Additive Insecticidal Effects of Chitosan/dsRNA Nanoparticles Targeting V-ATPaseD and Emamectin Benzoate–Lufenuron Formulations Against Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Insect Rearing and Maintenance
2.2. Insecticide and Reagents
2.3. Total RNA Isolation and cDNA Synthesis
2.4. Quantitative Real-Time PCR Analysis
2.5. Synthesis of dsRNA
2.6. RNAi by Injection
2.7. Preparation of Chitosan/dsRNA Nanocomplexes
2.8. RNAi by Feeding
2.9. RNAi by Topical Delivery
2.10. Bioassays with Chemical Insecticides
2.11. Combined Exposure to dsRNA Nanocomplex and Chemical Pesticide
2.12. Data Analysis
3. Results
3.1. RNAi of V-ATPaseD via dsRNA Injection
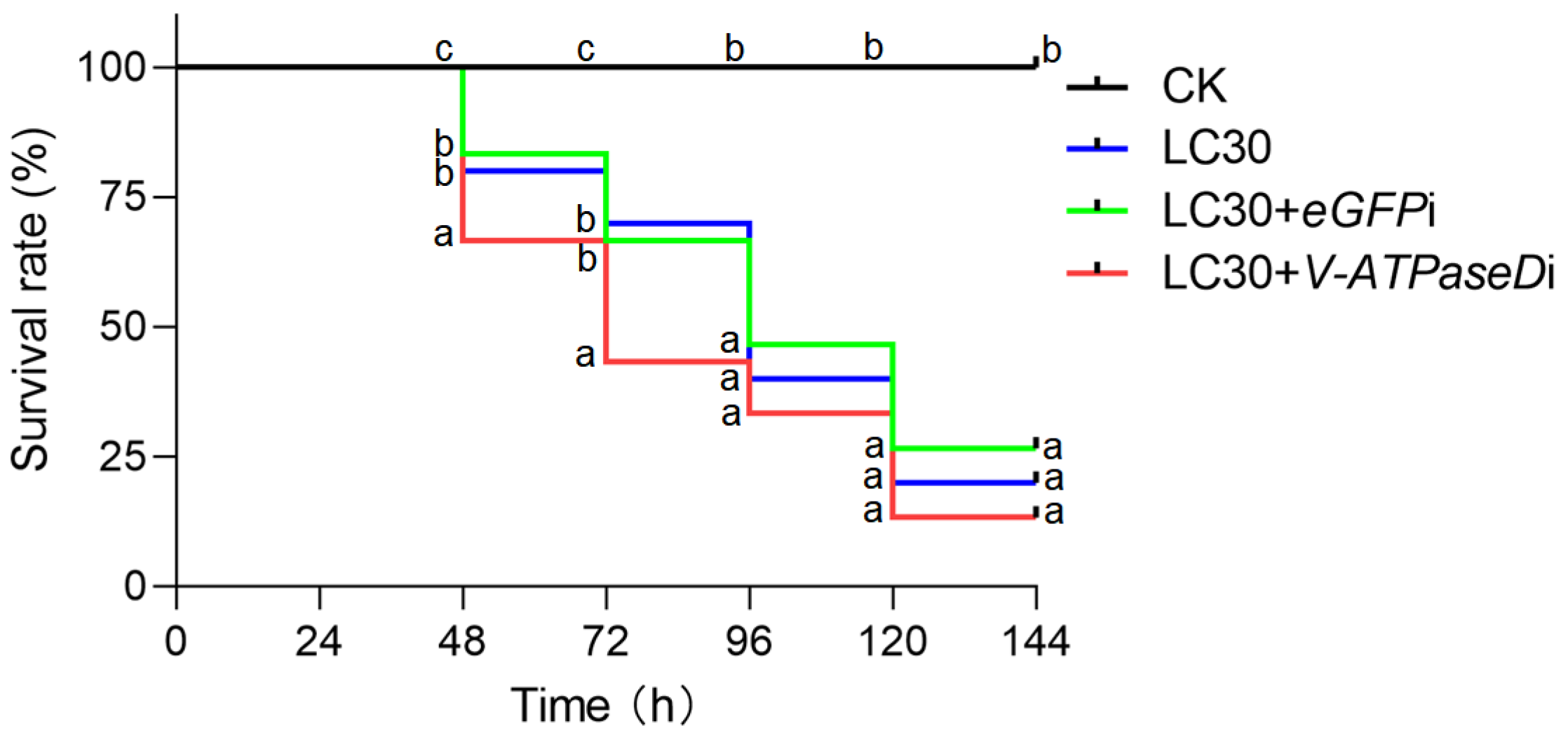
3.2. RNAi of V-ATPaseD via Topical Administration of Chitosan/dsRNA Nanocomplexes
3.3. RNAi of V-ATPaseD via Feeding Chitosan/dsRNA Nanocomplexes
3.4. Mortality and Growth Inhibition by Emamectin Benzoate–Lufenuron on S. frugiperda
3.5. Additive Effects of Chitosan/dsRNA-V-ATPaseD Nanocomplex and Emamectin Benzoate-Lufenuron Against S. frugiperda
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ashley, T.R.; Wiseman, B.R.; Davis, F.M.; Andrews, K.L. The fall armyworm: A bibliography. Fla. Entomol. 1989, 72, 152–202. [Google Scholar] [CrossRef]
- Montezano, D.G.; Specht, A.; Sosa-Gómez, D.R.; Roque-Specht, V.F.; Sousa-Silva, J.C.; Paula-Moraes, S.V.; Hunt, J.A.P.T.E. Host Plants of Spodoptera frugiperda (Lepidoptera: Noctuidae) in the Americas. Afr. Entomol. 2018, 26, 286–300. [Google Scholar] [CrossRef]
- Rane, R.; Walsh, T.K.; Lenancker, P.; Gock, A.; Dao, T.H.; Nguyen, V.L.; Khin, T.N.; Amalin, D.; Chittarath, K.; Faheem, M.; et al. Complex multiple introductions drive fall armyworm invasions into Asia and Australia. Sci Rep 2023, 13, 660. [Google Scholar] [CrossRef] [PubMed]
- Ntow, W.J.; Gijzen, H.J.; Kelderman, P.; Drechsel, P. Farmer perceptions and pesticide use practices in vegetable production in Ghana. Pest Manag. Sci. 2006, 62, 356–365. [Google Scholar] [CrossRef]
- Molina-Ochoa, J.; Carpenter, J.E.; Heinrichs, E.A.; Foster, J.E. Parasitoids and parasites of Spodoptera frugiperda (Lepidoptera: Noctuidae) in the Americas and Caribbean Basin: An inventory. Fla. Entomol. 2003, 86, 254–289. [Google Scholar] [CrossRef]
- Wan, J.; Huang, C.; Li, C.Y.; Zhou, H.X.; Ren, Y.L.; Li, Z.Y.; Xing, L.S.; Zhang, B.; Qiao, X.; Liu, B.; et al. Biology, invasion and management of the agricultural invader: Fall armyworm, Spodoptera frugiperda (Lepidoptera: Noctuidae). J. Integr. Agric. 2021, 20, 646–663. [Google Scholar] [CrossRef]
- Dandan, Z.W. The bioassay of Chinese domestic Bt-Cry1Ab and Bt-(Cry1Ab+Vip3Aa) maize against the fall armyworm, Spodoptera Frugiperda. Plant Prot. 2019, 45, 54–60. [Google Scholar]
- Silva, A.; Silva, L.; Malaquias, J.; Salustino, A.; Neto, D.; Pacheco, D.M.; Fragoso, D.; Pereira, E. Susceptibility of Fall Armyworm Field Populations to Vip3Aa/Cry Bt Maize in a Tropical Agricultural Region. Agronomy 2024, 14, 451. [Google Scholar] [CrossRef]
- Gurusamy, D.; Mogilicherla, K.; Palli, S. Chitosan nanoparticles help double-stranded RNA escape from endosomes and improve RNA interference in the fall armyworm, Spodoptera Frugiperda. Arch. Insect Biochem. Physiol. 2020, 104, e21677. [Google Scholar] [CrossRef]
- Ramesh kumar, D.; Gurusamy, D.; Palli, S. Protamine–Lipid–dsRNA Nanoparticles Improve RNAi Efficiency in the Fall Armyworm, Spodoptera Frugiperda. J. Agric. Food Chem. 2022, 70, 6634–6643. [Google Scholar]
- Rodrigues, T.B.; Mishra, S.K.; Sridharan, K.; Barnes, E.R.; Alyokhin, A.; Tuttle, R.; Kokulapalan, W.; Garby, D.; Skizim, N.J.; Tang, Y.-W.; et al. First Sprayable Double-Stranded RNA-Based Biopesticide Product Targets Proteasome Subunit Beta Type-5 in Colorado Potato Beetle (Leptinotarsa decemlineata). Front. Plant Sci. 2021, 12, 728652. [Google Scholar]
- Qadri, M.; Short, S.; Gast, K.; Hernandez, J.; Wong, A.C.-N. Microbiome Innovation in Agriculture: Development of Microbial Based Tools for Insect Pest Management. Front. Sustain. Food Syst. 2020, 4, 547751. [Google Scholar]
- Ma, Z.; Zheng, Y.; Chao, Z.; Chen, H.; Zhang, Y.; Yin, M.; Shen, J.; Yan, S. Visualization of the process of a nanocarrier-mediated gene delivery: Stabilization, endocytosis and endosomal escape of genes for intracellular spreading. J. Nanobiotechnology 2022, 20, 124. [Google Scholar]
- Gong, C.; Hasnain, A.; Wang, Q.; Liu, D.; Xu, Z.; Zhan, X.; Liu, X.; Pu, J.; Sun, M.; Wang, X. Eco-friendly deacetylated chitosan base siRNA biological-nanopesticide loading cyromazine for efficiently controlling Spodoptera Frugiperda. Int. J. Biol. Macromol. 2023, 241, 124575. [Google Scholar]
- Lyu, Z.; Chen, J.; Lyu, J.; Guo, P.; Liu, J.; Liu, J.; Zhang, W. Spraying double-stranded RNA targets UDP-N-acetylglucosamine pyrophosphorylase in the control of Nilaparvata Lugens. Int. J. Biol. Macromol. 2024, 271, 132455. [Google Scholar]
- Lv, B.; Zhang, X.; Wang, Y.; Wu, W.; Li, D.; Hu, Z. Discovery of the Chlorinated and Ammoniated Derivatives of Vanillin as Potential Insecticidal Candidates Targeting V-ATPase: Structure-Based Virtual Screening, Synthesis, and Bioassay. J. Agric. Food Chem. 2024, 72, 20872–20881. [Google Scholar]
- Jungreis, A.M.; Vaughan, G.L. Insensitivity of lepidopteran tissues to ouabain: Absence of ouabain binding and Na+ K+ ATPases in larval and adult midgut. J. Insect Physiol. 1977, 23, 503–509. [Google Scholar] [CrossRef]
- Forgac, M. Vacuolar ATPases: Rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 2007, 8, 917–929. [Google Scholar]
- Chang, Y.W.; Wang, Y.-C.; Zhang, X.-X.; Iqbal, J.; Du, Y.-Z. RNA Interference of Genes Encoding the Vacuolar-ATPase in Liriomyza trifolii. Insects 2021, 12, 41. [Google Scholar] [CrossRef]
- Martinez, Z.; De Schutter, K.; Van Damme, E.J.M.; Vogel, E.; Wynant, N.; Broeck, J.V.; Christiaens, O.; Smagghe, G. Accelerated delivery of dsRNA in lepidopteran midgut cells by a Galanthus nivalis lectin (GNA)-dsRNA-binding domain fusion protein. Pestic. Biochem. Physiol. 2021, 175, 104853. [Google Scholar]
- Wang, X.; Zhao, D.; Wang, Q.; Liu, Y.; Lu, X.; Guo, W. Identification and Functional Analysis of V-ATPaseA and C Genes in Hyphantria cunea. Insects 2024, 15, 515. [Google Scholar] [CrossRef] [PubMed]
- Köhler, H.R.; Triebskorn, R. Wildlife Ecotoxicology of Pesticides: Can We Track Effects to the Population Level and Beyond? Science 2013, 341, 759–765. [Google Scholar] [PubMed]
- Benowitz, K.M.; Allan, C.W.; Degain, B.A.; Li, X.; Fabrick, J.A.; Tabashnik, B.E.; Carrière, Y.; Matzkin, L.M.; Long, A. Novel genetic basis of resistance to Bt toxin Cry1Ac in Helicoverpa zea. Genet. 2022, 221, iyac037. [Google Scholar]
- Zuo, Y.; Wang, Z.; Ren, X.; Pei, Y.; Aioub, A.A.A.; Hu, Z.; Guedes, R.N.C. Evidence for Multiple Origins of Knockdown Resistance (kdr) in Spodoptera exigua (Hübna) (Lepidoptera: Noctuidae) From China. J. Econ. Entomol. 2022, 115, 1279–1284. [Google Scholar] [PubMed]
- Wang, Y.; Fu, Y.; Cheng, H.; Zhao, C.; Huang, Q.; Chang, M.; Qiu, W.; Shen, Y.; Li, D. lncR26319/miR-2834/EndophilinA axis regulates oogenesis of the silkworm, Bombyx mori. Insect Sci 2023, 30, 65–80. [Google Scholar]
- Zhou, L.; Meng, J.U.; Ruan, H.U.; Yang, C.I.; Zhang, C.U. Expression stability of candidate RT-PCR housekeeping genes in Spodoptera frugiperda (Lepidoptera: Noctuidae). Arch. Insect Biochem. Physiol. 2021, 108, e21831. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar]
- Sandal, S.; Singh, S.; Bansal, G.; Kaur, R.; Mogilicherla, K.; Pandher, S.; Roy, A.; Kaur, G.; Rathore, P.; Kalia, A. Nanoparticle-Shielded dsRNA Delivery for Enhancing RNAi Efficiency in Cotton Spotted Bollworm Earias vittella (Lepidoptera: Nolidae). Int. J. Mol. Sci. 2023, 24, 9161. [Google Scholar] [CrossRef]
- Gentleman, R.; Ihaka, R. R: A Language and Environment for Statistical Computing. Computing 2011, 1, 12–21. [Google Scholar]
- Ripley, B. Support Functions and Datasets for Venables and Ripley’s MASS [R package MASS Version 7.3-45]. R Package Version. 2015. Available online: https://cran.r-project.org/web/packages/MASS/index.html (accessed on 27 February 2025).
- Collins, M.; Forgac, M. Regulation and function of V-ATPases in physiology and disease. Biochim. Biophys. Acta (BBA) Biomembr. 2020, 1862, 183341. [Google Scholar]
- Eaton, A.; Merkulova, M.; Brown, D. The H+-ATPase (V-ATPase): From proton pump to signaling complex in health and disease. Am. J. Physiol. Cell Physiol. 2020, 320, C392–C414. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Z.; Wang, Z.; Zhang, L.-Z.; Zeng, Z.-J. A Comparison of RNA Interference via Injection and Feeding in Honey Bees. Insects 2022, 13, 928. [Google Scholar] [CrossRef] [PubMed]
- Qiao, H.; Zhao, J.; Wang, X.; Xiao, L.; Zhu-Salzman, K.; Lei, J.; Xu, D.; Xu, G.; Tan, Y.; Hao, D. An oral dsRNA delivery system based on chitosan induces G protein-coupled receptor kinase 2 gene silencing for Apolygus lucorum control. Pestic. Biochem. Physiol. 2023, 194, 105481. [Google Scholar]
- Kalliola, S.; Repo, E.; Sillanpää, M.; Arora, J.; He, J.; John, V. The stability of green nanoparticles in increased pH and salinity for applications in oil spill-treatment. Colloids Surf. A Physicochem. Eng. Asp. 2016, 493, 99–107. [Google Scholar] [CrossRef]
- Kolge, H.; Kadam, K.; Galande, S.; Lanjekar, V.; Ghormade, V. New Frontiers in Pest Control: Chitosan Nanoparticles-Shielded dsRNA as an Effective Topical RNAi Spray for Gram Podborer Biocontrol. ACS Appl. Bio Mater. 2021, 4, 5145–5157. [Google Scholar] [CrossRef]
- Christiaens, O.; Tardajos, M.; Martinez, Z.; Dash, M.; Dubruel, P.; Smagghe, G. Increased RNAi Efficacy in Spodoptera exigua via the Formulation of dsRNA With Guanylated Polymers. Front. Physiol. 2018, 9, 316. [Google Scholar] [CrossRef]




| Gene | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Utility |
|---|---|---|---|
| V-ATPaseD-dsRNA (MT707617.1) | GAAATTAATACGACTCACTATAGGCCTCCAAGTGAGGTTCCGTA | GAAATTAATACGACTCACTATAGGCAACTCGACCAGCAGTTTCA | Primers for synthesis of V-ATPaseD dsRNA |
| eGFP-dsRNA (MH070103.1) | GAAATTAATACGACTCACTATAGGGTACGGCGTGCAGTGCT | GAAATTAATACGACTCACTATAGGGTGATCGCGCTTCTCG | Primers for synthesis of eGFP dsRNA |
| V-ATPaseD (MT707617.1) | TCGCTTACATCATCTCCG | AACAGCAGGTCCTCGTCA | qRT-PCR primers for V-ATPaseD |
| β-actin (MN044625.1) | GATGTCGGGACGGGATA | TCATACGGCGAGTGCTT | qRT-PCR primers for β-actin |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, S.; Li, Z.; Zhao, X.; Zhang, D.; Ayra-Pardo, C.; Kan, Y.; Li, D. Additive Insecticidal Effects of Chitosan/dsRNA Nanoparticles Targeting V-ATPaseD and Emamectin Benzoate–Lufenuron Formulations Against Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae). Insects 2025, 16, 348. https://doi.org/10.3390/insects16040348
Guo S, Li Z, Zhao X, Zhang D, Ayra-Pardo C, Kan Y, Li D. Additive Insecticidal Effects of Chitosan/dsRNA Nanoparticles Targeting V-ATPaseD and Emamectin Benzoate–Lufenuron Formulations Against Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae). Insects. 2025; 16(4):348. https://doi.org/10.3390/insects16040348
Chicago/Turabian StyleGuo, Shigang, Zhongwei Li, Xuhui Zhao, Donghai Zhang, Camilo Ayra-Pardo, Yunchao Kan, and Dandan Li. 2025. "Additive Insecticidal Effects of Chitosan/dsRNA Nanoparticles Targeting V-ATPaseD and Emamectin Benzoate–Lufenuron Formulations Against Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae)" Insects 16, no. 4: 348. https://doi.org/10.3390/insects16040348
APA StyleGuo, S., Li, Z., Zhao, X., Zhang, D., Ayra-Pardo, C., Kan, Y., & Li, D. (2025). Additive Insecticidal Effects of Chitosan/dsRNA Nanoparticles Targeting V-ATPaseD and Emamectin Benzoate–Lufenuron Formulations Against Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae). Insects, 16(4), 348. https://doi.org/10.3390/insects16040348







