of the Mosquito, Anopheles gambiae. J. Exp. Biol. 2016, 219, 3945–3951. [Google Scholar] [CrossRef]
- Kim, M.S.; Byun, M.; Oh, B.H. Crystal Structure of Peptidoglycan Recognition Protein LB from Drosophila melanogaster. Nat. Immunol. 2003, 4, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, C.; Hudson, K.L.; Anderson, K.V. The Toll Gene of Drosophila, Required for Dorsal-Ventral Embryonic Polarity, Appears to Encode a Transmembrane Protein. Cell 1988, 52, 269–279. [Google Scholar] [CrossRef]
- Nüsslein-Volhard, C.; Lohs-Schardin, M.; Sander, K.; Cremer, C. A Dorso-Ventral Shift of Embryonic Primordia in a New Maternal-Effect Mutant of Drosophila. Nature 1980, 283, 474–476. [Google Scholar] [CrossRef]
- Gay, N.J.; Keith, F.J. Drosophila Toll and IL-1 Receptor. Nature 1991, 351, 355–356. [Google Scholar] [CrossRef]
- Tanaka, H.; Ishibashi, J.; Fujita, K.; Nakajima, Y.; Sagisaka, A.; Tomimoto, K.; Suzuki, N.; Yoshiyama, M.; Kaneko, Y.; Iwasaki, T. A Genome-Wide Analysis of Genes and Gene Families Involved in Innate Immunity of Bombyx mori. Insect Biochem. Mol. Biol. 2008, 38, 1087–1110. [Google Scholar] [CrossRef]
- Valanne, S.; Wang, J.; Rämet, M. The Drosophila Toll Signaling Pathway. J. Immunol. 2011, 186, 649–656. [Google Scholar] [CrossRef]
- Nonaka, S.; Kawamura, K.; Hori, A.; Salim, E.; Fukushima, K.; Nakanishi, Y.; Kuraishi, T. Characterization of Spz5 as a Novel Ligand for Drosophila Toll-1 Receptor. Biochem. Biophys. Res. Commun. 2018, 506, 510–515. [Google Scholar] [CrossRef]
- Aronstein, K.; Saldivar, E. Characterization of a Honey Bee Toll Related Receptor Gene Am18w and Its Potential Involvement in Antimicrobial Immune Defense. Apidologie 2005, 36, 3–14. [Google Scholar] [CrossRef]
- Christophides, G.K.; Zdobnov, E.; Barillas-Mury, C.; Birney, E.; Blandin, S.; Blass, C.; Brey, P.T.; Collins, F.H.; Danielli, A.; Dimopoulos, G.; et al. Immunity-Related Genes and Gene Families in Anopheles gambiae. Science 2002, 298, 159–165. [Google Scholar] [CrossRef]
- Liu, J.; Smagghe, G.; Swevers, L. Transcriptional Response of BmToll9-1 and RNAi Machinery Genes to Exogenous dsRNA in the Midgut of Bombyx mori. J. Insect Physiol. 2013, 59, 646–654. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Zhang, X.; Chen, X.; Cao, P.; Beerntsen, B.T.; Ling, E. BmToll9, an Arthropod Conservative Toll, Is Likely Involved in the Local Gut Immune Response in the Silkworm, Bombyx mori. Dev. Comp. Immunol. 2010, 34, 93–96. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, X.; He, Y.; Shuai, J.; Chen, X.; Ling, E. Expression of Antimicrobial Peptide Genes in Bombyx mori Gut Modulated by Oral Bacterial Infection and Development. Dev. Comp. Immunol. 2010, 34, 1191–1198. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Kolliopoulou, A.; Smagghe, G.; Swevers, L. Modulation of the Transcriptional Response of Innate Immune and RNAi Genes upon Exposure to dsRNA and LPS in Silkmoth-Derived Bm5 Cells Overexpressing BmToll9-1 Receptor. J. Insect Physiol. 2014, 66, 10–19. [Google Scholar] [CrossRef]
- Liu, J.; Yang, W.; Liao, W.; Huang, Y.; Chen, W.; Bu, X.; Huang, S.; Jiang, W.; Swevers, L. Immunological Function of Bombyx Toll9-2 in the Silkworm (Bombyx mori) Larval Midgut: Activation by Escherichia coli/Lipopolysaccharide and Regulation of Growth. Arch. Insect Biochem. Physiol. 2024, 116, e22130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, X.; Zhang, J.; Li, Y.; Wang, Y.; Song, Y.; Ren, F.; Yi, H.; Deng, X.; Zhong, Y.; et al. Toll9 from Bombyx mori Functions as a Pattern Recognition Receptor That Shares Features with Toll-like Receptor 4 from Mammals. Proc. Natl. Acad. Sci. USA 2021, 118, e2103021118. [Google Scholar] [CrossRef]
- Wang, G.; Xia, Q.; Cheng, D.; Duan, J.; Zhao, P.; Chen, J.; Zhu, L. Reference Genes Identified in the Silkworm Bombyx mori during Metamorphism Based on Oligonucleotide Microarray and Confirmed by qRT-PCR. Insect Sci. 2008, 15, 405–413. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Liang, Y.; Wang, T.; Yang, W.; Chen, Z.; Li, Q.; Swevers, L.; Liu, J. Silencing of the Immune Gene BmPGRP-L4 in the Midgut Affects the Growth of Silkworm (Bombyx mori) Larvae. Insect Mol. Biol. 2023, 32, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Lin, Y.; He, Y.; Li, Q.; Chen, W.; Lin, Q.; Swevers, L.; Liu, J. BmPGPR-L4 Is a Negative Regulator of the Humoral Immune Response in the Silkworm Bombyx mori. Arch. Insect Biochem. Physiol. 2024, 115, e22093. [Google Scholar] [CrossRef]
- Yu, B.; Sang, Q.; Pan, G.; Li, C.; Zhou, Z. A Toll-Spätzle Pathway in the Immune Response of Bombyx mori. Insects 2020, 11, 586. [Google Scholar] [CrossRef]
- Terenius, O.; Papanicolaou, A.; Garbutt, J.S.; Eleftherianos, I.; Huvenne, H.; Kanginakudru, S.; Albrechtsen, M.; An, C.; Aymeric, J.-L.; Barthel, A.; et al. RNA Interference in Lepidoptera: An Overview of Successful and Unsuccessful Studies and Implications for Experimental Design. J. Insect Physiol. 2011, 57, 231–245. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Keesey, I.W.; Hansson, B.S.; Knaden, M. Gut Microbiota Affects Development and Olfactory Behavior in Drosophila melanogaster. J. Exp. Biol. 2019, 222, jeb.192500. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.; Tang, J.; Ding, G.; Mashilingi, S.K.; Huang, J.; An, J. Gut Microbiota Is a Potential Factor in Shaping Phenotypic Variation in Larvae and Adults of Female Bumble Bees. Front. Microbiol. 2023, 14, 1117077. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Yao, Z.; Raza, M.F.; Cai, Z.; Zhang, H. Regulatory Mechanisms of Microbial Homeostasis in Insect Gut. Insect Sci. 2021, 28, 286–301. [Google Scholar] [CrossRef] [PubMed]
- Belvin, M.P.; Anderson, K.V. A Conserved Signaling Pathway: The Drosophila Toll-Dorsal Pathway. Annu. Rev. Cell Dev. Biol. 1996, 12, 393–416. [Google Scholar] [CrossRef]
- Shin, S.W.; Bian, G.; Raikhel, A.S. A Toll Receptor and a Cytokine, Toll5A and Spz1C, Are Involved in Toll Antifungal Immune Signaling in the Mosquito Aedes aegypti. J. Biol. Chem. 2006, 281, 39388–39395. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, Y.; Wang, Y.; Li, X.; Yang, R.S.; Yu, Z.; Qin, L. The Toll Signaling Pathway in the Chinese Oak Silkworm, Antheraea pernyi: Innate Immune Responses to Different Microorganisms. PLoS ONE 2016, 11, e0160200. [Google Scholar] [CrossRef]
- Aronstein, K.; Murray, K.D.; Saldivar, E. Transcriptional Responses in Honey Bee Larvae Infected with Chalkbrood Fungus. BMC Genom. 2010, 11, 391. [Google Scholar] [CrossRef]
- He, Y.J.; Lu, G.; Qi, Y.; Zhang, Y.; Zhang, X.; Huang, H.; Zhuo, J.; Sun, Z.; Yan, F.; Chen, J.; et al. Activation of Toll Immune Pathway in an Insect Vector Induced by a Plant Virus. Front. Immunol. 2021, 11, 613957. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhou, L.; Chen, F.; Peng, Y.; Lu, Z. Peptidoglycan Recognition Protein-S5 Functions as a Negative Regulator of the Antimicrobial Peptide Pathway in the Silkworm, Bombyx mori. Dev. Comp. Immunol. 2016, 61, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, W.; Xu, J.; Yang, W.; Li, Q.; Zhong, Y.; Cao, Y.; Yu, X.; Deng, X. Regulation of Antimicrobial Peptide Genes via Insulin-like Signaling Pathway in the Silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2018, 103, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Qie, X.; Yan, X.; Wang, W.; Liu, Y.; Zhang, L.; Hao, C.; Lu, Z.; Ma, L. Serpin-4 Negatively Regulates Prophenoloxidase Activation and Antimicrobial Peptide Synthesis in the Silkworm, Bombyx mori. Int. J. Mol. Sci. 2023, 25, 313. [Google Scholar] [CrossRef]
- Zhang, K.; Shen, L.; Wang, X.; Yang, H.; Zhang, X.; Pan, G.; Li, C.; Ji, H.; Abbas, M.N.; Li, C.; et al. Scavenger Receptor C Regulates Antimicrobial Peptide Expression by Activating Toll Signaling in Silkworm, Bombyx mori. Int. J. Biol. Macromol. 2021, 191, 396–404. [Google Scholar] [CrossRef]
- Ooi, J.Y.; Yagi, Y.; Hu, X.; Ip, Y.T. The Drosophila Toll-9 Activates a Constitutive Antimicrobial Defense. EMBO Rep. 2002, 3, 82–87. [Google Scholar] [CrossRef]
- Bettencourt, R.; Tanji, T.; Yagi, Y.; Ip, Y.T. Toll and Toll-9 in Drosophila Innate Immune Response. J. Endotoxin Res. 2004, 10, 261–268. [Google Scholar] [CrossRef]


| Gene | Accession Number | Primer Sequence (5′-3′) |
|---|---|---|
| Primer for qPCR | ||
| BmToll9-1 | PP496203 | F: CGCAGACCGTTGAGTACATG |
| R: CCAGACTGTCGTACCTTGGT | ||
| BmTIF4A | DQ443290 | F: TTCGTACTGGCTCTTCTCGT |
| R: CAAAGTTGATAGCAATTCCCT | ||
| BmTIF3s4 | DQ443238 | F: ACTTCAAGTTCAGGGCAGAT |
| R: TTAATTGTTTTGTGGAGGCT | ||
| Signaling | ||
| BmMyD88 | XM_028186400 | F: AACGGTCACGACTCGAACTC |
| R: TCTGCCCAGATTCTTCATCC | ||
| BmTube | XM_028173146 | F: GGCAGAAAGTTATGGCTTGG |
| R: ATCCTCAAATGCTCGCTGTT | ||
| BmPelle | XM_028182154 | F: ACATCAAGCCGGCTAACATC |
| R: ACCGTGAGACCTTCAGATGC | ||
| BmCactus | XM_028180230 | F: ACAGTCGTGCGTACATTTGG |
| R: CAGCCTCTCCCTATCGTCAA | ||
| BmRel | XM_028175224 | F: TCGAATACATCCCGGACTTC |
| R: TGGAAGGTCCTTTCTTGCTC | ||
| BmTollip-v | XM_028186930 | F: TGCTACTTCTGACGGTGTGG |
| R: AGGGCCACTTTGTGGTACTG | ||
| BmPellino | XM_028184930 | F: AGAGTCGCTCAGCACAACAA |
| R: CAATGTGGCTCCACACAGAT | ||
| BmTRAF2 | XM_028172769 | F: TCGCTCCTATGGGCATAACT |
| R: CCGCATGTTGTGATTACTGG | ||
| BmECSIT | XM_028171307 | F: ATGCCGCCTTAGCTAGAATG |
| R: GCCTTTGGGCAGTACGTCTA | ||
| Effector | ||
| BmAttacin1 (BmAtt1) | NM_001043541 | F: CAGTGAACTCGGATGGAACC R: GGCGCTGAGTACGTTCTTGT |
| BmCecropinA (BmCecA) | NM_001043997 | F: CCGTCATAGGGCAAGCGAAA R: AGCAATGACTGTGGTATGTCAA |
| BmCecropinD (BmCecD) | NM_001043368 | F: CTCCCGGCAACTTCTTCAA R: TTTGCCAGGGTGTCGACT |
| BmCecropinE (BmCecE) | XM_028187757 | F: CGGAACCGAGATGGAAGATT R: TGGTCCAGCCTTGATTATCC |
| BmDefensin (BmDef) | AB_367525 | F: GTTAAGTGCGGCGTTGACTG R: TGACAGGGAAAGTGGAAGGG |
| BmGloverin1 (BmGlv1) | AB_289654 | F: GCTGGGATAGAAGCATCAGC R: ACATCAGGCCTTCTGTGACC |
| BmGloverin2 (BmGlv2) | NM_001044218 | F: GGCTTACGGTACCAGGGTTT R: TGGCTTGTGCATTCTTGTTC |
| BmGloverin3 (BmGlv3) | NM_001099842 | F: GGCCAACAAGAACGCACAAG R: ATCAAGATCCCACACACCGG |
| BmGloverin4 (BmGlv4) | AB_289657 | F: GCAGATCTGGAATGACAGCA R: GTGACCGAATTCCTTCGAGA |
| BmMoricin (BmMor) | AB_006915 | F: TGTGGCAATGTCTCTGGTGT R: CTGGCGATATTGATGGCTCT |
| BmMoricin-like (BmMorLB) | HQ204039 | F: TCACTACATCTTCATACGCGAC R: TAGTTTATGTATTGGTTGTAGT |
| BmLebocin3 (BmLeb3) | NM_001126260 | F: CTCGATCCAAACCGAAGGTA R: CGGCTGGTCAAGTCCAGTAT |
| BmEnbocin (BmEnb) | FJ373019 | F: ACCTCGCACAACTAGTTCGG R: CCAACAGAACAAACCCACTCG |
| BmLysozyme (BmLys) | NM_001043983 | F: TAACGGCTCGAAGGACTACG R: GAGGTCGGAGCACTTAACGT |
| Lysozyme-like protein (BmLLP3) | XM_012696687 | F: GTTTAATCGAGCAGGGCAGC R: CACCCTTGCGACCTTCTTTG |
| BmPhenoloxidase inhibitor (BmPOI) | XR_001139981 | F: GGATACGTGACTGGAAATGCA R: GTCATAATCCACGGGTTTGTCC |
| Prophenoloxidase 1 (BmPPO1) | AF_178462 | F: AGTGGGAAGCCATTCTCCTT R: GCCAGGTTTCACTCCTTGAG |
| Prophenoloxidase 2 (BmPPO2) | XM_028180711 | F: CCATTCTTCTACCGCTGGCA R: CGGGTTCTCGAGCTCAGATC |
| Nitric oxide synthase1 (BmNOS1) | XM_012689821 | F: TCATCACCACTAGCGCATCC R: CCTTGTCCGTTCTGTGTCCT |
| Nitric oxide synthase2 (BmNOS2) | XM_012690766 | F: ACAACAGACGCCACATCCAT R: AAATTCGGGTAAGCGCTCGA |
| Primer for dsRNA synthesis | ||
| T7-BmToll9-1 | F: TAATACGACTCACTATAGG ACTATAGGCACAGGTCGGGT | |
| R: TAATACGACTCACTATAGG TCGTTGTCCCATTCGCTGAT | ||
| T7-GFP | F: TAATACGACTCACTATAGG TACGGCGTGCAGTGCT | |
| R: TAATACGACTCACTATAGG TGATCGCGCTTCTCG | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Chen, W.; Situ, J.; Li, J.; Chen, J.; Lai, M.; Huang, F.; Li, B. BmToll9-1 Is a Positive Regulator of the Immune Response in the Silkworm Bombyx mori. Insects 2024, 15, 643. https://doi.org/10.3390/insects15090643
Liu J, Chen W, Situ J, Li J, Chen J, Lai M, Huang F, Li B. BmToll9-1 Is a Positive Regulator of the Immune Response in the Silkworm Bombyx mori. Insects. 2024; 15(9):643. https://doi.org/10.3390/insects15090643
Chicago/Turabian StyleLiu, Jisheng, Weijian Chen, Jinrong Situ, Jiaxuan Li, Jiahua Chen, Minchun Lai, Fengyi Huang, and Baoqi Li. 2024. "BmToll9-1 Is a Positive Regulator of the Immune Response in the Silkworm Bombyx mori" Insects 15, no. 9: 643. https://doi.org/10.3390/insects15090643
APA StyleLiu, J., Chen, W., Situ, J., Li, J., Chen, J., Lai, M., Huang, F., & Li, B. (2024). BmToll9-1 Is a Positive Regulator of the Immune Response in the Silkworm Bombyx mori. Insects, 15(9), 643. https://doi.org/10.3390/insects15090643






