Predicting the Potential Global Distribution of the Plum Fruit Moth Grapholita funebrana Treitscheke Using Ensemble Models
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
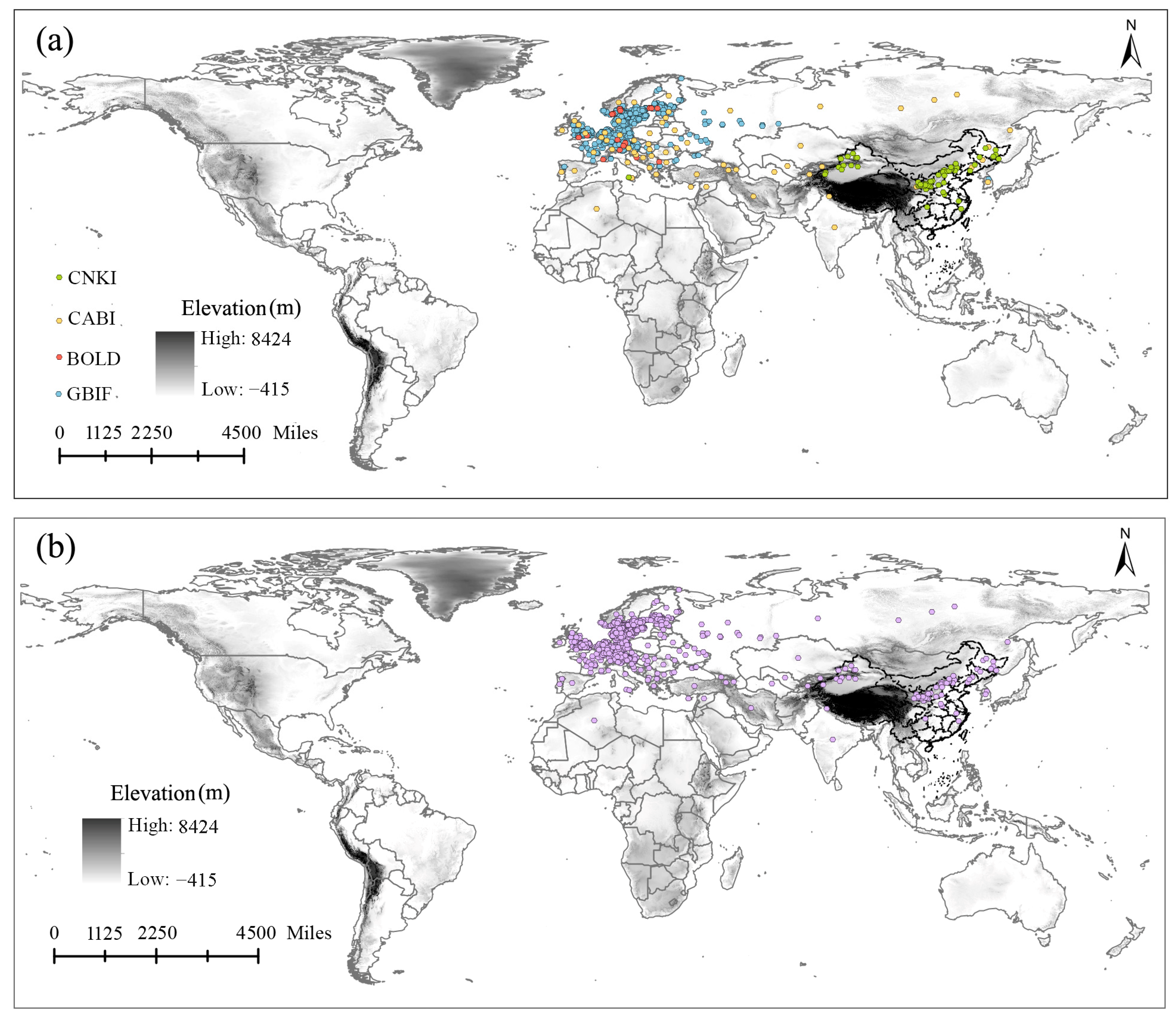
2.1. Occurrence Records
2.2. Environmental Variables
2.3. Model Fitting
2.4. Model Evaluation and Analyses
3. Results
3.1. Model Selection and Evaluation
3.2. The Current Potential Distributions under the Effects of Two Variable Combinations
3.3. Future Potential Distribution and Change Dynamics
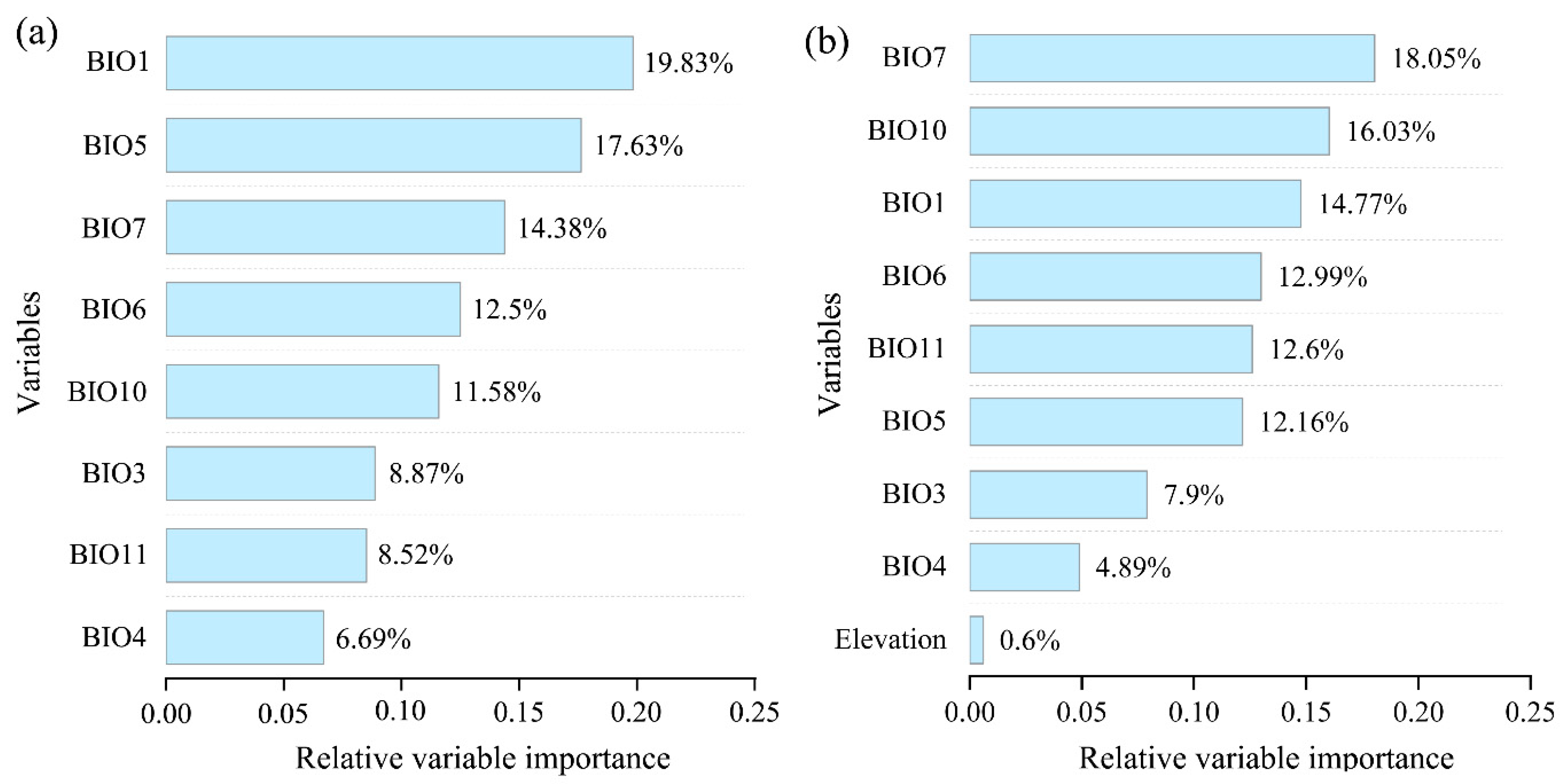
3.4. The Importance of Variables in Modeling
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Macfadyen, S.; McDonald, G.; Hill, M.P. From species distributions to climate change adaptation: Knowledge gaps in managing invertebrate pests in broad-acre grain crops. Agric. Ecosyst. Environ. 2018, 253, 208–219. [Google Scholar] [CrossRef]
- Barbet-Massin, M.; Jetz, W. A 40-year, continent-wide, multispecies assessment of relevant climate predictors for species distribution modelling. Divers. Distrib. 2014, 20, 1285–1295. [Google Scholar] [CrossRef]
- Parmesan, C. Influences of species, latitudes and methodologies on estimates of phenological response to global warming. Glob. Chang. Biol. 2007, 13, 1860–1872. [Google Scholar] [CrossRef]
- Elith, J.; Leathwick, J.R. Species distribution models: Ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 677–697. [Google Scholar] [CrossRef]
- Zhu, G.P.; Liu, G.Q.; Bu, W.J.; Gao, Y.B. Ecological niche modelling and its applications in biodiversity conservation. Biodivers. Sci. 2013, 21, 90–98. [Google Scholar]
- Tarroso, P.; Carvalho, S.B.; Brito, J.C. Simapse—Simulation maps for ecological niche modelling. Methods Ecol. Evol. 2012, 3, 787–791. [Google Scholar] [CrossRef]
- Drake, J.M.; Randin, C.; Guisan, A. Modelling ecological niches with support vector machines. J. Appl. Ecol. 2006, 43, 424–432. [Google Scholar] [CrossRef]
- Stockwell, D.; Peters, D. The GARP modelling system: Problems and solutions to automated spatial prediction. Int. J. Geogr. Inf. Sci. 1998, 13, 143–158. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modelling of species geographical distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef]
- Araújo, M.B.; New, M. Ensemble forecasting of species distributions. Trends Ecol. Evol. 2007, 22, 42–47. [Google Scholar] [CrossRef]
- Duffy, G.A.; Coetzee, B.W.; Latombe, G.; Akerman, A.H.; McGeoch, M.A.; Chown, S.L. Barriers to globally invasive species are weakening across the Antarctic. Divers. Distrib. 2017, 23, 982–996. [Google Scholar] [CrossRef]
- Li, W.B.; Teng, Y.; Zhang, M.Y.; Shen, Y.; Liu, J.W.; Qi, J.W.; Wang, X.C.; Wu, R.F.; Li, J.H.; Garber, P.A.; et al. Human activity and climate change accelerate the extinction risk to non-human primates in China. Glob. Chang. Biol. 2024, 30, e17114. [Google Scholar] [CrossRef]
- Ran, W.W.; Luo, G.M.; Zhao, Y.Q.; Li, C.; Dietrich, C.H.; Song, Y.H. Climate change may drive the distribution of tribe Zyginelline pests in China and the Indo-China Peninsula to shift towards higher latitude river-mountain systems. Pest Manag. Sci. 2024, 80, 613–626. [Google Scholar] [CrossRef]
- Song, H.; Zhang, X.; Wang, X.; Wang, Y.; Li, S.; Xu, Y. Not the expected poleward migration: Impact of climate change scenarios on the distribution of two endemic evergreen broad-leaved Quercus species in China. Sci. Total Environ. 2023, 889, 164273. [Google Scholar] [CrossRef]
- Tabor, J.A.; Koch, J.B. Ensemble models predict invasive bee habitat suitability will expand under future climate scenarios in Hawai’i. Insects 2021, 12, 443. [Google Scholar] [CrossRef]
- Zhao, H.X.; Xian, X.Q.; Yang, N.W.; Zhang, Y.J.; Liu, H.; Wan, F.H.; Guo, J.Y.; Liu, W.X. Insights from the biogeographic approach for biocontrol of invasive alien pests: Estimating the ecological niche overlap of three egg parasitoids against Spodoptera frugiperda in China. Sci. Total Environ. 2023, 862, 160785. [Google Scholar]
- Marchioro, C.A.; Krechemer, F.S. Prevention is better than cure: Integrating habitat suitability and invasion threat to assess global biological invasion risk by insect pests under climate change. Pest Manag. Sci. 2021, 77, 4510–4520. [Google Scholar] [CrossRef]
- Marchioro, C.A.; Krechemer, F.S. Reconstructing the biological invasion of Tuta absoluta: Evidence of niche shift and its consequences for invasion risk assessment. J. Pest Sci. 2024, 97, 127–141. [Google Scholar] [CrossRef]
- Rossi, J.P.; Rasplus, J.Y. Climate change and the potential distribution of the glassy-winged sharpshooter (Homalodisca vitripennis), an insect vector of Xylella fastidiosa. Sci. Total Environ. 2023, 860, 160375. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Peng, L.; He, Z.; Lu, Y.; Wang, F. Potential distribution of two invasive pineapple pests under climate change. Pest Manag. Sci. 2020, 76, 1652–1663. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Wang, Z.; Ding, W.; Xu, K.; Li, L.; Wang, Y.; Li, J.; Yang, M.; Liu, X.; et al. Modelling the current and future potential distribution of the bean bug Riptortus pedestris with increasingly serious damage to soybean. Pest Manag. Sci. 2022, 78, 4340–4352. [Google Scholar] [CrossRef] [PubMed]
- Komai, F. A taxonomic revision of the genus Grapholita and allied genera (Lepidoptera: Tortricidae) in the Palaearctic Region. Entomol. Scand. Suppl. 1999, 55, 110. [Google Scholar]
- Dueñas-López, M.A. Grapholita funebrana (Plum Fruit Moth); CABI Digital Library: Wallingford, UK, 2023. [Google Scholar] [CrossRef]
- Rizzo, R.; Farina, V.; Saiano, F.; Lombardo, A.; Ragusa, E.; Lo Verde, G. Do Grapholita funebrana infestation rely on specific plum fruit features? Insects 2019, 10, 444. [Google Scholar] [CrossRef]
- Jackson, D.M. Searching behavior and survival of 1st-instar codling moth. Ann. Entomol. Soc. Am. 1982, 75, 284–289. [Google Scholar] [CrossRef]
- Lo Verde, G.; Guarino, S.; Barone, S.; Rizzo, R. Can mating disruption be a possible route to control plum fruit moth in mediterranean environments? Insects 2020, 11, 589. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, R.; Lo Verde, G.; Lombardo, A. Effectiveness of spinosad and mineral oil for control of Grapholita funebrana Treitschke in organic plum orchards. New Medit 2012, 11, 70–72. [Google Scholar]
- CAPS. Grapholita funebrana. Cooperative Agricultural Pest Survey. CAPS Pest Datasheet. Purdue Universit. 2013. Available online: https://approvedmethods.ceris.purdue.edu/sheet/114 (accessed on 10 January 2023).
- Van der Geest, L.P.S.; Evenhuis, H.H. Tortricid Pests: Their Biology, Natural Enemies and Control; Elsevier Science Publishers: Amsterdam, The Netherlands, 1991; pp. xviii + 808. [Google Scholar]
- Negahban, M.; Sedaratian-Jahromi, A.; Ghanee-Jahromi, M.; Haghani, M. Monitoring of an Iranian population of Grapholita funebrana Treitschke, 1835 (Lepidoptera: Tortricidae) using sex pheromone traps: An applicable procedure for sustainable management. Entomofauna 2016, 37, 241–252. [Google Scholar]
- Ghizdavu, J. Test on control of the plum moth, Grapholitha funebrana Tr., using the synthetic sex attractant. (Essais de lutteconte le carpocapse des prunes, Grapholitha funebrana Tr., par l’emploi de l’attractif sexuel synthetique). Rev. Roum. Biol. Biol. Animale. 1982, 27, 145–154. [Google Scholar]
- Chang, J.; Pu, C.L.; Wang, Y.Z.; Zheng, R.T. Study on occurrence and control of Grapholita funebrana Treitschke in apricot orchard. Chin. J. Agric. Sci. 1991, 6, 31–33. [Google Scholar]
- Yang, L. The Species and Distribution of Fruit Borers in Xinjiang; Shihezi University: Shihezi, China, 2017. [Google Scholar]
- Zheng, Y.; Wu, R.X.; Dorn, S.; Chen, M.H. Diversity of tortricid moths in apple orchards: Evidence for a cryptic species of Grapholita (Lepidoptera: Tortricidae) from China. Bull. Entomol. Res. 2017, 107, 268–280. [Google Scholar] [CrossRef]
- Cao, L.J.; Song, W.; Chen, J.C.; Fan, X.L.; Hoffmann, A.A.; Wei, S.J. Population genomic signatures of the oriental fruit moth related to the Pleistocene climates. Commun. Biol. 2022, 5, 142. [Google Scholar] [CrossRef]
- Kirk, H.; Dorn, S.; Mazzi, D. Worldwide population genetic structure of the oriental fruit moth (Grapholita molesta), a globally invasive pest. BMC Ecol. 2013, 13, 12. [Google Scholar] [CrossRef] [PubMed]
- Torriani, M.V.; Mazzi, D.; Hein, S.; Dorn, S. Structured populations of the oriental fruit moth in an agricultural ecosystem. Mol. Ecol. 2010, 19, 2651–2660. [Google Scholar] [CrossRef] [PubMed]
- Wan, F.; Yin, C.; Tang, R.; Chen, M.; Wu, Q.; Huang, C.; Qian, W.; Rota-Stabelli, O.; Yang, N.; Wang, S.; et al. A chromosome-level genome assembly of Cydia pomonella provides insights into chemical ecology and insecticide resistance. Nat. Commun. 2019, 10, 4237. [Google Scholar] [CrossRef] [PubMed]
- EPPO. Grapholita funebrana (LASPFU). In EPPO Global Database; European and Mediterranean Plant Protection Organization: Paris, France, 2023; Available online: https://gd.eppo.int/taxon/LASPFU (accessed on 10 October 2023).
- Sillero, N.; Arenas-Castro, S.; Enriquez-Urzelai, U.; Vale, C.G.; Sousa-Guedes, D.; Martínez-Freiría, F.; Real, R.; Barbosa, A.M. Want to model a species niche? A step-by-step guideline on correlative ecological niche modelling. Ecol. Model. 2021, 456, 109671. [Google Scholar] [CrossRef]
- Thibaud, E.; Petitpierre, B.; Broennimann, O.; Davison, A.C.; Guisan, A. Measuring the relative effect of factors affecting species distribution model predictions. Meth. Ecol. Evol. 2014, 5, 947–955. [Google Scholar] [CrossRef]
- Aiello-Lammens, M.E.; Boria, R.A.; Radosavljevic, A.; Vilela, B.; Anderson, R.P. spThin: An r package for spatial thinning of species occurrence records for use in ecological niche models. Ecography 2015, 38, 541–545. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudík, M.; Elith, J.; Graham, C.H.; Lehmann, A.; Leathwick, J.; Ferrier, S. Sample selection bias and presence-only distribution models: Implications for background and pseudo-absence data. Ecol. Appl. 2009, 19, 181–197. [Google Scholar] [CrossRef]
- Zhu, G.P.; Liu, Q.; Gao, Y.B. Improving ecological niche model transferability to predict the potential distribution of invasive exotic species. Biodivers. Sci. 2014, 22, 223–230. [Google Scholar]
- Gaul, W.; Sadykova, D.; White, H.J.; León-Sánchez, L.; Caplat, P.; Emmerson, M.C.; Yearsley, J.M. Modelling the distribution of rare invertebrates by correcting class imbalance and spatial bias. Divers. Distrib. 2022, 28, 2171–2186. [Google Scholar] [CrossRef]
- Fick, S.E.; Hijmans, R.J. Worldclim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Dormann, C.F.; Elith, J.; Bacher, S.; Buchmann, C.; Carl, G.; Carré, G.; Jaime, R.; Marquéz, G.; Gruber, B.; Lafourcade, B.; et al. Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography 2013, 36, 27–46. [Google Scholar] [CrossRef]
- Wei, T.Y.; Simko, V. R Package ‘Corrplot’: Visualization of a Correlation Matrix (Version 0.92). 2021. Available online: https://github.com/taiyun/corrplot (accessed on 10 November 2023).
- Mendlik, T.; Gobiet, A. Selecting climate simulations for impact studies based on multivariate patterns of climate change. Clim. Chang. 2016, 135, 381–393. [Google Scholar] [CrossRef]
- Moss, R.H.; Edmonds, J.A.; Hibbard, K.A.; Manning, M.R.; Rose, S.K.; van Vuuren, D.P.; Carter, T.R.; Emori, S.; Kainuma, M.; Kram, T.; et al. The next generation of scenarios for climate change research and assessment. Nature 2010, 463, 747–756. [Google Scholar] [CrossRef]
- Naimi, B.; Araujo, M.B. sdm: A reproducible and extensible R platform for species distribution modelling. Ecography 2016, 39, 368–375. [Google Scholar] [CrossRef]
- Nelder, J.A.; Wedderburn, R.W. Generalized linear models. J. R. Stat. Soc. A Stat. 1972, 135, 370–384. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R. Generalized additive models: Some applications. J. Am. Stat. Assoc. 1987, 398, 371–386. [Google Scholar] [CrossRef]
- Busby, J.R. BIOCLIM—A bioclimate analysis and prediction system. Plant Prot. Q. Aust. 1991, 6, 8–9. [Google Scholar]
- Friedman, J.H. Multivariate adaptive regression splines. Ann. Statist. 1991, 19, 1–67. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Buja, A. Flexible discriminant analysis by optimal scoring. J. Am. Stat. Assoc. 1994, 89, 1255–1270. [Google Scholar] [CrossRef]
- Hearst, M.A.; Osuna, S.T.; Platt, J.; Scholkopf, B. Support vector machines. IEEE Intell. Syst. App. 1998, 13, 18–28. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Reinhartz-Berger, I. Towards automatization of domain modelling. Data Knowl. Eng. 2010, 69, 491–515. [Google Scholar] [CrossRef]
- Loh, W.Y. Classification and regression trees. Wires Data Min. Knowl. 2011, 1, 14–23. [Google Scholar] [CrossRef]
- Royle, J.A.; Chandler, R.B.; Yackulic, C.; Nichols, J.D. Likelihood analysis of species occurrence probability from presence-only data for modelling species distributions. Methods Ecol. Evol. 2012, 3, 545–554. [Google Scholar] [CrossRef]
- Engebretsen, S.; Bohlin, J. Statistical predictions with glmnet. Clin. Epigenet. 2019, 11, 123. [Google Scholar] [CrossRef]
- Barbet-Massin, M.; Jiguet, F.; Albert, C.H.; Thuiller, W. Selecting pseudo-absences for species distribution models: How, where and how many? Methods Ecol. Evol. 2012, 3, 327–338. [Google Scholar] [CrossRef]
- Lobo, J.M.; Jiménez-Valverde, A.; Real, R. AUC: A misleading measure of the performance of predictive distribution models. Glob. Ecol. Biogeogr. 2008, 172, 145–151. [Google Scholar] [CrossRef]
- Allouche, O.; Tsoar, A.; Kadmon, R. Assessing the accuracy of species distribution models: Prevalence, kappa and the true skill statistic (TSS). J. Appl. Ecol. 2006, 43, 1223–1232. [Google Scholar] [CrossRef]
- Peterson, A.T.; Papes, M.; Soberon, J. Rethinking receiver operating characteristic analysis applications in ecological niche modelling. Ecol. Model. 2008, 213, 63–72. [Google Scholar] [CrossRef]
- Zhu, G.P.; Fan, J.Y.; Wang, M.L.; Chen, M.; Qiao, H.J. The importance of the shape of receiver operating characteristic (ROC) curve in ecological niche model evaluation—Case study of Hlyphantria cunea. J. Biosaf. 2017, 26, 184–190. [Google Scholar]
- Pearce, J.; Ferrier, S. An evaluation of alternative algorithms for fitting species distribution models using logistic regression. Ecol. Model. 2000, 128, 127–147. [Google Scholar] [CrossRef]
- Liu, T.; Liu, H.; Tong, J.; Yang, Y. Habitat suitability of neotenic net-winged beetles (Coleoptera: Lycidae) in China using combined ecological models, with implications for biological conservation. Divers. Distrib. 2022, 28, 2806–2823. [Google Scholar] [CrossRef]
- Zhang, K.; Yao, L.; Meng, J.; Tao, J. Maxent modelling for predicting the potential geographical distribution of two peony species under climate change. Sci. Total Environ. 2018, 634, 1326–1334. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Data Analysis; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; pp. 189–201. [Google Scholar]
- Li, H. Microlepidoptera of Qinling Moutains (Insecta: Lepidoptera); Beijing Science Press: Beijing, China, 2012. [Google Scholar]
- Karsholt, O.; Razowski, J. (Eds.) The Lepidoptera of Europe: A Distributional Checklist; Apollo Books: Stenstrup, Denmark, 1996; 380p. [Google Scholar]
- Charmillot, P.J.; Vallier, R.; Tagini-Rosset, S. Plum fruit moth:(Grapholitha funebrana Tr.): A study of the cycle of development in relation to temperature sums and observations on the activity of adult moths. Mitt. Schweiz. Entomol. Ges. 1979, 52, 19–33. [Google Scholar]
- Danilevsky, A.S.; Kuznetsov, V.I. Insecta—Lepidoptera, 5(1): Tortricidae, tribe Laspeyresiini. (Nasekomye Cheshuekrylye, 5(1): Listovertki Tortricidae; Triba plodozhorki Laspeyresiini). Fauna SSSR, Novaya Seriya, 98; Akademia Nauk SSSR: Leningrad, Russia, 1968; 635p. [Google Scholar]
- Venette, R.C.; Davis, E.E.; DaCosta, M.; Heisler, H.; Larson, M. Mini Risk Assessment: Plum Fruit Moth, Cydia funebrana (Treitschke) [Lepidoptera: Tortricidae]; Department of Entomology, University of Minnesota: St. Paul, MN, USA, 2003; 25p. [Google Scholar]
- Sciarretta, A.; Trematerra, P.; Baumgärtner, J. Geostatistical analysis of Cydia funebrana (Lepidoptera: Tortricidae) pheromone trap catches at two spatial scales. Am. Entomol. 2001, 47, 174–181. [Google Scholar] [CrossRef]
- Liu, B.; Gao, X.; Ma, J.; Jiao, Z.; Xiao, J.; Hayat, M.A.; Wang, H. Modelling the present and future distribution of arbovirus vectors Aedes aegypti and Aedes albopictus under climate change scenarios in Mainland China. Sci. Total Environ. 2019, 664, 203–214. [Google Scholar] [CrossRef]
- Aidoo, O.F.; Souza, P.G.C.; Silva, R.S.; Júnior, P.A.S.; Picanço, M.C.; Heve, W.K.; Duker, R.Q.; Ablormeti, F.K.; Sétamou, M.; Borgemeister, C. Modelling climate change impacts on potential global distribution of Tamarixia radiata Waterston (Hymenoptera: Eulophidae). Sci. Total Environ. 2023, 864, 160962. [Google Scholar] [CrossRef]
- Liu, T.; Wang, J.; Hu, X.; Feng, J. Land-use change drives present and future distributions of fall armyworm, Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae). Sci. Total Environ. 2020, 706, 135872. [Google Scholar] [CrossRef]
- Santana, P.A., Jr.; Kumar, L.; Da Silva, R.S.; Pereira, J.L.; Picanco, M.C. Assessing the impact of climate change on the worldwide distribution of Dalbulus maidis (DeLong) using MaxEnt. Pest Manag. Sci. 2019, 75, 2706–2715. [Google Scholar] [CrossRef]
- Bebber, D.P.; Ramotowski, M.A.T.; Gurr, S.J. Crop pests and pathogens move polewards in a warming world. Nat. Clim. Chang. 2013, 3, 985–988. [Google Scholar] [CrossRef]
- Trisos, C.H.; Merow, C.; Pigot, A.L. The projected timing of abrupt ecological disruption from climate change. Nature 2020, 580, 496–501. [Google Scholar] [CrossRef] [PubMed]
- Bell, D.M.; Bradford, J.B.; Lauenroth, W.K. Early indicators of change: Divergent climate envelopes between tree life stages imply range shifts in the western United States. Glob. Ecol. Biogeogr. 2014, 23, 168–180. [Google Scholar] [CrossRef]
- Molinari, F. Notes on the biology and monitoring of Cydia funebrana (Treitschke). Bull. OILB/SROP 1995, 18, 39–42. [Google Scholar]
- Vernon, J.D.R. Observations on the biology and control of the plum fruit moth. Plant Pathol. 1971, 20, 106–110. [Google Scholar] [CrossRef]
- Butturini, A.; Tiso, R.; Molinari, F. Phenological forecasting model for Cydia funebrana. Bull. OEPP 2000, 30, 131–136. [Google Scholar] [CrossRef]
- Rauleder, H. Observations on the biology of the plum fruit moth (Cydia funebrana). Gesunde Pflanz. 2002, 54, 241–248. [Google Scholar]
- Azrag, A.G.A.; Pirk, C.W.; Yusuf, A.A.; Pinard, F.; Niassy, S.; Mosomtai, G.; Babin, R. Prediction of insect pest distribution as influenced by elevation: Combining field observations and temperature dependent development models for the coffee stink bug, Antestiopsis thunbergii (Gmelin). PLoS ONE 2018, 13, 18. [Google Scholar] [CrossRef]
- Smith, A.B.; Santos, M.J. Testing the ability of species distribution models to infer variable importance. Ecography 2020, 43, 801–813. [Google Scholar] [CrossRef]
- Meier, N.F. Results of the Work of the Laboratory of Biological Control at the Institute for Plant Protection; Lenin Academy of Agricultural Science: Leningrad, Russia, 1941; pp. 121–124. [Google Scholar]
- Wiackowski, S.K.; Wiackowska, I. Biological, microbiological and chemical control of the plum moth Laspeyresia funebrana Tr. (Lep. Tortricidae) in Poland. Entomophaga 1966, 11, 261–267. [Google Scholar] [CrossRef]
- Qu, Y.; Chen, X.; Monticelli, L.S.; Zhang, F.; Desneux, N.; Huijie, D.; Ramirez-Romero, R.; Wang, S. Parasitism performance of the parasitoid Trichogramma dendrolimi on the plum fruit moth Grapholitha funebrana. Entomol. Gen. 2020, 1, 40. [Google Scholar]
- Hassan, S.A. The mass rearing and utilization of Trichogramma to control lepidopterous pests: Achievements and outlook. Pestic. Sci. 1993, 37, 387–391. [Google Scholar] [CrossRef]
- Augustin, S.; Boonham, N.; De Kogel, W.J.; Donner, P.; Faccoli, M.; Lees, D.C.; Marini, L.; Mori, N.; Petrucco, T.E.; Quilici, S.; et al. A review of pest surveillance techniques for detecting quarantine pests in Europe. EPPO Bull. 2012, 42, 515–551. [Google Scholar] [CrossRef]
- Yokomi, R.; Delgado, J.K.; Unruh, T.R.; Barcenas, N.M.; Garczynski, S.F.; Walse, S.; Pérez de León, A.A.; Cooper, W.R. Molecular advances in larval fruit moth identification to facilitate fruit export from Western united states under systems approaches. Ann. Entomol. Soc. Am. 2022, 115, 105–112. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Huo, Y.; Wang, L.; Wang, J.; Zuo, S.; Pang, C.; Wang, Z.; Zhang, H.; Xu, K.; Ma, K. Predicting the Potential Global Distribution of the Plum Fruit Moth Grapholita funebrana Treitscheke Using Ensemble Models. Insects 2024, 15, 663. https://doi.org/10.3390/insects15090663
Yang M, Huo Y, Wang L, Wang J, Zuo S, Pang C, Wang Z, Zhang H, Xu K, Ma K. Predicting the Potential Global Distribution of the Plum Fruit Moth Grapholita funebrana Treitscheke Using Ensemble Models. Insects. 2024; 15(9):663. https://doi.org/10.3390/insects15090663
Chicago/Turabian StyleYang, Mingsheng, Yiqi Huo, Lei Wang, Jialu Wang, Shichao Zuo, Chaoyun Pang, Zhengbing Wang, Hongfei Zhang, Kedong Xu, and Keshi Ma. 2024. "Predicting the Potential Global Distribution of the Plum Fruit Moth Grapholita funebrana Treitscheke Using Ensemble Models" Insects 15, no. 9: 663. https://doi.org/10.3390/insects15090663
APA StyleYang, M., Huo, Y., Wang, L., Wang, J., Zuo, S., Pang, C., Wang, Z., Zhang, H., Xu, K., & Ma, K. (2024). Predicting the Potential Global Distribution of the Plum Fruit Moth Grapholita funebrana Treitscheke Using Ensemble Models. Insects, 15(9), 663. https://doi.org/10.3390/insects15090663





