β-1,3-Glucan Recognition Protein Can Inhibit the Proliferation of Bombyx mori Cytoplasmic Polyhedrosis Virus
Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Silkworms, Cell Culture, and Virus Propagation
2.2. Tissue Sample Collection for RNA-Seq Analysis
2.3. RNA Extraction and RNA-Seq Analysis
2.4. Quantitative Real-Time PCR (qPCR)
2.5. Construction of Overexpression Plasmids
2.6. dsRNA Synthesis
2.7. Cell Transfection
2.8. BmCPV Virion Purification
2.9. Overexpression of BmβGRP-3 and BmCPV Infection in BmN Cells
2.10. Construction of BmβGRP-3 Stable Overexpression Cell Line (BmN-βGRP)
2.11. RNA Interference (RNAi) of BmβGRP-3 and BmCPV Infection
2.12. Western Blotting Analysis
2.13. Statistical Analysis
3. Result
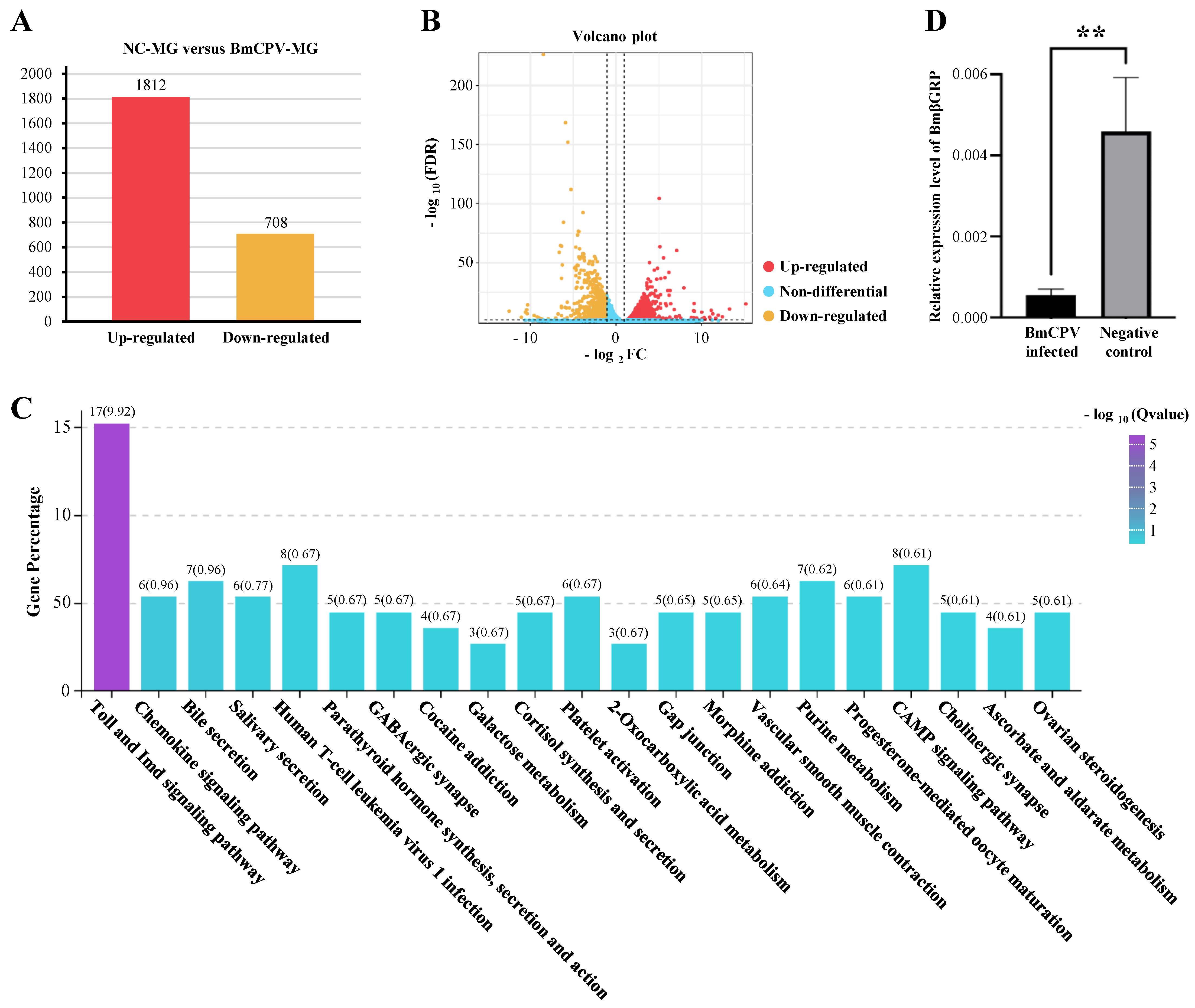
3.1. RNA-Seq, Data Analysis, and Validation
3.2. Overexpression of BmβGRP Suppresses BmCPV Proliferation in BmN Cells
3.3. Stable Overexpression of BmβGRP in BmN Cells Reduces Early Intracellular Proliferation of BmCPV Virus
3.4. Knock Down of BmβGRP Promotes BmCPV Proliferation in BmN Cells
3.5. BmβGRP Can Activate the ProPO Cascade, Toll Pathway, and Relish-Mediated Immune Cascade
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ochiai, M.; Ashida, M. Purification of a beta-1,3-glucan recognition protein in the prophenoloxidase activating system from hemolymph of the silkworm, Bombyx mori. J. Biol. Chem. 1988, 263, 12056–12062. [Google Scholar] [CrossRef]
- Chen, K.; Lu, Z. Immune responses to bacterial and fungal infections in the silkworm, Bombyx mori. Dev. Comp. Immunol. 2018, 83, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Xia, Y. Beta-1,3-Glucan recognition protein (betaGRP) is essential for resistance against fungal pathogen and opportunistic pathogenic gut bacteria in Locusta migratoria manilensis. Dev. Comp. Immunol. 2012, 36, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Takahasi, K.; Ochiai, M.; Horiuchi, M.; Kumeta, H.; Ogura, K.; Ashida, M.; Inagaki, F. Solution structure of the silkworm betaGRP/GNBP3 N-terminal domain reveals the mechanism for beta-1,3-glucan-specific recognition. Proc. Natl. Acad. Sci. USA 2009, 106, 11679–11684. [Google Scholar] [CrossRef] [PubMed]
- Kanagawa, M.; Satoh, T.; Ikeda, A.; Adachi, Y.; Ohno, N.; Yamaguchi, Y. Structural insights into recognition of triple-helical beta-glucans by an insect fungal receptor. J. Biol. Chem. 2011, 286, 29158–29165. [Google Scholar] [CrossRef]
- Jiang, H.; Ma, C.; Lu, Z.Q.; Kanost, M.R. Beta-1,3-glucan recognition protein-2 (betaGRP-2) from Manduca sexta; an acute-phase protein that binds beta-1,3-glucan and lipoteichoic acid to aggregate fungi and bacteria and stimulate prophenoloxidase activation. Insect Biochem. Mol. Biol. 2004, 34, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Fuchs, J.F.; Infanger, L.C.; Rocheleau, T.A.; Hillyer, J.F.; Chen, C.C.; Christensen, B.M. Mosquito innate immunity: Involvement of beta 1,3-glucan recognition protein in melanotic encapsulation immune responses in Armigeres subalbatus. Mol. Biochem. Parasitol. 2005, 139, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Rocheleau, T.A.; Fuchs, J.F.; Christensen, B.M. Beta 1,3-glucan recognition protein from the mosquito, Armigeres subalbatus, is involved in the recognition of distinct types of bacteria in innate immune responses. Cell Microbiol. 2006, 8, 1581–1590. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Kanost, M.R. A beta1,3-glucan recognition protein from an insect, Manduca sexta, agglutinates microorganisms and activates the phenoloxidase cascade. J. Biol. Chem. 2000, 275, 7505–7514. [Google Scholar] [CrossRef] [PubMed]
- Fabrick, J.A.; Baker, J.E.; Kanost, M.R. Innate immunity in a pyralid moth: Functional evaluation of domains from a beta-1,3-glucan recognition protein. J. Biol. Chem. 2004, 279, 26605–26611. [Google Scholar] [CrossRef] [PubMed]
- Mertz, B.; Gu, X.; Reilly, P.J. Analysis of functional divergence within two structurally related glycoside hydrolase families. Biopolymers 2009, 91, 478–495. [Google Scholar] [CrossRef]
- Davies, G.; Henrissat, B. Structures and mechanisms of glycosyl hydrolases. Structure 1995, 3, 853–859. [Google Scholar] [CrossRef]
- Henrissat, B.; Bairoch, A. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 1996, 316 Pt 2, 695–696. [Google Scholar] [CrossRef]
- Henrissat, B.; Bairoch, A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1993, 293 Pt 3, 781–788. [Google Scholar] [CrossRef]
- Henrissat, B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991, 280 Pt 2, 309–316. [Google Scholar] [CrossRef]
- Gottar, M.; Gobert, V.; Matskevich, A.A.; Reichhart, J.M.; Wang, C.; Butt, T.M.; Belvin, M.; Hoffmann, J.A.; Ferrandon, D. Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell 2006, 127, 1425–1437. [Google Scholar] [CrossRef]
- Manabe, N.; Yamaguchi, Y. 3D Structural Insights into beta-Glucans and Their Binding Proteins. Int. J. Mol. Sci. 2021, 22, 1578. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Wang, Y.; Guo, X.; Hartson, S.; Jiang, H. A pattern recognition serine proteinase triggers the prophenoloxidase activation cascade in the tobacco hornworm, Manduca sexta. J. Biol. Chem. 2004, 279, 34101–34106. [Google Scholar] [CrossRef] [PubMed]
- Fabrick, J.A.; Baker, J.E.; Kanost, M.R. cDNA cloning; purification; properties, and function of a beta-1,3-glucan recognition protein from a pyralid moth, Plodia interpunctella. Insect Biochem. Mol. Biol. 2003, 33, 579–594. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, L.B.; Ma, Y.; Liu, Y.X.; Cao, H.H.; Wang, Y.L.; Kong, X.; Huang, Z.H.; Zhu, H.D.; Wang, Y.X.; et al. Bombyx mori beta-1,3-Glucan Recognition Protein 4 (BmbetaGRP4) Could Inhibit the Proliferation of B. mori Nucleopolyhedrovirus through Promoting Apoptosis. Insects 2021, 12, 743. [Google Scholar] [CrossRef] [PubMed]
- Rio, D.C.; Ares, M., Jr.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, 2010, pdb-prot5439. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cui, Y.; Sun, J.; Zhou, Z.H. Multiple conformations of trimeric spikes visualized on a non-enveloped virus. Nat. Commun. 2022, 13, 550. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Zhang, Y.; Zhou, K.; Sun, J.; Zhou, Z.H. Conservative transcription in three steps visualized in a double-stranded RNA virus. Nat. Struct. Mol. Biol. 2019, 26, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.; Yan, J.; Wang, X.; Xie, Y.; Guo, N.; Swevers, L.; Sun, J. Peptidoglycan Recognition Protein S5 of Bombyx mori Facilitates the Proliferation of Bombyx mori Cypovirus 1. J Agric. Food Chem. 2023, 71, 6338–6347. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, J.; Kong, X.; Chen, N.; Zeng, X.; Wu, X. Bombyx mori nucleopolyhedrovirus Bm46 is essential for efficient production of infectious BV and nucleocapsid morphogenesis. Virus Res. 2020, 289, 198145. [Google Scholar] [CrossRef]
- Zhang, J.; Li, Y.; Zhao, S.; Wu, X. Identification of a functional region in Bombyx mori nucleopolyhedrovirus VP39 that is essential for nuclear actin polymerization. Virology 2020, 550, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wu, M. Pattern recognition receptors in health and diseases. Signal Transduct. Target. Ther. 2021, 6, 291. [Google Scholar] [PubMed]
- Zambon, R.A.; Nandakumar, M.; Vakharia, V.N.; Wu, L.P. The Toll pathway is important for an antiviral response in Drosophila. Proc. Natl. Acad. Sci. USA 2005, 102, 7257–7262. [Google Scholar] [CrossRef]
- Mussabekova, A.; Daeffler, L.; Imler, J.L. Innate and intrinsic antiviral immunity in Drosophila. Cell. Mol. Life Sci. 2017, 74, 2039–2054. [Google Scholar] [CrossRef]
- Seth, R.B.; Sun, L.; Chen, Z.J. Antiviral innate immunity pathways. Cell Res. 2006, 16, 141–147. [Google Scholar] [CrossRef]
- Kingsolver, M.B.; Huang, Z.; Hardy, R.W. Insect antiviral innate immunity: Pathways, effectors, and connections. J. Mol. Biol. 2013, 425, 4921–4936. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.; Jan, E.; Sarnow, P.; Schneider, D. The Imd pathway is involved in antiviral immune responses in Drosophila. PLoS ONE 2009, 4, e7436. [Google Scholar] [CrossRef]
- Zhao, P.; Lu, Z.; Strand, M.R.; Jiang, H. Antiviral, anti-parasitic, and cytotoxic effects of 5,6-dihydroxyindole (DHI), a reactive compound generated by phenoloxidase during insect immune response. Insect Biochem. Mol. Biol. 2011, 41, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.; Imler, J.L. Sensing and signalling viral infection in drosophila. Dev. Comp. Immunol. 2021, 117, 103985. [Google Scholar] [CrossRef]
- Mendez-Lopez, T.T.; Carrero, J.C.; Lanz-Mendoza, H.; Ochoa-Zarzosa, A.; Mukherjee, K.; Contreras-Garduno, J. Metabolism and immune memory in invertebrates: Are they dissociated? Front. Immunol. 2024, 15, 1379471. [Google Scholar] [CrossRef]
- Lanz-Mendoza, H.; Contreras-Garduno, J. Innate immune memory in invertebrates: Concept and potential mechanisms. Dev. Comp. Immunol. 2022, 127, 104285. [Google Scholar] [CrossRef] [PubMed]
- Lanz-Mendoza, H.; Galvez, D.; Contreras-Garduno, J. The plasticity of immune memory in invertebrates. J. Exp. Biol. 2024, 227 (Suppl. S1), jeb246158. [Google Scholar] [CrossRef] [PubMed]




| Labels of Primer | Primer Pair (5′–3′) | Note |
|---|---|---|
| VP1-qPCR-F | ACCATTAACGCTGCTGGTGA | Product’s Length: 116 bp NCBI ID: AY388398.1 |
| VP1-qPCR-R | GCTCCGGCTAGTGGTACATC | |
| RP49-qPCR-F | CAGGCGGTTCAAGGGTCAATAC | Product’s Length: 213 bp NCBI ID: AY769302.1 |
| RP49-qPCR-R | TGCTGGGCTCTTTCCACGA | |
| BmRelish-qPCR-F | TTCGGTGGAATGGGTATCAT | Product’s Length: 165 bp NCBI ID: NM_001102465.1 |
| BmRelish-qPCR-R | GCTGAACTTCAAACGCACAA | |
| BmCecA-qPCR-F | GCCCAGGTGGAAACTCTTCA | Product’s Length: 99 bp NCBI ID: NM_001043997.1 |
| BmCecA-qPCR-R | GCTTGCCCTATGACGGCTAT | |
| BmCecB-qPCR-F | ATCCTTCGTCTTCGCTCTGG | Product’s Length: 106 bp NCBI ID: D11114.1 |
| BmCecB-qPCR-R | ACGGATGTTCCTGCCCATTT | |
| BmDredd-qPCR-F | TGTGCGAGGGCACATTTTTG | Product’s Length: 176 bp NCBI ID: AB292816.1 |
| BmDredd-qPCR-R | CCAAGGGCGAGTCAGTTGTT | |
| BmFADD-qPCR-F | AACAGGTACAAACTGGCGGG | Product’s Length: 142 bp NCBI ID: NM_001202536.1 |
| BmFADD-qPCR-R | TTGTGGATCTGCTCGTTGCT | |
| BmMyd88-qPCR-F | ACACAGAATTGTTCCCAGGCT | Product’s Length: 128 bp NCBI ID: XM_004921516.5 |
| BmMyd88-qPCR-R | GGCTCTCCGATCTGAACTCG | |
| BmPPAE-qPCR-F | TGTTTGCTGTCCATGCAACG | Product’s Length: 175 bp NCBI ID: NM_001043367.1 |
| BmPPAE-qPCR-R | TGGCGCAGTAACACAGCATA | |
| BmPPO-qPCR-F | GCAGCAGCGTAATGGGAATG | Product’s Length: 148 bp NCBI ID: EU569724.1 |
| BmPPO-qPCR-R | CGGATTGTGTTGTTGCCAGG |
| Labels of Primer | Primer Pair (5′–3′) | Note |
|---|---|---|
| mCherry-F | GGCGGCCGCTCGAGTCTAGAATGGTGAGCAAGGGCGAGGA | Construct the Overexpression Plasmid pIZT-mCherry-egfp |
| mCherry-R | TTCGAACCGCGGGCCCTCTAGACGTTACTTGTACAGCTCGTCCATGC | |
| βGRP-F | AGCGAATTTAAAGCTTGGTACCGAGCTCATGTTCTTCAAAATTATTATAT | Construct the Overexpression Plasmid pIZT-βGRP-egfp |
| βGRP-R | GGAGAGGGTTAGGGATAGGCTTACCACTAGTAAGTGCATAAACTCTGACG | |
| dsRNA-Red-F | TAATACGACTCACTATAGGGAAGCTGAAGGTGACCAAGG | Synthesis dsRNA dsRed |
| dsRNA-Red-R | TAATACGACTCACTATAGGGTGGTGTAGTCCTCGTTGTGG | |
| dsRNA-βGRP-F | TAATACGACTCACTATAGGGGATAACGGCGAATGGACTGT | Synthesis dsRNA dsβGRP |
| dsRNA-βGRP-R | TAATACGACTCACTATAGGGGGTCATAACTGGCGGAAGAA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Yan, J.; Xie, Y.; Wang, X.; Ren, F.; Bian, H.; Sun, J. β-1,3-Glucan Recognition Protein Can Inhibit the Proliferation of Bombyx mori Cytoplasmic Polyhedrosis Virus. Insects 2025, 16, 431. https://doi.org/10.3390/insects16040431
Zhang Y, Yan J, Xie Y, Wang X, Ren F, Bian H, Sun J. β-1,3-Glucan Recognition Protein Can Inhibit the Proliferation of Bombyx mori Cytoplasmic Polyhedrosis Virus. Insects. 2025; 16(4):431. https://doi.org/10.3390/insects16040431
Chicago/Turabian StyleZhang, Yinong, Jiming Yan, Yukai Xie, Xiong Wang, Feifei Ren, Haixu Bian, and Jingchen Sun. 2025. "β-1,3-Glucan Recognition Protein Can Inhibit the Proliferation of Bombyx mori Cytoplasmic Polyhedrosis Virus" Insects 16, no. 4: 431. https://doi.org/10.3390/insects16040431
APA StyleZhang, Y., Yan, J., Xie, Y., Wang, X., Ren, F., Bian, H., & Sun, J. (2025). β-1,3-Glucan Recognition Protein Can Inhibit the Proliferation of Bombyx mori Cytoplasmic Polyhedrosis Virus. Insects, 16(4), 431. https://doi.org/10.3390/insects16040431






