Abstract
Ebola virus (EBOV), member of genus Ebolavirus, family Filoviridae, have a non-segmented, single-stranded RNA that contains seven genes: (a) nucleoprotein (NP), (b) viral protein 35 (VP35), (c) VP40, (d) glycoprotein (GP), (e) VP30, (f) VP24, and (g) RNA polymerase (L). All genes encode for one protein each except GP, producing three pre-proteins due to the transcriptional editing. These pre-proteins are translated into four products, namely: (a) soluble secreted glycoprotein (sGP), (b) Δ-peptide, (c) full-length transmembrane spike glycoprotein (GP), and (d) soluble small secreted glycoprotein (ssGP). Further, shed GP is released from infected cells due to cleavage of GP by tumor necrosis factor α-converting enzyme (TACE). This review presents a detailed discussion on various functional aspects of all EBOV proteins and their residues. An introduction to ebolaviruses and their life cycle is also provided for clarity of the available analysis. We believe that this review will help understand the roles played by different EBOV proteins in the pathogenesis of the disease. It will help in targeting significant protein residues for therapeutic and multi-protein/peptide vaccine development.
1. Introduction
The genus Ebolavirus belongs to the family Filoviridae and consists of six identified species. Schematic taxonomy classification of Filoviridae according to the International Committee on Taxonomy of Viruses (ICTV) is presented in Figure 1 [1,2]. Recently, a new genus, Dianlovirus, has been proposed by Yang and co-workers, including the virus circulating in Chinese bats, named Měnglà virus (MLAV) (Figure 1) [3,4]. According to the current terminology, the Ebola virus (EBOV) is a Zaire ebolavirus species in the Ebolavirus genus. Most of the members of the genus Ebolavirus, except Reston virus (RESTV) and Bombali virus (BOMV), cause severe and frequently fatal hemorrhagic fever in humans and non-human primates (NHPs). In contrast, BOMV infects bats exclusively while RESTV is known to be pathogenic for humanized mice, though no human infection has been detected as yet [1,2,5].
Figure 1.
Taxonomical classification of ebolaviruses.
EBOV has a thread-like shape virion, which can be changed to circular or filamentous [6]. The filamentous shape could appear as long, short, branched, unbranched, or forming “6” and “U” configurations [7]. The viral genome is 19 kb long, linear, non-segmented negative sense (NNS), single-stranded RNA, encoding seven genes [8,9] (Table 1). Each gene, except GP, contains a single open reading frame (ORF). In contrast, the GP gene consists of three overlapping ORFs [10,11]. During the assembly, viral RNA forms a ribonucleoprotein (RNP) complex with NP, L, VP30, VP35 and VP24 [12], which appears as a helical nucleocapsid (NC) [13,14]. NC protects the viral RNA from degradation by endonucleases and hosts immune response [14,15].

Table 1.
Ebola virus genes and their products.
In the last 45 years, ebolavirus outbreaks with varying fatalities have been documented mainly in Africa, resulting in over 15,200 deaths [16]. The rising frequency of episodes has led to better disease management measures and vaccine development efforts worldwide [17,18,19,20,21]. On 19 December 2019, the Ervebo vaccine, based on recombinant vesicular stomatitis virus (VSV) expressing EBOV GP, received approval from the Food and Drug Administration (FDA) as the first licensed vaccine against EBOV [22,23]. However, multiple obstacles, such as high frequency side effects, difficulties to manufacture, high cost, low immunogenicity, and lack of a global outreach, interfere with efficacy of EBOV outbreak control [24,25]. This review will provide a comprehensive analysis of all EBOV proteins functions and enlist the protein residues involved. This review will identify therapeutic and multi-protein/peptide vaccine development targets by understanding viral proteins’ role in the replication cycle.
2. Ebola Virus Life Cycle
A schematic depiction of the viral life cycle is presented in Figure 2.
Figure 2.
A simple diagrammatic representation of various steps in the EBOV life cycle. 1. Attachment—EBOV can interact with different host cell receptors, and none of the receptors is indispensable for attachment. In the Figure, DC-SIGN receptor is shown as an example. 2. Uptake—Uptake mainly occurs by micropinocytosis, as shown, though other methods such as clathrin-mediated endocytosis and caveolin-mediated endocytosis are also contemplated. 3. Entry—GP1 proteolysis inside endosome enables viral interaction with obligate host receptor cholesterol transporter Niemann-Pick C1 (NPC1; shown in red color). 4. Release—After membrane fusion, the viral genome is released in the host cell cytoplasm. 5. Transcription and Replication—Primary transcription occurs in the host cell cytoplasm followed by a translation. Antigenome is used as a template for synthesis of progeny genomes. 6. Transport—Various proteins are transported near the plasma membrane. 7. Assembly and Budding—VP40 plays a crucial role in assembly, virus-like particle (VLP) formation and budding.
2.1. Attachment and Entry
To enter the host cell, EBOV can use several attachment factors, such as human folate receptor- α [38], β1 integrins [39], TYRO3 receptor tyrosine kinase family members [40], T-cell immunoglobulin, and mucin domain 1 (TIM1) [41]. Additionally, various lectins, such as dendritic cell-specific intercellular adhesion molecule-3-grabbing non-integrin (DC-SIGN), liver/lymph node-specific ICAM-3 grabbing non-integrin (L- SIGN), and human macrophage galactose and N-acetylgalactosamine-specific C-type lectin (hMGL) [42,43,44] were shown to serve as entry receptors. None of these receptors are indispensable for EBOV attachment [45,46,47], which could include explainability of the virus to target various cell types.
Upon binding to the receptor, EBOV enters the host cells via three mechanisms: (a) Macropinocytosis [48], (b) Clathrin-mediated endocytosis [49,50], and (c) caveolin-mediated endocytosis [51]. The internalization mechanism appears related to the shape of the virus. Currently, macropinocytosis is believed to be the primary uptake mechanism [52,53,54], while a combination of different mechanisms is also suggested [55,56,57,58].
EBOV GP consists of two subunits, GP1 and GP2 (discussed later). After uptake, proteolysis of EBOV GP1 appears significant for viral entry [47]. The mechanism of proteolysis varies depending on the host cell type [59] and can be carried out by cathepsin B, cathepsin L [60,61], as well as thermolysin [62]. This EBOV GP1 proteolysis is essential for viral interaction with the obligate host receptor cholesterol transporter Niemann-Pick C1 (NPC1), a step critical for viral entry [63,64]. This interaction initiates the fusion of viral and host cell membrane, leading to the release of viral RNP into the cytoplasm (Figure 2) [46].
2.2. Transcription and Replication
NP encapsidates both, filoviral genome, as well as anti-genome [65]. NP associated RNA acts as a template for viral RNA transcription and replication [66]. Primary transcription and translation take place in the host cell cytoplasm (Figure 2). Accumulation of NP and other EBOV proteins results in the formation of inclusion bodies, which serve as additional sites for transcription and replication [67,68,69]. Replication is initiated at a promoter region of viral RNA that flanks the transcription initiation sequence of the first EBOV gene [70]. Many host factors, such as DNA topoisomerase I (TOP1) [71], RNA-binding protein Staufen 1 [72], and RNA splicing and export factors Nuclear RNA export factor 1 (NXF1) and TEDx-box helicase 39B (DDX39B) [73], are essential for transcription and replication. Importantly, transcription and replication is regulated by the phosphorylated state of VP30 (discussed later).
Transcriptional editing of the GP gene [29] results in the generation of three transcripts, pre-sGP, pre-GP, and pre-ssGP (Figure 3), which are translated into pre-sGP pre-GP0 and pre-ssGP, respectively [74]. Pre-sGP is cleaved post-translationally by furin into sGP and Δ-peptide [30]. Pre-GP0 forms GP post-translationally, cleaving furin into GP1 and GP2 subunits linked by disulfide bonds (Figure 3) [31,75,76]. Shed GP is released from infected cells due to cleavage of GP at aa 637 by tumor necrosis factor α-converting enzyme (TACE) (Figure 3) [77]. GP is heavily glycosylated (discussed later), a phenomenon significant for EBOV pathogenesis.
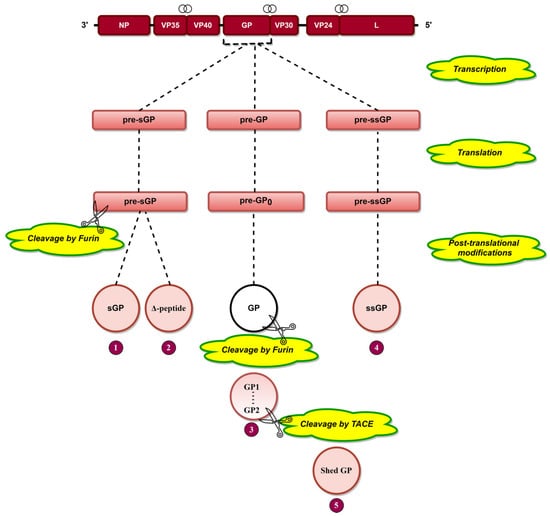
Figure 3.
The arrangement of EBOV genes is presented at the top, where the symbol  indicates the overlapping genes. Transcriptional editing of the GP gene by L protein results in three RNA transcripts: pre-sGP, pre-GP, and pre-ssGP. Translation of these three transcripts results in three pre-proteins, namely, pre-sGP, pre-GP0, and pre-ssGP, respectively. Pre-sGP is cleaved post-translationally by furin into sGP and Δ-peptide. pre-GP0 and pre-ssGP result in full-length transmembrane spike glycoprotein (GP) and soluble small-secreted glycoprotein (ssGP). GP is cleaved by furin into GP1 and GP2 subunits held together by disulfide bonds. Shed GP is released from infected cells due to cleavage of GP by tumor necrosis factor α-converting enzyme (TACE).
indicates the overlapping genes. Transcriptional editing of the GP gene by L protein results in three RNA transcripts: pre-sGP, pre-GP, and pre-ssGP. Translation of these three transcripts results in three pre-proteins, namely, pre-sGP, pre-GP0, and pre-ssGP, respectively. Pre-sGP is cleaved post-translationally by furin into sGP and Δ-peptide. pre-GP0 and pre-ssGP result in full-length transmembrane spike glycoprotein (GP) and soluble small-secreted glycoprotein (ssGP). GP is cleaved by furin into GP1 and GP2 subunits held together by disulfide bonds. Shed GP is released from infected cells due to cleavage of GP by tumor necrosis factor α-converting enzyme (TACE).
 indicates the overlapping genes. Transcriptional editing of the GP gene by L protein results in three RNA transcripts: pre-sGP, pre-GP, and pre-ssGP. Translation of these three transcripts results in three pre-proteins, namely, pre-sGP, pre-GP0, and pre-ssGP, respectively. Pre-sGP is cleaved post-translationally by furin into sGP and Δ-peptide. pre-GP0 and pre-ssGP result in full-length transmembrane spike glycoprotein (GP) and soluble small-secreted glycoprotein (ssGP). GP is cleaved by furin into GP1 and GP2 subunits held together by disulfide bonds. Shed GP is released from infected cells due to cleavage of GP by tumor necrosis factor α-converting enzyme (TACE).
indicates the overlapping genes. Transcriptional editing of the GP gene by L protein results in three RNA transcripts: pre-sGP, pre-GP, and pre-ssGP. Translation of these three transcripts results in three pre-proteins, namely, pre-sGP, pre-GP0, and pre-ssGP, respectively. Pre-sGP is cleaved post-translationally by furin into sGP and Δ-peptide. pre-GP0 and pre-ssGP result in full-length transmembrane spike glycoprotein (GP) and soluble small-secreted glycoprotein (ssGP). GP is cleaved by furin into GP1 and GP2 subunits held together by disulfide bonds. Shed GP is released from infected cells due to cleavage of GP by tumor necrosis factor α-converting enzyme (TACE).
2.3. Assembly and Budding
At the late stage of transcription, the RNP complex, GP, and VP40 are transported to the cell surface via different mechanisms. Transport of RNP employs actin [78,79], while GP is carried to the cell surface via secretory pathway, where it is glycosylated [80], as well as cleaved into GP1 and GP2 subunits [76]. VP40 trafficking requires interaction with IQ motif containing GTPase activating protein 1(IQGAP1) [81], coat protein complex II (COPII) [82], microtubules [83,84], or actin [85,86].
3. Ebola Virus Proteins and Their Functions
3.1. Nucleoprotein (NP)
EBOV NP is a multifunctional protein, contributing to NC and RNP formation [12,13,14]. It was shown that aa 1–600 (Figure 4) are crucial for NC formation and viral replication [87]. Additionally, aa 1–450 are involved in RNA encapsidation/ssRNA binding [12,13,88] and NP oligomerization [13,87,89] (Table 2). NP oligomerization facilitates NP-ssRNA interaction, which is essential for NC formation [89,90]. A recent study indicated the significance of aa 111 in NP oligomerization, viral transcription, and replication [91]. Another study highlighted the significance of NP C-terminal domain (CTD) aa 641–739 (Figure 4) information of inclusion bodies and infectious virus-like particle (VLP) production [92]. Interestingly, laboratory data indicate that only point mutations in NP and L are required for virus adaptation to different species [93].
Figure 4.
A schematic representation of EBOV nucleoprotein (NP). NP aa 1–450 are involved in RNA encapsidation, NP oligomerization, nucleocapsid (NC) as well as NP-VP35 interaction (along with aa 481–500). aa 562–567 and 581–591 consist of LxxIxE and SxPxLE motifs, respectively, interacting with different host proteins. PPxPxY motif amongst aa 600–617 is significant for NP-VP30 interaction. Interaction of these three motifs with their respective targets is significant to regulate viral transcription.

Table 2.
A summary of various critical functions performed by various EBOV proteins and their amino acids involved.
Additionally, various post-translational modifications of NP are documented. It was shown that NP undergoes O-glycosylation and sialylation, which are significant for NC formation [87,94] as they facilitate direct NP-VP35 and NP-VP24 interaction [94]. NP-VP35 interaction involves aa 1–450 of NP (Table 2) [88,90]. This interaction regulates viral RNA synthesis by chaperoning NP in a monomer state, preventing its binding to ssRNA [88,90]. Recently, NP central domain aa 481–500 (Figure 4) were also suggested to be essential for NP-VP35 interaction [92]. Additionally, NP aa 2–150 and 601–739 were shown to be involved in NP-VP40 interaction, which is significant for the recruitment of NP into VLP [95]. Additionally, the PPxPxY motif, especially aa residues 600–617 (Figure 4), is responsible for NP-VP30 interaction, which is essential for viral RNA transcription [96,97].
Recent studies indicate that NP could interact with host cell proteins to facilitate virus transcription and replication. It was shown that NP recruits host factor carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase in an RNA-independent manner to facilitate EBOV genome transcription and replication [98]. Additionally, LxxIxE motif of NP (aa 562–567) interacts with host PP2A-B56 phosphatase, which dephosphorylates NP-bound-VP30 and enables viral transcription [99]. Another NP motif, SxPxLE (aa 581–591) (Figure 4), recruits host SET and MYND domain-containing protein 3 (SMYD3), which regulates viral transcription by increasing the NP-VP30 interaction in a dose-dependent manner [100]. Additionally, Wendt et al. proposed that NP could recruit host nuclear RNA export factor 1 (NXF1), a component of nuclear mRNA export pathway, to facilitate viral mRNA transport from inclusion bodies [101]. Another host protein, HSP70, was reported to maintain NP stability, enabling viral replication, as NP degraded in its absence [102]. NP also interacts with RUVBL (RuvB-like) 1 and RUVBL2 proteins in an RNA-independent manner forming the R2TP complex required for capsid assembly [103].
The critical NP residues discussed above are summarized in Table 2. It could be suggested that NP is essential for viral replication and transcription. Additionally, NP is vital for RNA encapsidation, NC formation and capsid assembly. NP, in part, completes these functions via interaction with EBOV (except GP) and host proteins. Therefore, a therapeutic or peptide/protein vaccine candidate targeting critical NP aa and motifs (Table 2, Figure 4) could hinder NC formation and NP-host protein interactions, abrogating virus transcription and virus assembly.
3.2. Viral Protein 35 (VP35)
Tetrameric VP35 is functionally analogous to other NNS RNA viruses [8,27,134]. VP35 is crucial for viral transcription and replication [134] and possesses NTPase and helicase activities, suggesting that it could affect transcription via NTP hydrolysis and NTP-dependent unwinding of RNA helices, respectively [135]. VP35 also contributes to genome packaging [136] and nucleocapsid assembly [13,14] as it binds the monomeric state of NP to prevent premature and non-specific encapsidation of viral RNA.
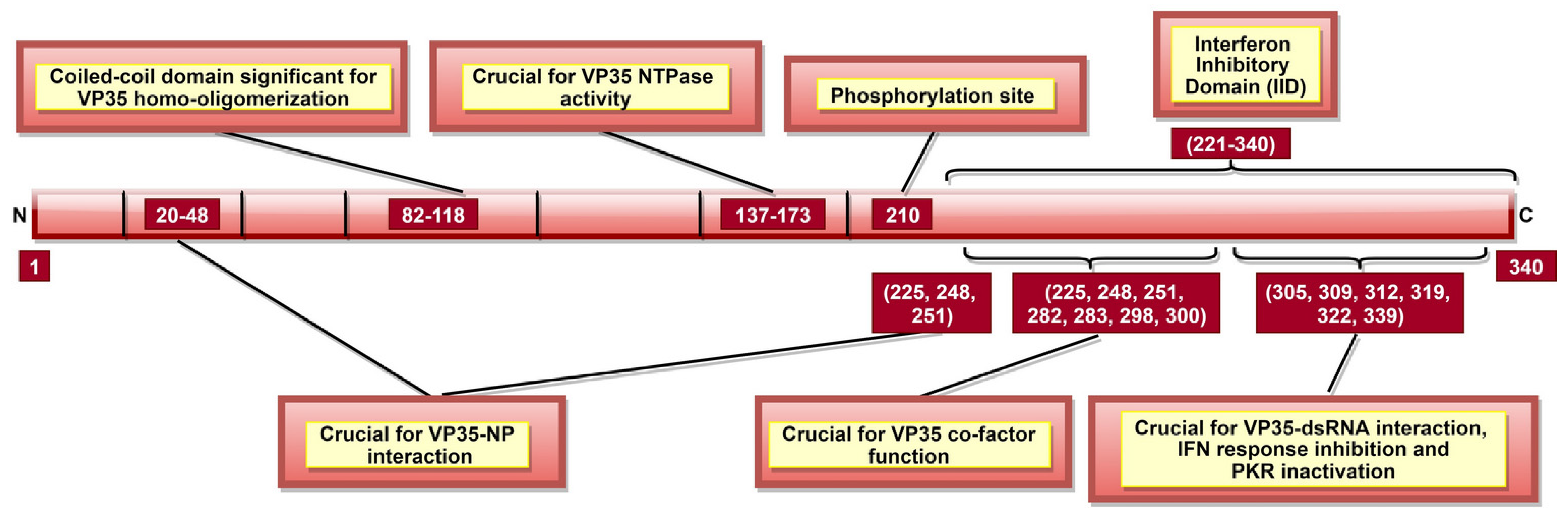
VP35 is crucial for host immune response evasion, where host anti-viral defense is inhibited in multiple ways. It can suppress host interferon (IFN) response in both dsRNA-binding-dependent and dsRNA-binding-independent manners. In this respect, the VP35 CTD region, in particular aa 221–340, were shown to function as IFN inhibitory domain (IID) [109] or RNA-binding domain (RBD) [110] (Figure 5). It was demonstrated that specific residues within IID (Table 2, Figure 5) are required to bind VP35 to viral dsRNA [107,110]. This interaction is crucial for the IFN inhibiting function of VP35, as it blocks viral dsRNA recognition by retinoic-acid-inducible gene I (RIG-I) and melanoma differentiation-associated protein 5 (MDA-5), the intracellular pattern recognition receptors [109].
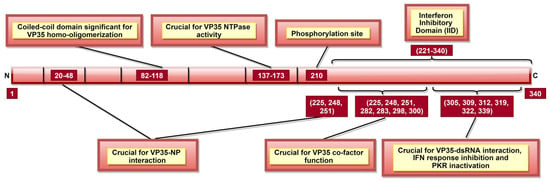
Figure 5.
A schematic representation of EBOV viral protein 35 (VP35). VP35 aa 137–173 are significant for VP35 NTPase activity, which affects viral transcription and rep-lication. Interferon inhibitory domain (IID; aa 221–340) consists of aa 225, 248, and 251 which are involved in two roles: VP35-NP interaction (along with N-terminal aa 20–48) and VP35 co-factor function (along with IID aa 282, 283, 298, and 300). Phosphorylation of aa 210 regulates VP35-NP interaction, as well as viral transcription and replication.
It was shown that VP35 could also inhibit IFN regulatory factor 3 (IRF-3) dimerization, phosphorylation and nuclear localization [137]. VP35 can do it by impairing the ability of TANK-binding kinase 1 (TBK-1) and IκB kinase epsilon (IKKε) kinases to interact with IRF-3 [138]. VP35 can also suppress IFN transcription by increasing SUMOylation of IRF-7 via interaction with protein inhibitors of activated STATs 1 (PIAS1) [139]. IID of VP35, especially aa 239, 312, and 322, also blocks the protein activator of IFN-induced protein kinase (PACT), thus, preventing activation of PACT-induced RIG-I ATPase [106]. Further, VP35 aa 304–340 (Figure 5) can inactivate protein kinase R (PKR; an antiviral protein), which enables continuous viral protein synthesis [108].
Interaction of VP35 with other viral proteins is significant for multiple purposes. VP35-L interaction requires VP35 homo-oligomerization, completed by N-terminal aa 82–118 (Figure 5) [105]. VP35 could also function as a non-enzymatic co-factor for the L protein [107], where several IID constituent residues are critical (Table 2, Figure 5) [104]. It appears that the co-factor function is independent of IID binding to dsRNA [107], but the latter can still modulate VP35-NP interaction [104]. VP35 residues 20–48, 225, 248, and 251 (Table 2) are significant for VP35-NP complex formation and regulate viral RNA synthesis [88,104]. A recent study reported the role of VP35 phosphorylation, especially at aa 210 (Figure 5), in the regulation of VP35-NP interaction, as well as viral transcription and replication [140].
To conclude, VP35 plays a chief role in host immune evasion by blocking viral dsRNA recognition by host immune receptors, inhibiting IRF-3 and TBK-1/ IKKε complex formation, increasing IRF-7 SUMOylation, preventing RIG-I ATPase activation and inactivating PKR (Table 2, Figure 5). Moreover, VP35 is suggested to play a role in NC formation, viral transcription, replication, and genome packaging. Therefore, targeting VP35 might enable the host to mount a more robust immune response and disturb the viral structural integrity.
3.3. VP40
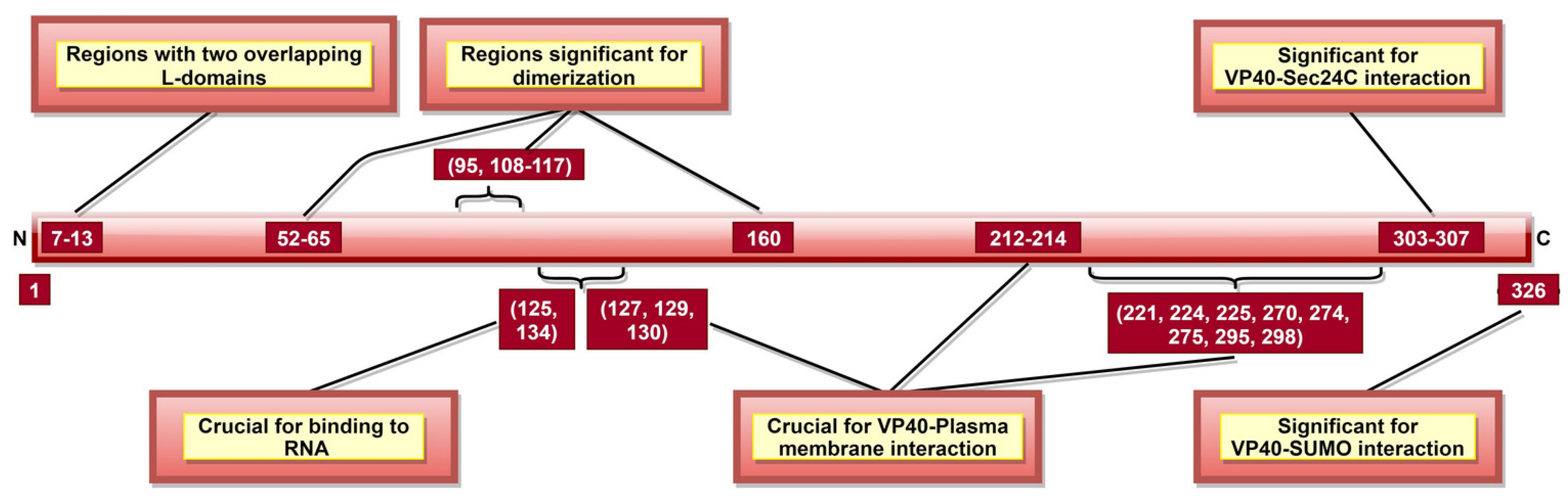
VP40, the most abundantly expressed protein [28], is essential for viral assembly and budding [141]. VP40 aa 292–295 (Table 2) were reported as critical for VLP production and controlled inhibition of viral transcription, as a mutation in 292–295 aa sequence altered these functions [116]. A recent study suggested that aa 326 (Figure 6) is involved in SUMO–VP40 interaction which is significant for VP40 stability [142]. Additionally, VP40 contains two late budding domains (L-domains) located at aa 7–10 (PTAP) and aa 10–13 (PPEY) (Figure 6) [143], interacting with host proteins. PTAP makes a complex with tumor susceptibility gene 101 protein (tsg101) [144], while PPEY binds to ubiquitin ligase, neuronal precursor cell-expressed developmentally downregulated 4 (Nedd4) [145], as well as to ITCH E3 ubiquitin ligase [146]. The PTAPtsg101 interaction helps recruit VP40 into lipid raft domains on the plasma membrane [143], while the PPEY–Nedd4 complex covalently ubiquitinates viral matrix proteins, which is required for virus budding [145]. These data strongly suggest that L-domains are essential for budding [147] but limited in viral replication [148].
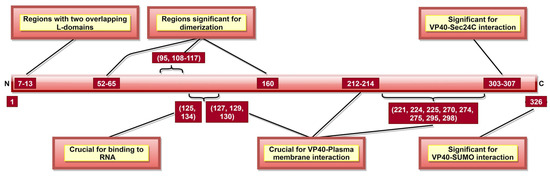
Figure 6.
A schematic representation of EBOV viral protein 40 (VP40). VP40 aa 7–13 consists of two late budding domains (L-domains), namely PTAP (aa 7–10) and PPEY (aa 10–13). Both L-domains are crucial to viral budding. VP40 aa 52–65, 95, 108–117, and 160 are involved in dimerization, while aa 303–307 enables VP40–Sec24C interaction. Both the dimerization state and the interaction with Sec24C are significant for proper cellular trafficking of VP40. The binding of octameric VP40 to ssRNA (via VP40 aa 125 and 134) negatively regulates transcription. aa 326 provides stability to VP40 via interaction with host SUMO protein.
VP40 is classified as a transformer protein [149,150], as it could obtain multiple conformational states: dimers, hexamers, filaments and octamers [151]. VP40 dimerizes in solution with the help of aa 95 and 160 [111], as well as NTD aa 52–65 and 108–117 [112] (Table 2, Figure 6). Dimerized confirmation is essential for proper trafficking of VP40, where aa 303–307 (Figure 6) form VP40-Sec24C complex facilitating intracellular transport to the plasma membrane [82]. Dimers can also assemble into filaments via CTD interactions between two dimers, where aa 241 and 307 (Table 2) are shown to be critical. This state is essential for proper matrix assembly and budding [112].
NTD aa 127, 129 and 130 [114] and aa 212–214 [115] (Figure 6) are required for proper VP40 localization to the plasma membrane, oligomerization and budding. The interaction with the plasma membrane is mediated by various VP40 CTD residues (Table 2, Figure 6) [112]. Additionally, aa 213, 295, and 298 facilitate deep penetration of VP40 into the plasma membrane [152]. The VP40 dimers at the plasma membrane are then oligomerized into linear VP40 hexamers [112,153]. Both deep penetration and hexamer formation are critical to viral assembly and budding [112,152]. A recent study highlighted the significance of aa 191 for effective VP40 localization to the plasma membrane, oligomerization and VLP formation [154].
In addition to dimers, filaments, and hexamers, VP40 can form an octameric ring configuration where NTD plays the leading role [112]. This configuration and aa 125 and 134 (Figure 6) were essential for VP40-ssRNA binding [112,113]. The latter is implicated in the negative regulation of transcription [111].
These data indicate that the main function of VP40 is in viral assembly and budding (Table 2, Figure 6). Therefore, targeting critical VP40 aa might help suppress viral spread after infection by hindering the budding process. Additionally, it is suggested to play a role in viral transcription inhibition and interaction with the host cell plasma membrane.
3.4. Glycoprotein
3.4.1. GP
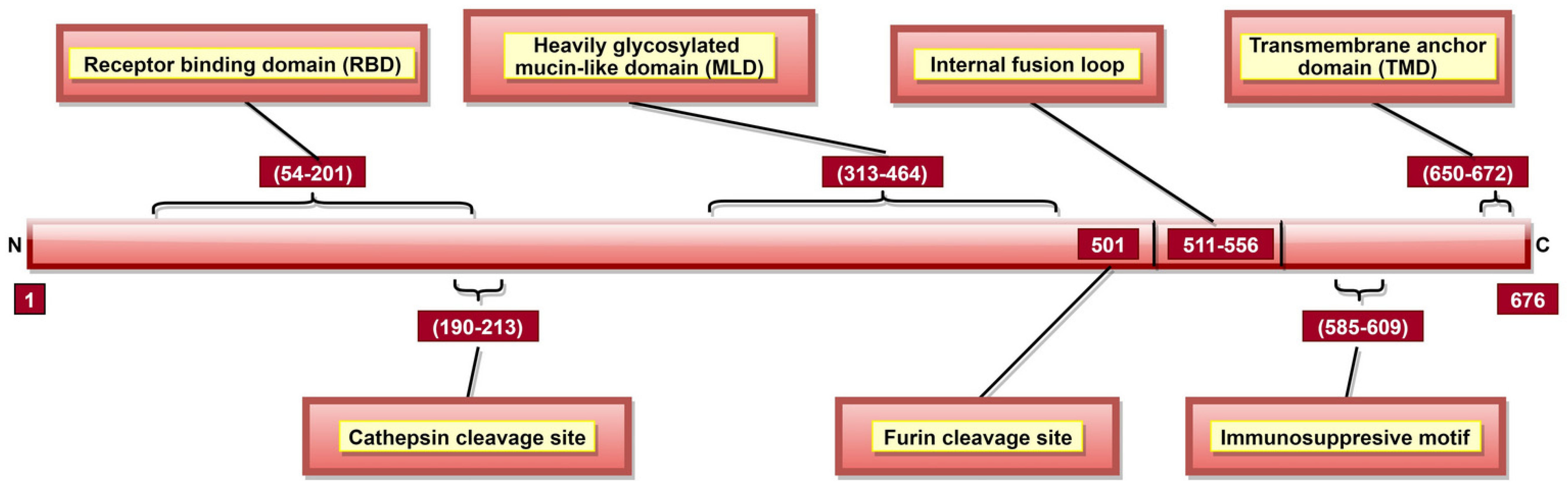
GP is a type I transmembrane fusogenic protein, which is translated only 20% of time by the GP gene as it can be cytotoxic to certain cell types [29,31]. As the only EBOV protein present on the viral surface, GP is responsible for pathogenic differences of ebolaviruses [123]. In 2010, it was reported that GP could cause “steric occlusion”, a phenomenon which interferes with immune recognition of HLA class I and II molecules [155]. In other studies, it was shown that aa 159, 160, 162, 170 [120], and 214–270 [119] (Table 2) provide protein stability, while aa 55, 57, 63, and 64 are involved in membrane fusion-mediated conformational changes [123]. GP has a cathepsin cleavage site at aa 190–213 (Table 2, Figure 7) [123,124], which is proteolyzed inside the endosome, a step critical for viral infection [60,61]. GP is post-translationally cleaved by furin at aa residue 501 (Figure 7), resulting in GP1 and GP2 subunits linked by disulfide bonds [31,75,76]. These heterodimers form trimeric viral peplomer [31].
Figure 7.
A schematic representation of EBOV glycoprotein (GP). GP aa 54–201 form the receptor-binding domain (RBD) or receptor binding site (RBS) responsible for attachment to host cell-surface receptors. Cathepsin cleavage site is present in aa 190–213, and proteolysis via cathepsins is significant for viral infectivity. GP is cleaved at aa 501 by furin into GP1 and GP2 subunits. The immunosuppressive motif (aa 585–609) plays a role in bystander lymphocyte apoptosis and cytokine dysregulation. The transmembrane anchor domain (TMD; aa 650–672) helps tether GP onto the viral surface.
GP1 mediates attachment to host cells using the receptor-binding site (RBS) located at aa 54–201 (Figure 7) [122]. Studies have identified multiple aa (Table 2) as critical for viral entry [117,118,119,120,121,156]. Among these, aa 64, 95 [121] and 114, 115, 140 [124] are involved in direct contact with host cell receptors while aa 43, 54, 56, 60, 61 and 79 contribute to post-binding steps of viral entry [121]. A Mucin-like domain (MLD) was identified in aa 313–464 (Figure 7) [120], which does not directly impact viral entry [32] but can stimulate host dendritic cells by activation of mitogen-activated protein kinase (MAPK) and nuclear factor kappa B (NF-κB) pathways [157].
GP2 contributes to the fusion of viral and host cell membranes [158]. An internal fusion loop at aa 511–556 position (Figure 7) [120] consists of hydrophobic residues (Table 2) inserted into the target cell membrane [123]. GP2 also has a transmembrane anchor domain (TMD; aa 650–672) (Figure 7) [31] which helps to tether GP onto the viral surface [74]. Interestingly, GP2 also has the anti-tetherin activity, promoting VP40-mediated viral budding by the host cell and disabling the immune response stimulation via NF-κB signaling [159,160]. An immunosuppressive motif (aa 585–609; Figure 7) located near the C-terminal [29,31] can cause lymphocyte apoptosis, as well as cytokine dysregulation during EVD [126].
Post-translational N-linked glycosylation of GP results in a thick coating of oligosaccharides, protecting viral GP against host humoral immune response [123] and promoting protein expression and function [161]. In total, 15 N-linked and 80 O-linked glycosylation sites have been identified in GP1 [32,161]. GP2 has 2 N-linked glycosylation sites (aa 563 and 618; Table 2) [75] critical for GP processing, oligomerization and confirmation [125]. Once a threshold amount of viral GP is present on the host cell plasma membrane, the highly glycosylated MLD masks specific cell-surface proteins, such as major histocompatibility complex (MHC) class I, αVβ3, etc., in a cell-dependent manner [155,162,163,164]. MHC class I masking protects the virus from host CD8+ T cell recognition [155]. Additionally, MLD masks self GP epitopes, shielding GP from the host immune recognition [155,162].
3.4.2. Soluble Secreted Glycoprotein (sGP)
sGP is the primary GP gene product, sharing 295 N-terminal aa with the full-length transmembrane spike glycoprotein (GP) [31]. Six N-glycosylation sites (aa 40, 204, 228, 238, 257, and 268) [165] and one C-mannosylation site (aa 288) [166] have been identified in sGP. It is a non-structural protein (NSP) [4], though a structural role was contemplated in a study, wherein sGP substituted for GP1, producing a functional sGP-GP2 protein [167]. A potential contribution of sGP in viral dissemination was suggested by Bradley et al., demonstrating inhibition of pro-inflammatory cytokines production from non-infected macrophages and impairing chemotaxis of activated macrophages [168]. sGP also has an anti-inflammatory capacity to restore the endothelial cell barrier function dysregulated by GP [165,169]. It was also shown that sGP contributes to host immune evasion by acting as a decoy for anti-GP antibodies [170]. In a recent study, detection of serum sGP was suggested as a biomarker for Ebola virus disease (EVD) diagnosis as large quantities of this protein are found in blood at the early stages of the disease [171].
3.4.3. Δ-Peptide
Δ-peptide is a non-structural, secretory product of the GP gene, composed of 40 aa [30]. It is a O-glycosylated, sialylated peptide [30] rich in cationic and aromatic residues [172]. It can inhibit viral entry into filovirus-permissive cells, preventing superinfection [172,173]. In 2015, Gallaher et al. reported the role of Δ-peptide as a viroporin, based on presence of a lytic sequence motif [172,174]. In another study, Δ-peptide was shown to indiscriminately permeabilized mammalian cells in culture, supporting its role as a viroporin [175].
3.4.4. Shed GP
Like sGP, shed GP is believed to function as “antibody sinks” [32] as they compete with GP for antibody binding [176]. It is also believed that shed GP helps to reduce the cellular cytotoxicity caused by GP [177]. Additionally, shed GP activates non-infected dendritic cells and macrophages [177], leading to massive cytokine production and increased vascular permeability [177].
3.4.5. Soluble Small Secreted Glycoprotein (ssGP)
ssGP is an NSP composed of 298 aa [32] (Table 1), with 295 N-terminal aa identical to sGP and GP [10]. It is secreted as a N-glycosylated homodimer formed by intermolecular disulfide bond at aa 53 [10]. Function of ssGP still remains unknown [171].
To summarize, GP, as the only protein on the viral surface, is responsible for viral entry, i.e., attachment to host cells and fusion of viral and host cell membrane. GP–NPC1 interaction is an indispensable step for viral infection, and, therefore, targeting GP shall hinder the viral infection. Additionally, GP assists in viral budding, while GP and shed GP contribute to the cytokine storm. Further, GP, sGP, and shed GP enable host immune evasion, while sGP contributes to viral spread and may be used as a biomarker for early diagnosis of EVD. Therefore, targeting various GP gene products might be beneficial in the development of an effective vaccine.
3.5. VP30
VP30 is a structural, hexameric phosphoprotein composed of three dimers [33,129]. Dimers are formed by aa 142–264, while aa 94–112 (Figure 8) are required to produce hexameric form [129,178]. VP30 aa 27–40 (Figure 8) form a disordered, non-hydrophobic, arginine-rich region interacting with viral RNA [127]. VP30 is indispensable for RNA transcription initiation [128]. Interestingly, mutations at aa 179, 180, and 183 render VP30 incapable of transcription initiation, suggesting their importance in this process [129].
Figure 8.
A schematic representation of EBOV viral protein 30 (VP30). aa 68–95 comprise of zinc-binding site. Zinc is important for VP30’s transcriptional functions and VP30–RNA interaction via VP30 RNA binding site (aa 27–40). aa 143 and 146 are involved in three roles: (a) phosphorylation sites, (b) dimerization (aa 142–264), and (c) VP30–NP interaction (aa 140–266).
Factors impacting the role of VP30 in transcription initiation are RNA secondary structure formation, VP30–NP interaction, zinc-binding and VP30 phosphorylation. The NP gene transcription starts signal forms a stem-loop like secondary structure, essential for VP30-dependent transcription initiation [179]. The VP30 aa 140–266 (Table 2, Figure 8) are responsible for VP30–NP complex formation [96,97,129], and a threshold level of interaction is required for optimal viral transcription, below and beyond which transcriptional activity is flawed [96]. Further, VP30 role in regulating viral RNA transcription requires a zinc-binding site located at aa 68–95 (Figure 8) [128].
VP30 phosphorylation mainly occurs at serine (aa 29–31, 42, 44, and 46) and threonine (aa 52, 143, and 146) residues (Figure 8) [33,180]. Low or un-phosphorylated VP30 seems responsible for transcription initiation of all seven genes in the EBOV genome [181,182]. Whether low or un-phosphorylated VP30 is required as a transcription factor depends on the virus replication stage. Additionally, if low phosphorylated, a constant phosphorylation/dephosphorylation within the same VP30 molecule is required for effective transcription initiation [183]. Amongst the phosphorylation sites, aa 29 seems to be the most critical as it can solely execute all VP30 transcription functions [183]. Complete VP30 phosphorylation at all serine residues between aa 29–46 abrogates transcription initiation function [184].
The impact of phosphorylation on VP30-NP interaction was extensively debated over the past two decades [33,181,182]. The latest consensus states that phosphorylation leads to a more robust interaction with NP which allows VP30 to be associated with NC and enter newly synthesized virus particles, wherein the un-phosphorylated VP30 can initiate transcription [182,183]. Overall, it is accepted that the phosphorylation state of VP30 is dynamic and modulated by the virus to achieve an intricate balance between transcription and replication processes, which happen simultaneously during the EBOV life cycle [181,184].
The above discussion suggests that VP30 chiefly plays a role in viral transcription initiation via its zinc-binding, NP interaction and phosphorylation characteristics (Table 2, Figure 8). Abrogating this function shall hinder the production of multiple RNA copies. Therefore, VP30 is a plausible candidate for therapeutic and vaccine development studies targeting primary transcription in the host cell cytoplasm.
3.6. VP24
VP24 makes up approximately 7.5% of total EBOV proteins and is one of the five proteins involved in EBOV NC formation. VP24 aa 169–173 (Table 2, Figure 9) interact with NP, which is indispensable for NC formation and completion of the EBOV replication cycle [133]. Additionally, VP24, together with VP35 and NP, is responsible for packaging NC into virions as it induces favorable conformational changes in the NC. This structural change also signals the end of replication and the beginning of the egress phase [133,185].
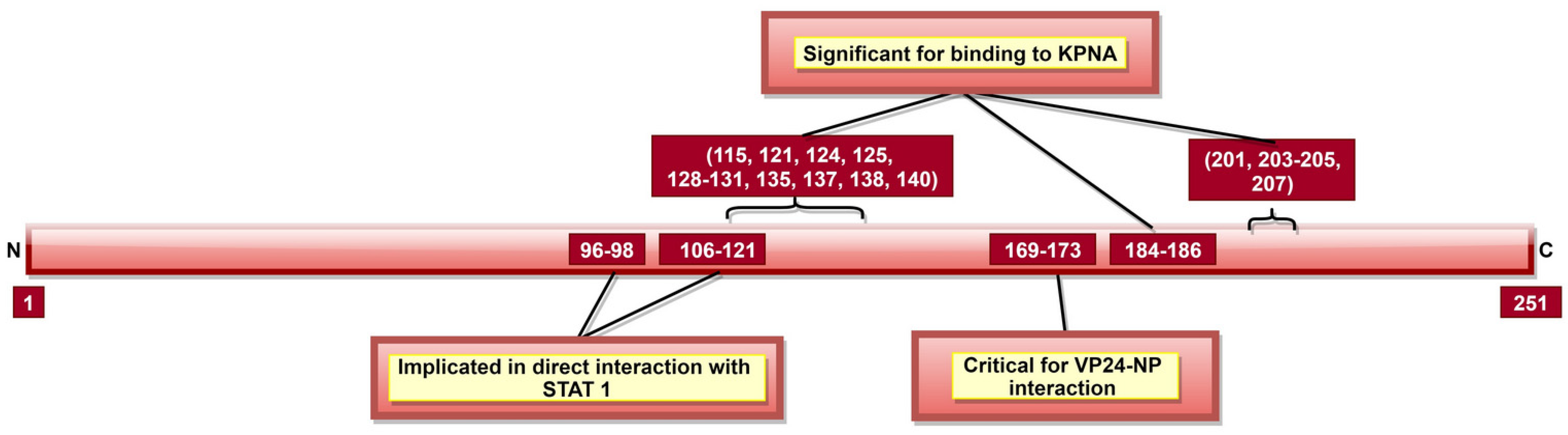
Figure 9.
A schematic representation of EBOV viral protein 24 (VP24). VP24 aa 169–173 facilitate interaction with NP, which is crucial for nucleocapsid (NC) formation, EBOV replication cycle and egress phase. Tyrosine-phosphorylated STAT1, hnRNP C1/C2 and VP24 interact with KPNA at the same position; therefore, upon VP24–KPNA interaction, transport of tyrosine-phosphorylated STAT1 to the nucleus is stopped as well as translocation of hnRNP C1/C2 from the cytoplasm to the nucleus is partially prevented.
Like VP35, VP24 inhibits host immune response using several mechanisms [186]. VP24 inhibits IFN responses by blocking p38 phosphorylation and hampering the p38 MAPK pathway [187]. It can also block activation of NF-κB, which has multiple IFN responsive genes targeted downstream [188]. IFN inducible pathways could also be inhibited by arresting the nuclear translocation of tyrosine-phosphorylated STAT1 via VP24 interaction with NPI-1 subfamily members of karyopherin-α (KPNA, also called importin-α) [132]. Additionally, the role of aa 96–98 and 106–121 (Figure 9) indirect interaction of VP24 with un-phosphorylated STAT1 was reported [130,131]. VP24 aa 142–147 and 26–50 (Table 2) were reported as necessary to facilitate binding to KPNA1 [130,132]. Multiple other aa (Table 2, Figure 9) were also shown responsible for VP24–KPNA5 interaction [131]. Further, it was reported that oxidative stress effects the VP24 modulation of the host response, facilitating recovery of infected cells from stress [189].
It appears that the central role of VP24 is in host immune evasion, carried out by inhibiting p38 MAPK and NF-κB pathway activation. VP24 also seems critical to viral replication and is involved in budding viral initiation. Therefore, targeting these residues (Table 2) during vaccine development may hinder viral replication.
3.7. L Protein
L, a part of the RNP complex [36], is the largest, multi-subunit and multifunctional EBOV protein composed of 2212 aa [36] (Table 1). There are five domains in L protein (Figure 10), namely, (a) RNA-dependent RNA polymerase (RdRp) domain with transcription/replication and polyadenylation activity, (b) capping domain with polyribonucleotidyl transferase (PRNTase) activity, (c) connector domain (CD) with an organizational role, (d) a methyltransferase domain with MTase activity, and e) a small C-terminal domain (Table 3, Figure 10) [190]. In addition, EBOV L residues 1–450 contain a homo-oligomerization domain (Figure 10) [105]. L protein aa 1–380 (Figure 10) are involved in L-VP35 interaction, which happens in a non-competitive manner and does not require L homo-oligomerization [105]. The L-VP35 binding enables re-localization of L into viral inclusion bodies. In addition to VP35, The RESTV L protein was shown to interact with VP30 [191].
Figure 10.
A schematic representation of EBOV L protein. It has five domains, namely, (a) RdRp domain responsible for polyadenylation and transcription/replication (via GDNQ motif), (b) capping domain with PRNTase activity which caps mRNA co-transcriptionally via transfer of a GDP molecule, (c) connector domain, (d) methyltransferase domain involved in methylation of capped mRNA, as well as cap-independent methylation of internal mRNA adenosine, and (e) C-terminal domain responsible for the regulation of methylation activities of methyltransferase domain. L aa 1–380 are involved in L-VP35 interaction, enabling L’s re-localization into viral inclusion bodies.

Table 3.
Various domains of L protein and their functions.
L protein is essential for virus replication, yet little is known about its other functions, mainly due to its large size and lack of specific antibodies [192,193]. The current understanding of this protein’s function comes from studies using vesicular stomatitis virus (VSV), as L protein is highly conserved and homologous amongst Mononegavirales members [65,190,194,195]. Therefore, the L domains functions presented here are based on reports made using VSV L and not always the EBOV.
A GDNQ motif has been reported as a catalytic center in several negative sense RNA polymerases. In 2017, The EBOV RdRp domain was also reported to consist of the GDNQ motif within aa residues 741–744 (Figure 10), which is crucial for viral replication and transcription [193]. The RdRp domain is involved in the polyadenylation of L mRNA [196]. The capping of viral transcripts occurs co-transcriptionally via the transfer of a GDP molecule by PRNTase to 5′ phosphate of mRNA [197]. Then, the first nucleotide and guanosine of the capped mRNA become methylated at 2′-O and N-7 positions, respectively, by MTase activity [198]. Additionally, a cap-independent, internal adenosine 2′-O methylation activity of methyltransferase domain in SUDV L protein (aa 1693–2036) was demonstrated [194,199]. The small CTD of SUDV L protein located at aa 2037–2210 plays a critical role in RNA recruitment for methylation [194]. It also regulates cap methylation (aa 2043, 2067, 2068, 2118, 2189, and 2193) and internal methylation (aa 2043, 2067, 2068, 2112, 2113, 2118, 2189, and 2193) activities of methyltransferase domain [194].
4. Conclusions and Future Perspectives
It is imperative to understand proteins functions with respect to the EBOV life cycle and pathogenesis to identify practical and specific protein therapeutic targets. This review summarizes data on aa residues of EBOV proteins involved in essential functions, such as viral entry, host immune evasion, replication, transcription, and budding. A cross-reactive, multi-protein/peptide vaccine candidate developed by utilizing the information presented in this review could help curb various characteristic EVD symptoms and affect different stages of the EVD life cycle.
Multiple proteins and aa sites were highlighted in our review, which have a high potential for vaccine development. For instance, NC formation and viral replication could be hindered by collectively targeting NP aa 110–400 and VP24 aa 169–173. Replication and its regulation could also be affected by targeting VP35 aa 210 and L aa 741–744. Viral transcription could be affected by targeting NP aa 562–567, NP aa 600–617, and VP30 aa 140–266, while transcription regulation can be blocked by collectively targeting NP aa 240–383, VP35 aa 20–48, and VP30 aa 29–183. Moreover, targeting NP aa stretch 550–600 might disable multiple NP-host-protein interactions significant for transcription and regulation. Viral ingress may be blocked by targeting GP aa 54–213 as it affects GP stability, interaction with obligate host receptor (NPC1) and membrane-fusion mediated conformational changes. A critical step for viral spread is host immune evasion, accomplished with the help of VP35, sGP, shed GP and VP24 proteins. This could be prevented by targeting specific regions of these proteins, such as VP30 aa 221–340 and VP24 aa 120–190. Moreover, as aa 1–295 are identical in GP, sGP, and ssGP; therefore, therapeutic or vaccine candidates against GP aa 54–213 could prove effective against sGP (primary GP gene product), as well as ssGP. This will be effective in preventing viral entry, as well as in curbing host immune evasion. Three proteins contribute to viral egress: VP40, GP, and VP24. GP2 displays anti-tetherin activity, while VP24 causes favorable structural changes in NC to signal viral egress initiation. As VP40 is primarily involved in assembly and budding, targeting aa 212–275, a region significant for localization to and interaction with the plasma membrane (Table 2), may hinder the budding process.
Multiple studies have analyzed EBOV proteins viz., NP [20,200,201,202], VP35 [203,204], VP40 [203], GP [21,205,206,207,208,209,210,211,212,213], and VP24 [214,215] potential as therapeutics and vaccine candidates. In December 2019, Ervebo, a recombinant vesicular stomatitis virus (VSV) vector-based vaccine, was approved by FDA as the first vaccine against the Ebola virus [216]. Currently, nearly 14 potent Ebola vaccine candidates are in various phases of clinical trials [217]. Still, the presence of non-immunogenic, unwanted components in traditional vaccines, allergic, toxic, or autoimmune reactions, the requirement for BSL-4 facility and probability of a reversal of attenuated virus state [218,219,220] impede EBOV vaccine development efforts. Clearly, GP has been the major focus for vaccine development, while less focus was placed on other viral proteins. Therefore, further studies will facilitate our understanding of the EBOV proteins potentials to protect from infection. Key motifs of these proteins might be analyzed to find potential epitopes. These identified epitopes might be estimated for the generation of T and B cell immune response and their binding affinity with human leukocyte antigen (HLA). Further, immunodominant epitopes belonging to different EBOV proteins may be linked to form one multi-peptide vaccine construct. The immunogenic potential of the construct might be validated in different animal models, before considering it for clinical trials. Targeting the key domains of EBOV proteins outlined in this review or their combination could be an approach for development of future EBOV vaccine which is globally effective.
Author Contributions
Conceptualization, M.B.; validation, M.B.; formal analysis, A.R.; writing—original draft preparation, S.J.; writing—review and editing, E.M., M.B. and S.K. All authors have read and agreed to the published version of the manuscript.
Funding
This work did not receive any financial support.
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
This study was supported by DNA Research Center Autonomous Nonprofit Organization. This work is part of the Kazan Federal University Strategic Academic Leadership Program. In addition, we would like to thank the scientific community.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Filoviridae. Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/negative-sense-rna-viruses/mononegavirales/w/filoviridae (accessed on 20 January 2021).
- Jacob, S.T.; Crozier, I.; Fischer, W.A., 2nd; Hewlett, A.; Kraft, C.S.; Vega, M.A.; Soka, M.J.; Wahl, V.; Griffiths, A.; Bollinger, L.; et al. Ebola virus disease. Nat. Rev. Dis. Primers 2020, 6, 13. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.L.; Tan, C.W.; Anderson, D.E.; Jiang, R.D.; Li, B.; Zhang, W.; Zhu, Y.; Lim, X.F.; Zhou, P.; Liu, X.L.; et al. Characterization of a filovirus (Měnglà virus) from Rousettus bats in China. Nat. Microbiol. 2019, 4, 390–395. [Google Scholar] [CrossRef]
- Feldmann, H.; Sprecher, A.; Geisbert, T.W. Ebola. N. Engl. J. Med. 2020, 382, 1832–1842. [Google Scholar] [CrossRef]
- Escudero-Pérez, B.; Ruibal, P.; Rottstegge, M.; Lüdtke, A.; Port, J.R.; Hartmann, K.; Gómez-Medina, S.; Müller-Guhl, J.; Nelson, E.V.; Krasemann, S.; et al. Comparative pathogenesis of Ebola virus and Reston virus infection in humanized mice. JCI Insight 2019, 4, e126070. [Google Scholar] [CrossRef] [PubMed]
- Geisbert, T.W.; Jahrling, P.B. Differentiation of filoviruses by electron microscopy. Virus Res. 1995, 39, 129–150. [Google Scholar] [CrossRef]
- Filoviridae. Available online: https://www.cdc.gov/vhf/virus-families/filoviridae.html (accessed on 17 March 2021).
- Grifoni, A.; Lo Presti, A.; Giovanetti, M.; Montesano, C.; Amicosante, M.; Colizzi, V.; Lai, A.; Zehender, G.; Cella, E.; Angeletti, S.; et al. Genetic diversity in Ebola virus: Phylogenetic and in silico structural studies of Ebola viral proteins. Asian Pac. J. Trop. Med. 2016, 9, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.; Geisbert, T.; Feldmann, H. Filoviridae: Marburg and Ebola Viruses; Lippincott Williams and Williams: Philadelphia, PA, USA, 2007. [Google Scholar]
- Mehedi, M.; Falzarano, D.; Seebach, J.; Hu, X.; Carpenter, M.S.; Schnittler, H.J.; Feldmann, H. A new Ebola virus nonstructural glycoprotein expressed through RNA editing. J. Virol. 2011, 85, 5406–5414. [Google Scholar] [CrossRef] [PubMed]
- Ikegami, T.; Calaor, A.B.; Miranda, M.E.; Niikura, M.; Saijo, M.; Kurane, I.; Yoshikawa, Y.; Morikawa, S. Genome structure of Ebola virus subtype Reston: Differences among Ebola subtypes. Brief report. Arch. Virol. 2001, 146, 2021–2027. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Yang, P.; Li, G.; Liu, B.; Wang, W.; Liu, X.; Xia, B.; Yang, C.; Lou, Z.; Guo, Y.; et al. Insight into the Ebola virus nucleocapsid assembly mechanism: Crystal structure of Ebola virus nucleoprotein core domain at 1.8 A resolution. Protein Cell 2015, 6, 351–362. [Google Scholar] [CrossRef]
- Kirchdoerfer, R.N.; Saphire, E.O.; Ward, A.B. Cryo-EM structure of the Ebola virus nucleoprotein-RNA complex. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2019, 75, 340–347. [Google Scholar] [CrossRef]
- Bharat, T.A.; Noda, T.; Riches, J.D.; Kraehling, V.; Kolesnikova, L.; Becker, S.; Kawaoka, Y.; Briggs, J.A. Structural dissection of Ebola virus and its assembly determinants using cryo-electron tomography. Proc. Natl. Acad. Sci. USA 2012, 109, 4275–4280. [Google Scholar] [CrossRef]
- Sun, Y.; Guo, Y.; Lou, Z. A versatile building block: The structures and functions of negative-sense single-stranded RNA virus nucleocapsid proteins. Protein Cell 2012, 3, 893–902. [Google Scholar] [CrossRef]
- Ebola (Ebola Virus Disease). Available online: https://www.cdc.gov/vhf/ebola/history/chronology.html (accessed on 18 May 2021).
- Gross, L.; Lhomme, E.; Pasin, C.; Richert, L.; Thiebaut, R. Ebola vaccine development: Systematic review of pre-clinical and clinical studies, and meta-analysis of determinants of antibody response variability after vaccination. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2018, 74, 83–96. [Google Scholar] [CrossRef]
- Sharma, R.; Jangid, K.; Anuradha. Ebola Vaccine: How Far are we? J. Clin. Diagn. Res. JCDR 2017, 11, DE01–DE04. [Google Scholar] [CrossRef] [PubMed]
- Matz, K.M.; Marzi, A.; Feldmann, H. Ebola vaccine trials: Progress in vaccine safety and immunogenicity. Expert Rev. Vaccines 2019, 18, 1229–1242. [Google Scholar] [CrossRef]
- Jain, S.; Baranwal, M. Conserved peptide vaccine candidates containing multiple Ebola nucleoprotein epitopes display interactions with diverse HLA molecules. Med. Microbiol. Immunol. 2019, 208, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Baranwal, M. Computational analysis in designing T cell epitopes enriched peptides of Ebola glycoprotein exhibiting strong binding interaction with HLA molecules. J. Theor. Biol. 2019, 465, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Ollmann Saphire, E. A Vaccine against Ebola Virus. Cell 2020, 181, 6. [Google Scholar] [CrossRef]
- ERVEBO. Available online: https://www.fda.gov/vaccines-blood-biologics/ervebo (accessed on 14 July 2020).
- Iversen, P.L.; Kane, C.D.; Zeng, X.; Panchal, R.G.; Warren, T.K.; Radoshitzky, S.R.; Kuhn, J.H.; Mudhasani, R.R.; Cooper, C.L.; Shurtleff, A.C.; et al. Recent successes in therapeutics for Ebola virus disease: No time for complacency. Lancet Infect. Dis. 2020, 20, E231–E237. [Google Scholar] [CrossRef]
- Fausther-Bovendo, H.; Kobinger, G. Vaccine innovation spurred by the long wait for an Ebola virus vaccine. Lancet Infect. Dis. 2021, 21, 440–441. [Google Scholar] [CrossRef]
- UniProtKB—P18272 (NCAP_EBOZM). Available online: https://www.uniprot.org/uniprot/P18272 (accessed on 9 December 2020).
- Bruhn, J.F.; Kirchdoerfer, R.N.; Urata, S.M.; Li, S.; Tickle, I.J.; Bricogne, G.; Saphire, E.O. Crystal Structure of the Marburg Virus VP35 Oligomerization Domain. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Madara, J.J.; Han, Z.; Ruthel, G.; Freedman, B.D.; Harty, R.N. The multifunctional Ebola virus VP40 matrix protein is a promising therapeutic target. Future Virol. 2015, 10, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.; Trappier, S.G.; Mahy, B.W.; Peters, C.J.; Nichol, S.T. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc. Natl. Acad. Sci. USA 1996, 93, 3602–3607. [Google Scholar] [CrossRef]
- Volchkova, V.A.; Klenk, H.D.; Volchkov, V.E. Delta-peptide is the carboxy-terminal cleavage fragment of the nonstructural small glycoprotein sGP of Ebola virus. Virology 1999, 265, 164–171. [Google Scholar] [CrossRef]
- Sanchez, A.; Yang, Z.Y.; Xu, L.; Nabel, G.J.; Crews, T.; Peters, C.J. Biochemical analysis of the secreted and virion glycoproteins of Ebola virus. J. Virol. 1998, 72, 6442–6447. [Google Scholar] [CrossRef]
- Cook, J.D.; Lee, J.E. The secret life of viral entry glycoproteins: Moonlighting in immune evasion. PLoS Pathog. 2013, 9, e1003258. [Google Scholar] [CrossRef]
- Modrof, J.; Mühlberger, E.; Klenk, H.D.; Becker, S. Phosphorylation of VP30 impairs ebola virus transcription. J. Biol. Chem. 2002, 277, 33099–33104. [Google Scholar] [CrossRef]
- UniProtKB—Q05322 (VP24_EBOZM). Available online: https://www.uniprot.org/uniprot/Q05322 (accessed on 14 May 2020).
- Elliott, L.H.; Kiley, M.P.; McCormick, J.B. Descriptive analysis of Ebola virus proteins. Virology 1985, 147, 169–176. [Google Scholar] [CrossRef]
- Ayub, G.; Waheed, Y. Sequence analysis of the L protein of the Ebola 2014 outbreak: Insight into conserved regions and mutations. Mol. Med. Rep. 2016, 13, 4821–4826. [Google Scholar] [CrossRef] [PubMed]
- Volchkov, V.E.; Volchkova, V.A.; Chepurnov, A.A.; Blinov, V.M.; Dolnik, O.; Netesov, S.V.; Feldmann, H. Characterization of the L gene and 5′ trailer region of Ebola virus. J. Gen. Virol. 1999, 80 Pt 2, 355–362. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.Y.; Empig, C.J.; Welte, F.J.; Speck, R.F.; Schmaljohn, A.; Kreisberg, J.F.; Goldsmith, M.A. Folate receptor-alpha is a cofactor for cellular entry by Marburg and Ebola viruses. Cell 2001, 106, 117–126. [Google Scholar] [CrossRef]
- Takada, A.; Watanabe, S.; Ito, H.; Okazaki, K.; Kida, H.; Kawaoka, Y. Downregulation of beta1 integrins by Ebola virus glycoprotein: Implication for virus entry. Virology 2000, 278, 20–26. [Google Scholar] [CrossRef]
- Shimojima, M.; Takada, A.; Ebihara, H.; Neumann, G.; Fujioka, K.; Irimura, T.; Jones, S.; Feldmann, H.; Kawaoka, Y. Tyro3 family-mediated cell entry of Ebola and Marburg viruses. J. Virol. 2006, 80, 10109–10116. [Google Scholar] [CrossRef]
- Kondratowicz, A.S.; Lennemann, N.J.; Sinn, P.L.; Davey, R.A.; Hunt, C.L.; Moller-Tank, S.; Meyerholz, D.K.; Rennert, P.; Mullins, R.F.; Brindley, M.; et al. T-cell immunoglobulin and mucin domain 1 (TIM-1) is a receptor for Zaire Ebolavirus and Lake Victoria Marburgvirus. Proc. Natl. Acad. Sci. USA 2011, 108, 8426–8431. [Google Scholar] [CrossRef] [PubMed]
- Takada, A.; Fujioka, K.; Tsuiji, M.; Morikawa, A.; Higashi, N.; Ebihara, H.; Kobasa, D.; Feldmann, H.; Irimura, T.; Kawaoka, Y. Human macrophage C-type lectin specific for galactose and N-acetylgalactosamine promotes filovirus entry. J. Virol. 2004, 78, 2943–2947. [Google Scholar] [CrossRef] [PubMed]
- Simmons, G.; Reeves, J.D.; Grogan, C.C.; Vandenberghe, L.H.; Baribaud, F.; Whitbeck, J.C.; Burke, E.; Buchmeier, M.J.; Soilleux, E.J.; Riley, J.L.; et al. DC-SIGN and DC-SIGNR bind ebola glycoproteins and enhance infection of macrophages and endothelial cells. Virology 2003, 305, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, C.P.; Lasala, F.; Carrillo, J.; Muñiz, O.; Corbí, A.L.; Delgado, R. C-type lectins DC-SIGN and L-SIGN mediate cellular entry by Ebola virus in cis and in trans. J. Virol. 2002, 76, 6841–6844. [Google Scholar] [CrossRef]
- Hoenen, T.; Groseth, A.; Feldmann, H. Therapeutic strategies to target the Ebola virus life cycle. Nat. Rev. Microbiol. 2019, 17, 593–606. [Google Scholar] [CrossRef]
- Emanuel, J.; Marzi, A.; Feldmann, H. Filoviruses: Ecology, Molecular Biology, and Evolution. Adv. Virus Res. 2018, 100, 189–221. [Google Scholar] [CrossRef]
- Groseth, A.; Hoenen, T. Forty Years of Ebolavirus Molecular Biology: Understanding a Novel Disease Agent Through the Development and Application of New Technologies. Methods Mol. Biol. 2017, 1628, 15–38. [Google Scholar] [CrossRef]
- Quinn, K.; Brindley, M.A.; Weller, M.L.; Kaludov, N.; Kondratowicz, A.; Hunt, C.L.; Sinn, P.L.; McCray, P.B., Jr.; Stein, C.S.; Davidson, B.L.; et al. Rho GTPases modulate entry of Ebola virus and vesicular stomatitis virus pseudotyped vectors. J. Virol. 2009, 83, 10176–10186. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Hope, T.J.; Young, J.A. Differential requirements for clathrin endocytic pathway components in cellular entry by Ebola and Marburg glycoprotein pseudovirions. Virology 2011, 419, 1–9. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Warfield, K.L.; Ruthel, G.; Bavari, S.; Aman, M.J.; Hope, T.J. Ebola virus uses clathrin-mediated endocytosis as an entry pathway. Virology 2010, 401, 18–28. [Google Scholar] [CrossRef]
- Empig, C.J.; Goldsmith, M.A. Association of the caveola vesicular system with cellular entry by filoviruses. J. Virol. 2002, 76, 5266–5270. [Google Scholar] [CrossRef]
- Saeed, M.F.; Kolokoltsov, A.A.; Albrecht, T.; Davey, R.A. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010, 6, e1001110. [Google Scholar] [CrossRef]
- Nanbo, A.; Imai, M.; Watanabe, S.; Noda, T.; Takahashi, K.; Neumann, G.; Halfmann, P.; Kawaoka, Y. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010, 6, e1001121. [Google Scholar] [CrossRef] [PubMed]
- Mulherkar, N.; Raaben, M.; de la Torre, J.C.; Whelan, S.P.; Chandran, K. The Ebola virus glycoprotein mediates entry via a non-classical dynamin-dependent macropinocytic pathway. Virology 2011, 419, 72–83. [Google Scholar] [CrossRef]
- Aleksandrowicz, P.; Marzi, A.; Biedenkopf, N.; Beimforde, N.; Becker, S.; Hoenen, T.; Feldmann, H.; Schnittler, H.J. Ebola virus enters host cells by macropinocytosis and clathrin-mediated endocytosis. J. Infect. Dis. 2011, 204 (Suppl. 3), S957–S967. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A. Analysis of filovirus entry into vero e6 cells, using inhibitors of endocytosis, endosomal acidification, structural integrity, and cathepsin (B and L) activity. J. Infect. Dis. 2007, 196 (Suppl. 2), S251–S258. [Google Scholar] [CrossRef]
- Hunt, C.L.; Kolokoltsov, A.A.; Davey, R.A.; Maury, W. The Tyro3 receptor kinase Axl enhances macropinocytosis of Zaire ebolavirus. J. Virol. 2011, 85, 334–347. [Google Scholar] [CrossRef] [PubMed]
- Hunt, C.L.; Lennemann, N.J.; Maury, W. Filovirus entry: A novelty in the viral fusion world. Viruses 2012, 4, 258–275. [Google Scholar] [CrossRef]
- Martinez, O.; Johnson, J.; Manicassamy, B.; Rong, L.; Olinger, G.G.; Hensley, L.E.; Basler, C.F. Zaire Ebola virus entry into human dendritic cells is insensitive to cathepsin L inhibition. Cell. Microbiol. 2010, 12, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Chandran, K.; Sullivan, N.J.; Felbor, U.; Whelan, S.P.; Cunningham, J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science 2005, 308, 1643–1645. [Google Scholar] [CrossRef] [PubMed]
- Schornberg, K.; Matsuyama, S.; Kabsch, K.; Delos, S.; Bouton, A.; White, J. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J. Virol. 2006, 80, 4174–4178. [Google Scholar] [CrossRef] [PubMed]
- Brecher, M.; Schornberg, K.L.; Delos, S.E.; Fusco, M.L.; Saphire, E.O.; White, J.M. Cathepsin cleavage potentiates the Ebola virus glycoprotein to undergo a subsequent fusion-relevant conformational change. J. Virol. 2012, 86, 364–372. [Google Scholar] [CrossRef]
- Carette, J.E.; Raaben, M.; Wong, A.C.; Herbert, A.S.; Obernosterer, G.; Mulherkar, N.; Kuehne, A.I.; Kranzusch, P.J.; Griffin, A.M.; Ruthel, G.; et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature 2011, 477, 340–343. [Google Scholar] [CrossRef]
- Côté, M.; Misasi, J.; Ren, T.; Bruchez, A.; Lee, K.; Filone, C.M.; Hensley, L.; Li, Q.; Ory, D.; Chandran, K.; et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature 2011, 477, 344–348. [Google Scholar] [CrossRef]
- Martin, B.; Canard, B.; Decroly, E. Filovirus proteins for antiviral drug discovery: Structure/function bases of the replication cycle. Antivir. Res. 2017, 141, 48–61. [Google Scholar] [CrossRef]
- Ruigrok, R.W.; Crépin, T.; Kolakofsky, D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr. Opin. Microbiol. 2011, 14, 504–510. [Google Scholar] [CrossRef]
- Hoenen, T.; Shabman, R.S.; Groseth, A.; Herwig, A.; Weber, M.; Schudt, G.; Dolnik, O.; Basler, C.F.; Becker, S.; Feldmann, H. Inclusion bodies are a site of ebolavirus replication. J. Virol. 2012, 86, 11779–11788. [Google Scholar] [CrossRef]
- Lier, C.; Becker, S.; Biedenkopf, N. Dynamic phosphorylation of Ebola virus VP30 in NP-induced inclusion bodies. Virology 2017, 512, 39–47. [Google Scholar] [CrossRef]
- Nanbo, A.; Watanabe, S.; Halfmann, P.; Kawaoka, Y. The spatio-temporal distribution dynamics of Ebola virus proteins and RNA in infected cells. Sci. Rep. 2013, 3, 1206. [Google Scholar] [CrossRef]
- Weik, M.; Enterlein, S.; Schlenz, K.; Mühlberger, E. The Ebola virus genomic replication promoter is bipartite and follows the rule of six. J. Virol. 2005, 79, 10660–10671. [Google Scholar] [CrossRef]
- Takahashi, K.; Halfmann, P.; Oyama, M.; Kozuka-Hata, H.; Noda, T.; Kawaoka, Y. DNA topoisomerase 1 facilitates the transcription and replication of the Ebola virus genome. J. Virol. 2013, 87, 8862–8869. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Pietzsch, C.; Ramanathan, P.; Santos, R.I.; Ilinykh, P.A.; Garcia-Blanco, M.A.; Bukreyev, A.; Bradrick, S.S. Staufen1 Interacts with Multiple Components of the Ebola Virus Ribonucleoprotein and Enhances Viral RNA Synthesis. mBio 2018, 9, e01771-18. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.; Chiramel, A.I.; Schmidt, M.L.; Chen, Y.C.; Whitt, N.; Watt, A.; Dunham, E.C.; Shifflett, K.; Traeger, S.; Leske, A.; et al. A genome-wide siRNA screen identifies a druggable host pathway essential for the Ebola virus life cycle. Genome Med. 2018, 10, 58. [Google Scholar] [CrossRef] [PubMed]
- Ning, Y.J.; Deng, F.; Hu, Z.; Wang, H. The roles of ebolavirus glycoproteins in viral pathogenesis. Virol. Sin. 2017, 32, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Jeffers, S.A.; Sanders, D.A.; Sanchez, A. Covalent modifications of the ebola virus glycoprotein. J. Virol. 2002, 76, 12463–12472. [Google Scholar] [CrossRef]
- Volchkov, V.E.; Feldmann, H.; Volchkova, V.A.; Klenk, H.D. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc. Natl. Acad. Sci. USA 1998, 95, 5762–5767. [Google Scholar] [CrossRef]
- Dolnik, O.; Volchkova, V.; Garten, W.; Carbonnelle, C.; Becker, S.; Kahnt, J.; Ströher, U.; Klenk, H.D.; Volchkov, V. Ectodomain shedding of the glycoprotein GP of Ebola virus. EMBO J. 2004, 23, 2175–2184. [Google Scholar] [CrossRef]
- Schudt, G.; Dolnik, O.; Kolesnikova, L.; Biedenkopf, N.; Herwig, A.; Becker, S. Transport of Ebolavirus Nucleocapsids Is Dependent on Actin Polymerization: Live-Cell Imaging Analysis of Ebolavirus-Infected Cells. J. Infect. Dis. 2015, 212 (Suppl. 2), S160-166. [Google Scholar] [CrossRef] [PubMed]
- Takamatsu, Y.; Kolesnikova, L.; Becker, S. Ebola virus proteins NP, VP35, and VP24 are essential and sufficient to mediate nucleocapsid transport. Proc. Natl. Acad. Sci. USA 2018, 115, 1075–1080. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, H.; Nichol, S.T.; Klenk, H.D.; Peters, C.J.; Sanchez, A. Characterization of filoviruses based on differences in structure and antigenicity of the virion glycoprotein. Virology 1994, 199, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Qu, Y.; Liu, Y.; Jambusaria, R.; Han, Z.; Ruthel, G.; Freedman, B.D.; Harty, R.N. Host IQGAP1 and Ebola virus VP40 interactions facilitate virus-like particle egress. J. Virol. 2013, 87, 7777–7780. [Google Scholar] [CrossRef]
- Yamayoshi, S.; Noda, T.; Ebihara, H.; Goto, H.; Morikawa, Y.; Lukashevich, I.S.; Neumann, G.; Feldmann, H.; Kawaoka, Y. Ebola virus matrix protein VP40 uses the COPII transport system for its intracellular transport. Cell Host Microbe 2008, 3, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Ruthel, G.; Demmin, G.L.; Kallstrom, G.; Javid, M.P.; Badie, S.S.; Will, A.B.; Nelle, T.; Schokman, R.; Nguyen, T.L.; Carra, J.H.; et al. Association of ebola virus matrix protein VP40 with microtubules. J. Virol. 2005, 79, 4709–4719. [Google Scholar] [CrossRef]
- Noda, T.; Ebihara, H.; Muramoto, Y.; Fujii, K.; Takada, A.; Sagara, H.; Kim, J.H.; Kida, H.; Feldmann, H.; Kawaoka, Y. Assembly and budding of Ebolavirus. PLoS Pathog. 2006, 2, e99. [Google Scholar] [CrossRef]
- Adu-Gyamfi, E.; Digman, M.A.; Gratton, E.; Stahelin, R.V. Single-particle tracking demonstrates that actin coordinates the movement of the Ebola virus matrix protein. Biophys. J. 2012, 103, L41–L43. [Google Scholar] [CrossRef]
- Han, Z.; Harty, R.N. Packaging of actin into Ebola virus VLPs. Virol. J. 2005, 2, 92. [Google Scholar] [CrossRef]
- Watanabe, S.; Noda, T.; Kawaoka, Y. Functional mapping of the nucleoprotein of Ebola virus. J. Virol. 2006, 80, 3743–3751. [Google Scholar] [CrossRef]
- Leung, D.W.; Borek, D.; Luthra, P.; Binning, J.M.; Anantpadma, M.; Liu, G.; Harvey, I.B.; Su, Z.; Endlich-Frazier, A.; Pan, J.; et al. An Intrinsically Disordered Peptide from Ebola Virus VP35 Controls Viral RNA Synthesis by Modulating Nucleoprotein-RNA Interactions. Cell Rep. 2015, 11, 376–389. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Wu, C.; Shi, L.; Luthra, P.; Pintilie, G.D.; Johnson, B.; Porter, J.R.; Ge, P.; Chen, M.; Liu, G.; et al. Electron Cryo-microscopy Structure of Ebola Virus Nucleoprotein Reveals a Mechanism for Nucleocapsid-like Assembly. Cell 2018, 172, 966–978.e12. [Google Scholar] [CrossRef]
- Kirchdoerfer, R.N.; Abelson, D.M.; Li, S.; Wood, M.R.; Saphire, E.O. Assembly of the Ebola Virus Nucleoprotein from a Chaperoned VP35 Complex. Cell Rep. 2015, 12, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.E.; Diehl, W.E.; Cai, Y.; Finch, C.L.; Akusobi, C.; Kirchdoerfer, R.N.; Bollinger, L.; Schaffner, S.F.; Brown, E.A.; Saphire, E.O.; et al. Reporter Assays for Ebola Virus Nucleoprotein Oligomerization, Virion-Like Particle Budding, and Minigenome Activity Reveal the Importance of Nucleoprotein Amino Acid Position 111. Viruses 2020, 12, 105. [Google Scholar] [CrossRef]
- Miyake, T.; Farley, C.M.; Neubauer, B.E.; Beddow, T.P.; Hoenen, T.; Engel, D.A. Ebola Virus Inclusion Body Formation and RNA Synthesis Are Controlled by a Novel Domain of Nucleoprotein Interacting with VP35. J. Virol. 2020, 94, e02100–e02119. [Google Scholar] [CrossRef] [PubMed]
- Pappalardo, M.; Reddin, I.G.; Cantoni, D.; Rossman, J.S.; Michaelis, M.; Wass, M.N. Changes associated with Ebola virus adaptation to novel species. Bioinformatics 2017, 33, 1911–1915. [Google Scholar] [CrossRef]
- Huang, Y.; Xu, L.; Sun, Y.; Nabel, G.J. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol. Cell 2002, 10, 307–316. [Google Scholar] [CrossRef]
- Noda, T.; Watanabe, S.; Sagara, H.; Kawaoka, Y. Mapping of the VP40-binding regions of the nucleoprotein of Ebola virus. J. Virol. 2007, 81, 3554–3562. [Google Scholar] [CrossRef]
- Kirchdoerfer, R.N.; Moyer, C.L.; Abelson, D.M.; Saphire, E.O. The Ebola Virus VP30-NP Interaction Is a Regulator of Viral RNA Synthesis. PLoS Pathog. 2016, 12, e1005937. [Google Scholar] [CrossRef]
- Xu, W.; Luthra, P.; Wu, C.; Batra, J.; Leung, D.W.; Basler, C.F.; Amarasinghe, G.K. Ebola virus VP30 and nucleoprotein interactions modulate viral RNA synthesis. Nat. Commun. 2017, 8, 15576. [Google Scholar] [CrossRef]
- Brandt, J.; Wendt, L.; Bodmer, B.S.; Mettenleiter, T.C.; Hoenen, T. The Cellular Protein CAD is Recruited into Ebola Virus Inclusion Bodies by the Nucleoprotein NP to Facilitate Genome Replication and Transcription. Cells 2020, 9, 1126. [Google Scholar] [CrossRef]
- Kruse, T.; Biedenkopf, N.; Hertz, E.P.T.; Dietzel, E.; Stalmann, G.; López-Méndez, B.; Davey, N.E.; Nilsson, J.; Becker, S. The Ebola Virus Nucleoprotein Recruits the Host PP2A-B56 Phosphatase to Activate Transcriptional Support Activity of VP30. Mol. Cell 2018, 69, 136–145.e6. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; He, Z.; Yuan, Y.; Huang, F.; Luo, B.; Zhang, J.; Pan, T.; Zhang, H.; Zhang, J. Host factor SMYD3 is recruited by Ebola virus nucleoprotein to facilitate viral mRNA transcription. Emerg. Microbes Infect. 2019, 8, 1347–1360. [Google Scholar] [CrossRef] [PubMed]
- Wendt, L.; Brandt, J.; Bodmer, B.S.; Reiche, S.; Schmidt, M.L.; Traeger, S.; Hoenen, T. The Ebola Virus Nucleoprotein Recruits the Nuclear RNA Export Factor NXF1 into Inclusion Bodies to Facilitate Viral Protein Expression. Cells 2020, 9, 187. [Google Scholar] [CrossRef] [PubMed]
- García-Dorival, I.; Wu, W.; Armstrong, S.D.; Barr, J.N.; Carroll, M.W.; Hewson, R.; Hiscox, J.A. Elucidation of the Cellular Interactome of Ebola Virus Nucleoprotein and Identification of Therapeutic Targets. J. Proteome Res. 2016, 15, 4290–4303. [Google Scholar] [CrossRef] [PubMed]
- Morwitzer, M.J.; Tritsch, S.R.; Cazares, L.H.; Ward, M.D.; Nuss, J.E.; Bavari, S.; Reid, S.P. Identification of RUVBL1 and RUVBL2 as Novel Cellular Interactors of the Ebola Virus Nucleoprotein. Viruses 2019, 11, 372. [Google Scholar] [CrossRef]
- Prins, K.C.; Binning, J.M.; Shabman, R.S.; Leung, D.W.; Amarasinghe, G.K.; Basler, C.F. Basic residues within the ebolavirus VP35 protein are required for its viral polymerase cofactor function. J. Virol. 2010, 84, 10581–10591. [Google Scholar] [CrossRef]
- Trunschke, M.; Conrad, D.; Enterlein, S.; Olejnik, J.; Brauburger, K.; Muhlberger, E. The L-VP35 and L-L interaction domains reside in the amino terminus of the Ebola virus L protein and are potential targets for antivirals. Virology 2013, 441, 135–145. [Google Scholar] [CrossRef]
- Luthra, P.; Ramanan, P.; Mire, C.E.; Weisend, C.; Tsuda, Y.; Yen, B.; Liu, G.; Leung, D.W.; Geisbert, T.W.; Ebihara, H.; et al. Mutual Antagonism between the Ebola Virus VP35 Protein and the RIG-I Activator PACT Determines Infection Outcome. Cell Host Microbe 2013, 14, 74–84. [Google Scholar] [CrossRef]
- Leung, D.W.; Prins, K.C.; Borek, D.M.; Farahbakhsh, M.; Tufariello, J.M.; Ramanan, P.; Nix, J.C.; Helgeson, L.A.; Otwinowski, Z.; Honzatko, R.B.; et al. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat. Struct. Mol. Biol. 2010, 17, 165–172. [Google Scholar] [CrossRef]
- Schümann, M.; Gantke, T.; Mühlberger, E. Ebola virus VP35 antagonizes PKR activity through its C-terminal interferon inhibitory domain. J. Virol. 2009, 83, 8993–8997. [Google Scholar] [CrossRef]
- Leung, D.W.; Ginder, N.D.; Fulton, D.B.; Nix, J.; Basler, C.F.; Honzatko, R.B.; Amarasinghe, G.K. Structure of the Ebola VP35 interferon inhibitory domain. Proc. Natl. Acad. Sci. USA 2009, 106, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Kimberlin, C.R.; Bornholdt, Z.A.; Li, S.; Woods, V.L., Jr.; MacRae, I.J.; Saphire, E.O. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc. Natl. Acad. Sci. USA 2010, 107, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Hoenen, T.; Biedenkopf, N.; Zielecki, F.; Jung, S.; Groseth, A.; Feldmann, H.; Becker, S. Oligomerization of Ebola Virus VP40 Is Essential for Particle Morphogenesis and Regulation of Viral Transcription. J. Virol. 2010, 84, 7053. [Google Scholar] [CrossRef] [PubMed]
- Bornholdt, Z.A.; Noda, T.; Abelson, D.M.; Halfmann, P.; Wood, M.R.; Kawaoka, Y.; Saphire, E.O. Structural rearrangement of ebola virus VP40 begets multiple functions in the virus life cycle. Cell 2013, 154, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Hoenen, T.; Volchkov, V.; Kolesnikova, L.; Mittler, E.; Timmins, J.; Ottmann, M.; Reynard, O.; Becker, S.; Weissenhorn, W. VP40 octamers are essential for Ebola virus replication. J. Virol. 2005, 79, 1898–1905. [Google Scholar] [CrossRef] [PubMed]
- Adu-Gyamfi, E.; Soni, S.P.; Jee, C.S.; Digman, M.A.; Gratton, E.; Stahelin, R.V. A loop region in the N-terminal domain of Ebola virus VP40 is important in viral assembly, budding, and egress. Viruses 2014, 6, 3837–3854. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, S.E.; Johnson, R.F.; Zhang, Y.A.; Sunyer, J.O.; Harty, R.N. Role for amino acids 212KLR214 of Ebola virus VP40 in assembly and budding. J. Virol. 2007, 81, 11452–11460. [Google Scholar] [CrossRef] [PubMed]
- Urata, S.; Ishikawa, T.; Yasuda, J. Roles of YIGL sequence of Ebola virus VP40 on genome replication and particle production. J. Gen. Virol. 2019, 100, 1099–1111. [Google Scholar] [CrossRef]
- Mpanju, O.M.; Towner, J.S.; Dover, J.E.; Nichol, S.T.; Wilson, C.A. Identification of two amino acid residues on Ebola virus glycoprotein 1 critical for cell entry. Virus Res. 2006, 121, 205–214. [Google Scholar] [CrossRef]
- Manicassamy, B.; Wang, J.; Jiang, H.; Rong, L. Comprehensive analysis of ebola virus GP1 in viral entry. J. Virol. 2005, 79, 4793–4805. [Google Scholar] [CrossRef] [PubMed]
- Brindley, M.A.; Hughes, L.; Ruiz, A.; McCray, P.B., Jr.; Sanchez, A.; Sanders, D.A.; Maury, W. Ebola virus glycoprotein 1: Identification of residues important for binding and postbinding events. J. Virol. 2007, 81, 7702–7709. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Fusco, M.L.; Hessell, A.J.; Oswald, W.B.; Burton, D.R.; Saphire, E.O. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature 2008, 454, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Manicassamy, B.; Caffrey, M.; Rong, L. Characterization of the receptor-binding domain of Ebola glycoprotein in viral entry. Virol. Sin. 2011, 26, 156–170. [Google Scholar] [CrossRef]
- Kuhn, J.H.; Radoshitzky, S.R.; Guth, A.C.; Warfield, K.L.; Li, W.; Vincent, M.J.; Towner, J.S.; Nichol, S.T.; Bavari, S.; Choe, H.; et al. Conserved receptor-binding domains of Lake Victoria marburgvirus and Zaire ebolavirus bind a common receptor. J. Biol. Chem. 2006, 281, 15951–15958. [Google Scholar] [CrossRef]
- Lee, J.E.; Saphire, E.O. Ebolavirus glycoprotein structure and mechanism of entry. Future Virol. 2009, 4, 621–635. [Google Scholar] [CrossRef]
- Dube, D.; Brecher, M.B.; Delos, S.E.; Rose, S.C.; Park, E.W.; Schornberg, K.L.; Kuhn, J.H.; White, J.M. The primed ebolavirus glycoprotein (19-kilodalton GP1,2): Sequence and residues critical for host cell binding. J. Virol. 2009, 83, 2883–2891. [Google Scholar] [CrossRef]
- Wang, B.; Wang, Y.; Frabutt, D.A.; Zhang, X.; Yao, X.; Hu, D.; Zhang, Z.; Liu, C.; Zheng, S.; Xiang, S.H.; et al. Mechanistic understanding of N-glycosylation in Ebola virus glycoprotein maturation and function. J. Biol. Chem. 2017, 292, 5860–5870. [Google Scholar] [CrossRef] [PubMed]
- Yaddanapudi, K.; Palacios, G.; Towner, J.S.; Chen, I.; Sariol, C.A.; Nichol, S.T.; Lipkin, W.I. Implication of a retrovirus-like glycoprotein peptide in the immunopathogenesis of Ebola and Marburg viruses. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2006, 20, 2519–2530. [Google Scholar] [CrossRef]
- John, S.P.; Wang, T.; Steffen, S.; Longhi, S.; Schmaljohn, C.S.; Jonsson, C.B. Ebola Virus VP30 Is an RNA Binding Protein. J. Virol. 2007, 81, 8967. [Google Scholar] [CrossRef]
- Modrof, J.; Becker, S.; Mühlberger, E. Ebola virus transcription activator VP30 is a zinc-binding protein. J. Virol. 2003, 77, 3334–3338. [Google Scholar] [CrossRef]
- Hartlieb, B.; Muziol, T.; Weissenhorn, W.; Becker, S. Crystal structure of the C-terminal domain of Ebola virus VP30 reveals a role in transcription and nucleocapsid association. Proc. Natl. Acad. Sci. USA 2007, 104, 624–629. [Google Scholar] [CrossRef]
- Zhang, A.P.; Bornholdt, Z.A.; Liu, T.; Abelson, D.M.; Lee, D.E.; Li, S.; Woods, V.L., Jr.; Saphire, E.O. The ebola virus interferon antagonist VP24 directly binds STAT1 and has a novel, pyramidal fold. PLoS Pathog. 2012, 8, e1002550. [Google Scholar] [CrossRef]
- Xu, W.; Edwards, M.R.; Borek, D.M.; Feagins, A.R.; Mittal, A.; Alinger, J.B.; Berry, K.N.; Yen, B.; Hamilton, J.; Brett, T.J.; et al. Ebola virus VP24 targets a unique NLS binding site on karyopherin alpha 5 to selectively compete with nuclear import of phosphorylated STAT1. Cell Host Microbe 2014, 16, 187–200. [Google Scholar] [CrossRef]
- Mateo, M.; Reid, S.P.; Leung, L.W.; Basler, C.F.; Volchkov, V.E. Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signaling. J. Virol. 2010, 84, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- Banadyga, L.; Hoenen, T.; Ambroggio, X.; Dunham, E.; Groseth, A.; Ebihara, H. Ebola virus VP24 interacts with NP to facilitate nucleocapsid assembly and genome packaging. Sci. Rep. 2017, 7, 7698. [Google Scholar] [CrossRef] [PubMed]
- Mühlberger, E.; Weik, M.; Volchkov, V.E.; Klenk, H.D.; Becker, S. Comparison of the transcription and replication strategies of marburg virus and Ebola virus by using artificial replication systems. J. Virol. 1999, 73, 2333–2342. [Google Scholar] [CrossRef] [PubMed]
- Shu, T.; Gan, T.; Bai, P.; Wang, X.; Qian, Q.; Zhou, H.; Cheng, Q.; Qiu, Y.; Yin, L.; Zhong, J.; et al. Ebola virus VP35 has novel NTPase and helicase-like activities. Nucleic Acids Res. 2019, 47, 5837–5851. [Google Scholar] [CrossRef]
- Johnson, R.F.; McCarthy, S.E.; Godlewski, P.J.; Harty, R.N. Ebola virus VP35-VP40 interaction is sufficient for packaging 3E-5E minigenome RNA into virus-like particles. J. Virol. 2006, 80, 5135–5144. [Google Scholar] [CrossRef] [PubMed]
- Basler, C.F.; Mikulasova, A.; Martinez-Sobrido, L.; Paragas, J.; Mühlberger, E.; Bray, M.; Klenk, H.D.; Palese, P.; García-Sastre, A. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 2003, 77, 7945–7956. [Google Scholar] [CrossRef]
- Prins, K.C.; Cárdenas, W.B.; Basler, C.F. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKKepsilon and TBK-1. J. Virol. 2009, 83, 3069–3077. [Google Scholar] [CrossRef]
- Chang, T.H.; Kubota, T.; Matsuoka, M.; Jones, S.; Bradfute, S.B.; Bray, M.; Ozato, K. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 2009, 5, e1000493. [Google Scholar] [CrossRef]
- Ivanov, A.; Ramanathan, P.; Parry, C.; Ilinykh, P.A.; Lin, X.; Petukhov, M.; Obukhov, Y.; Ammosova, T.; Amarasinghe, G.K.; Bukreyev, A.; et al. Global phosphoproteomic analysis of Ebola virions reveals a novel role for VP35 phosphorylation-dependent regulation of genome transcription. Cell. Mol. Life Sci. CMLS 2020, 77, 2579–2603. [Google Scholar] [CrossRef] [PubMed]
- Feldmann, H.; Klenk, H.D.; Sanchez, A. Molecular biology and evolution of filoviruses. Unconv. Agents Unclassif. Viruses 1993, 7, 81–100. [Google Scholar] [CrossRef]
- Baz-Martínez, M.; El Motiam, A.; Ruibal, P.; Condezo, G.N.; de la Cruz-Herrera, C.F.; Lang, V.; Collado, M.; San Martín, C.; Rodríguez, M.S.; Muñoz-Fontela, C.; et al. Regulation of Ebola virus VP40 matrix protein by SUMO. Sci. Rep. 2016, 6, 37258. [Google Scholar] [CrossRef] [PubMed]
- Licata, J.M.; Simpson-Holley, M.; Wright, N.T.; Han, Z.; Paragas, J.; Harty, R.N. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: Involvement of host proteins TSG101 and VPS-4. J. Virol. 2003, 77, 1812–1819. [Google Scholar] [CrossRef] [PubMed]
- Martin-Serrano, J.; Zang, T.; Bieniasz, P.D. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 2001, 7, 1313–1319. [Google Scholar] [CrossRef] [PubMed]
- Harty, R.N.; Brown, M.E.; Wang, G.; Huibregtse, J.; Hayes, F.P. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: Implications for filovirus budding. Proc. Natl. Acad. Sci. USA 2000, 97, 13871–13876. [Google Scholar] [CrossRef]
- Han, Z.; Sagum, C.A.; Bedford, M.T.; Sidhu, S.S.; Sudol, M.; Harty, R.N. ITCH E3 Ubiquitin Ligase Interacts with Ebola Virus VP40 To Regulate Budding. J. Virol. 2016, 90, 9163–9171. [Google Scholar] [CrossRef]
- Freed, E.O. Viral Late Domains. J. Virol. 2002, 76, 4679. [Google Scholar] [CrossRef]
- Neumann, G.; Ebihara, H.; Takada, A.; Noda, T.; Kobasa, D.; Jasenosky, L.D.; Watanabe, S.; Kim, J.H.; Feldmann, H.; Kawaoka, Y. Ebola Virus VP40 Late Domains Are Not Essential for Viral Replication in Cell Culture. J. Virol. 2005, 79, 10300. [Google Scholar] [CrossRef] [PubMed]
- Gc, J.B.; Johnson, K.A.; Husby, M.L.; Frick, C.T.; Gerstman, B.S.; Stahelin, R.V.; Chapagain, P.P. Interdomain salt-bridges in the Ebola virus protein VP40 and their role in domain association and plasma membrane localization. Protein Sci. A Publ. Protein Soc. 2016, 25, 1648–1658. [Google Scholar] [CrossRef] [PubMed]
- Knauer, S.H.; Artsimovitch, I.; Rösch, P. Transformer proteins. Cell Cycle 2012, 11, 4289–4290. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Knauer, S.H.; Rösch, P.; Artsimovitch, I. Transformation: The next level of regulation. RNA Biol. 2012, 9, 1418–1423. [Google Scholar] [CrossRef][Green Version]
- Adu-Gyamfi, E.; Soni, S.P.; Xue, Y.; Digman, M.A.; Gratton, E.; Stahelin, R.V. The Ebola virus matrix protein penetrates into the plasma membrane: A key step in viral protein 40 (VP40) oligomerization and viral egress. J. Biol. Chem. 2013, 288, 5779–5789. [Google Scholar] [CrossRef]
- Adu-Gyamfi, E.; Digman, M.A.; Gratton, E.; Stahelin, R.V. Investigation of Ebola VP40 assembly and oligomerization in live cells using number and brightness analysis. Biophys. J. 2012, 102, 2517–2525. [Google Scholar] [CrossRef]
- Johnson, K.A.; Pokhrel, R.; Budicini, M.R.; Gerstman, B.S.; Chapagain, P.P.; Stahelin, R.V. A Conserved Tryptophan in the Ebola Virus Matrix Protein C-Terminal Domain Is Required for Efficient Virus-Like Particle Formation. Pathogens 2020, 9, 402. [Google Scholar] [CrossRef]
- Francica, J.R.; Varela-Rohena, A.; Medvec, A.; Plesa, G.; Riley, J.L.; Bates, P. Steric shielding of surface epitopes and impaired immune recognition induced by the ebola virus glycoprotein. PLoS Pathog. 2010, 6, e1001098. [Google Scholar] [CrossRef]
- Hoffmann, M.; Crone, L.; Dietzel, E.; Paijo, J.; González-Hernández, M.; Nehlmeier, I.; Kalinke, U.; Becker, S.; Pöhlmann, S. A Polymorphism within the Internal Fusion Loop of the Ebola Virus Glycoprotein Modulates Host Cell Entry. J. Virol. 2017, 91. [Google Scholar] [CrossRef]
- Martinez, O.; Valmas, C.; Basler, C.F. Ebola virus-like particle-induced activation of NF-kappaB and Erk signaling in human dendritic cells requires the glycoprotein mucin domain. Virology 2007, 364, 342–354. [Google Scholar] [CrossRef]
- Malashkevich, V.N.; Schneider, B.J.; McNally, M.L.; Milhollen, M.A.; Pang, J.X.; Kim, P.S. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-A resolution. Proc. Natl. Acad. Sci. USA 1999, 96, 2662–2667. [Google Scholar] [CrossRef]
- Kühl, A.; Banning, C.; Marzi, A.; Votteler, J.; Steffen, I.; Bertram, S.; Glowacka, I.; Konrad, A.; Stürzl, M.; Guo, J.T.; et al. The Ebola virus glycoprotein and HIV-1 Vpu employ different strategies to counteract the antiviral factor tetherin. J. Infect. Dis. 2011, 204 (Suppl. 3), S850–S860. [Google Scholar] [CrossRef]
- Cocka, L.J.; Bates, P. Identification of Alternatively Translated Tetherin Isoforms with Differing Antiviral and Signaling Activities. PLoS Pathog. 2012, 8, e1002931. [Google Scholar] [CrossRef]
- Lennemann, N.J.; Rhein, B.A.; Ndungo, E.; Chandran, K.; Qiu, X.; Maury, W. Comprehensive functional analysis of N-linked glycans on Ebola virus GP1. mBio 2014, 5, e00862-13. [Google Scholar] [CrossRef]
- Reynard, O.; Borowiak, M.; Volchkova, V.A.; Delpeut, S.; Mateo, M.; Volchkov, V.E. Ebolavirus glycoprotein GP masks both its own epitopes and the presence of cellular surface proteins. J. Virol. 2009, 83, 9596–9601. [Google Scholar] [CrossRef]
- Sullivan, N.J.; Peterson, M.; Yang, Z.Y.; Kong, W.P.; Duckers, H.; Nabel, E.; Nabel, G.J. Ebola virus glycoprotein toxicity is mediated by a dynamin-dependent protein-trafficking pathway. J. Virol. 2005, 79, 547–553. [Google Scholar] [CrossRef]
- Francica, J.R.; Matukonis, M.K.; Bates, P. Requirements for cell rounding and surface protein down-regulation by Ebola virus glycoprotein. Virology 2009, 383, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Falzarano, D.; Krokhin, O.; Wahl-Jensen, V.; Seebach, J.; Wolf, K.; Schnittler, H.J.; Feldmann, H. Structure-function analysis of the soluble glycoprotein, sGP, of Ebola virus. Chembiochem. A Eur. J. Chem. Biol. 2006, 7, 1605–1611. [Google Scholar] [CrossRef] [PubMed]
- Falzarano, D.; Krokhin, O.; Van Domselaar, G.; Wolf, K.; Seebach, J.; Schnittler, H.J.; Feldmann, H. Ebola sGP--the first viral glycoprotein shown to be C-mannosylated. Virology 2007, 368, 83–90. [Google Scholar] [CrossRef]
- Iwasa, A.; Shimojima, M.; Kawaoka, Y. sGP serves as a structural protein in Ebola virus infection. J. Infect. Dis. 2011, 204 (Suppl. 3), S897–S903. [Google Scholar] [CrossRef] [PubMed]
- Bradley, J.H.; Harrison, A.; Corey, A.; Gentry, N.; Gregg, R.K. Ebola virus secreted glycoprotein decreases the anti-viral immunity of macrophages in early inflammatory responses. Cell. Immunol. 2018, 324, 24–32. [Google Scholar] [CrossRef]
- Wahl-Jensen, V.M.; Afanasieva, T.A.; Seebach, J.; Ströher, U.; Feldmann, H.; Schnittler, H.J. Effects of Ebola virus glycoproteins on endothelial cell activation and barrier function. J. Virol. 2005, 79, 10442–10450. [Google Scholar] [CrossRef]
- Mohan, G.S.; Li, W.; Ye, L.; Compans, R.W.; Yang, C. Antigenic subversion: A novel mechanism of host immune evasion by Ebola virus. PLoS Pathog. 2012, 8, e1003065. [Google Scholar] [CrossRef]
- Zhu, W.; Banadyga, L.; Emeterio, K.; Wong, G.; Qiu, X. The Roles of Ebola Virus Soluble Glycoprotein in Replication, Pathogenesis, and Countermeasure Development. Viruses 2019, 11, 999. [Google Scholar] [CrossRef]
- Gallaher, W.R.; Garry, R.F. Modeling of the Ebola virus delta peptide reveals a potential lytic sequence motif. Viruses 2015, 7, 285–305. [Google Scholar] [CrossRef] [PubMed]
- Radoshitzky, S.R.; Warfield, K.L.; Chi, X.; Dong, L.; Kota, K.; Bradfute, S.B.; Gearhart, J.D.; Retterer, C.; Kranzusch, P.J.; Misasi, J.N.; et al. Ebolavirus delta-peptide immunoadhesins inhibit marburgvirus and ebolavirus cell entry. J. Virol. 2011, 85, 8502–8513. [Google Scholar] [CrossRef] [PubMed]
- Hyser, J.M.; Collinson-Pautz, M.R.; Utama, B.; Estes, M.K. Rotavirus Disrupts Calcium Homeostasis by NSP4 Viroporin Activity. mBio 2010, 1, e00265-10. [Google Scholar] [CrossRef]
- He, J.; Melnik, L.I.; Komin, A.; Wiedman, G.; Fuselier, T.; Morris, C.F.; Starr, C.G.; Searson, P.C.; Gallaher, W.R.; Hristova, K.; et al. Ebola Virus Delta Peptide Is a Viroporin. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Ito, H.; Watanabe, S.; Takada, A.; Kawaoka, Y. Ebola virus glycoprotein: Proteolytic processing, acylation, cell tropism, and detection of neutralizing antibodies. J. Virol. 2001, 75, 1576–1580. [Google Scholar] [CrossRef]
- Escudero-Perez, B.; Volchkova, V.A.; Dolnik, O.; Lawrence, P.; Volchkov, V.E. Shed GP of Ebola virus triggers immune activation and increased vascular permeability. PLoS Pathog. 2014, 10, e1004509. [Google Scholar] [CrossRef]
- Hartlieb, B.; Modrof, J.; Mühlberger, E.; Klenk, H.D.; Becker, S. Oligomerization of Ebola virus VP30 is essential for viral transcription and can be inhibited by a synthetic peptide. J. Biol. Chem. 2003, 278, 41830–41836. [Google Scholar] [CrossRef]
- Weik, M.; Modrof, J.; Klenk, H.D.; Becker, S.; Mühlberger, E. Ebola virus VP30-mediated transcription is regulated by RNA secondary structure formation. J. Virol. 2002, 76, 8532–8539. [Google Scholar] [CrossRef] [PubMed]
- Ilinykh, P.A.; Tigabu, B.; Ivanov, A.; Ammosova, T.; Obukhov, Y.; Garron, T.; Kumari, N.; Kovalskyy, D.; Platonov, M.O.; Naumchik, V.S.; et al. Role of protein phosphatase 1 in dephosphorylation of Ebola virus VP30 protein and its targeting for the inhibition of viral transcription. J. Biol. Chem. 2014, 289, 22723–22738. [Google Scholar] [CrossRef]
- Martinez, M.J.; Volchkova, V.A.; Raoul, H.; Alazard-Dany, N.; Reynard, O.; Volchkov, V.E. Role of VP30 Phosphorylation in the Ebola Virus Replication Cycle. J. Infect. Dis. 2011, 204, S934–S940. [Google Scholar] [CrossRef]
- Biedenkopf, N.; Hartlieb, B.; Hoenen, T.; Becker, S. Phosphorylation of Ebola virus VP30 influences the composition of the viral nucleocapsid complex: Impact on viral transcription and replication. J. Biol. Chem. 2013, 288, 11165–11174. [Google Scholar] [CrossRef]
- Biedenkopf, N.; Lier, C.; Becker, S. Dynamic Phosphorylation of VP30 Is Essential for Ebola Virus Life Cycle. J. Virol. 2016, 90, 4914–4925. [Google Scholar] [CrossRef] [PubMed]
- Martínez, M.J.; Biedenkopf, N.; Volchkova, V.; Hartlieb, B.; Alazard-Dany, N.; Reynard, O.; Becker, S.; Volchkov, V. Role of Ebola Virus VP30 in Transcription Reinitiation. J. Virol. 2008, 82, 12569–12573. [Google Scholar] [CrossRef]
- Watt, A.; Moukambi, F.; Banadyga, L.; Groseth, A.; Callison, J.; Herwig, A.; Ebihara, H.; Feldmann, H.; Hoenen, T. A novel life cycle modeling system for Ebola virus shows a genome length-dependent role of VP24 in virus infectivity. J. Virol. 2014, 88, 10511–10524. [Google Scholar] [CrossRef] [PubMed]
- Ilinykh, P.A.; Lubaki, N.M.; Widen, S.G.; Renn, L.A.; Theisen, T.C.; Rabin, R.L.; Wood, T.G.; Bukreyev, A. Different Temporal Effects of Ebola Virus VP35 and VP24 Proteins on Global Gene Expression in Human Dendritic Cells. J. Virol. 2015, 89, 7567–7583. [Google Scholar] [CrossRef]
- Halfmann, P.; Neumann, G.; Kawaoka, Y. The Ebolavirus VP24 protein blocks phosphorylation of p38 mitogen-activated protein kinase. J. Infect. Dis. 2011, 204 (Suppl. 3), S953–S956. [Google Scholar] [CrossRef] [PubMed]
- Guito, J.C.; Albariño, C.G.; Chakrabarti, A.K.; Towner, J.S. Novel activities by ebolavirus and marburgvirus interferon antagonists revealed using a standardized in vitro reporter system. Virology 2017, 501, 147–165. [Google Scholar] [CrossRef] [PubMed]
- Basler, C.F. Innate immune evasion by filoviruses. Virology 2015, 479–480, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Li, Z.; Jenni, S.; Rahmeh, A.A.; Morin, B.M.; Grant, T.; Grigorieff, N.; Harrison, S.C.; Whelan, S.P.J. Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy. Cell 2015, 162, 314–327. [Google Scholar] [CrossRef] [PubMed]
- Groseth, A.; Charton, J.E.; Sauerborn, M.; Feldmann, F.; Jones, S.M.; Hoenen, T.; Feldmann, H. The Ebola virus ribonucleoprotein complex: A novel VP30-L interaction identified. Virus Res. 2009, 140, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Cantoni, D.; Rossman, J.S. Ebolaviruses: New roles for old proteins. PLoS Negl. Trop. Dis. 2018, 12, e0006349. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.L.; Hoenen, T. Characterization of the catalytic center of the Ebola virus L polymerase. PLoS Negl. Trop. Dis. 2017, 11, e0005996. [Google Scholar] [CrossRef]
- Valle, C.; Martin, B.; Debart, F.; Vasseur, J.-J.; Imbert, I.; Canard, B.; Coutard, B.; Decroly, E. The C-terminal domain of the Sudan ebolavirus L protein is essential for RNA binding and methylation. J. Virol. 2020, 94, e00520-20. [Google Scholar] [CrossRef]
- Ogino, T.; Green, T.J. RNA Synthesis and Capping by Non-segmented Negative Strand RNA Viral Polymerases: Lessons From a Prototypic Virus. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Poch, O.; Blumberg, B.M.; Bougueleret, L.; Tordo, N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: Theoretical assignment of functional domains. J. Gen. Virol. 1990, 71 Pt 5, 1153–1162. [Google Scholar] [CrossRef]
- Ogino, T.; Banerjee, A.K. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell 2007, 25, 85–97. [Google Scholar] [CrossRef]
- Rahmeh, A.A.; Li, J.; Kranzusch, P.J.; Whelan, S.P. Ribose 2′-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J. Virol. 2009, 83, 11043–11050. [Google Scholar] [CrossRef] [PubMed]
- Martin, B.; Coutard, B.; Guez, T.; Paesen, G.C.; Canard, B.; Debart, F.; Vasseur, J.J.; Grimes, J.M.; Decroly, E. The methyltransferase domain of the Sudan ebolavirus L protein specifically targets internal adenosines of RNA substrates, in addition to the cap structure. Nucleic Acids Res. 2018, 46, 7902–7912. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.T.; Islam, M.O. A Highly Conserved GEQYQQLR Epitope Has Been Identified in the Nucleoprotein of Ebola Virus by Using an In Silico Approach. Adv. Bioinform. 2015, 2015, 278197. [Google Scholar] [CrossRef]
- Simmons, G.; Lee, A.; Rennekamp, A.J.; Fan, X.; Bates, P.; Shen, H. Identification of murine T-cell epitopes in Ebola virus nucleoprotein. Virology 2004, 318, 224–230. [Google Scholar] [CrossRef]
- Tsuda, Y.; Caposio, P.; Parkins, C.J.; Botto, S.; Messaoudi, I.; Cicin-Sain, L.; Feldmann, H.; Jarvis, M.A. A replicating cytomegalovirus-based vaccine encoding a single Ebola virus nucleoprotein CTL epitope confers protection against Ebola virus. PLoS Negl. Trop. Dis. 2011, 5, e1275. [Google Scholar] [CrossRef]
- Becquart, P.; Mahlakoiv, T.; Nkoghe, D.; Leroy, E.M. Identification of continuous human B-cell epitopes in the VP35, VP40, nucleoprotein and glycoprotein of Ebola virus. PLoS ONE 2014, 9, e96360. [Google Scholar] [CrossRef]
- Binning, J.M.; Wang, T.; Luthra, P.; Shabman, R.S.; Borek, D.M.; Liu, G.; Xu, W.; Leung, D.W.; Basler, C.F.; Amarasinghe, G.K. Development of RNA aptamers targeting Ebola virus VP35. Biochemistry 2013, 52, 8406–8419. [Google Scholar] [CrossRef]
- Ahmad, B.; Ashfaq, U.A.; Rahman, M.U.; Masoud, M.S.; Yousaf, M.Z. Conserved B and T cell epitopes prediction of ebola virus glycoprotein for vaccine development: An immuno-informatics approach. Microb. Pathog. 2019, 132, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Babirye, P.; Musubika, C.; Kirimunda, S.; Downing, R.; Lutwama, J.J.; Mbidde, E.K.; Weyer, J.; Paweska, J.T.; Joloba, M.L.; Wayengera, M. Identity and validity of conserved B cell epitopes of filovirus glycoprotein: Towards rapid diagnostic testing for Ebola and possibly Marburg virus disease. BMC Infect. Dis. 2018, 18, 498. [Google Scholar] [CrossRef]
- Balzarini, J. Targeting the glycans of glycoproteins: A novel paradigm for antiviral therapy. Nat. Rev. Microbiol. 2007, 5, 583–597. [Google Scholar] [CrossRef]
- Bukreyev, A.; Marzi, A.; Feldmann, F.; Zhang, L.; Yang, L.; Ward, J.M.; Dorward, D.W.; Pickles, R.J.; Murphy, B.R.; Feldmann, H.; et al. Chimeric human parainfluenza virus bearing the Ebola virus glycoprotein as the sole surface protein is immunogenic and highly protective against Ebola virus challenge. Virology 2009, 383, 348–361. [Google Scholar] [CrossRef]
- Gaudinski, M.R.; Coates, E.E.; Novik, L.; Widge, A.; Houser, K.V.; Burch, E.; Holman, L.A.; Gordon, I.J.; Chen, G.L.; Carter, C.; et al. Safety, tolerability, pharmacokinetics, and immunogenicity of the therapeutic monoclonal antibody mAb114 targeting Ebola virus glycoprotein (VRC 608): An open-label phase 1 study. Lancet 2019, 393, 889–898. [Google Scholar] [CrossRef]
- Keck, Z.Y.; Enterlein, S.G.; Howell, K.A.; Vu, H.; Shulenin, S.; Warfield, K.L.; Froude, J.W.; Araghi, N.; Douglas, R.; Biggins, J.; et al. Macaque Monoclonal Antibodies Targeting Novel Conserved Epitopes within Filovirus Glycoprotein. J. Virol. 2016, 90, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Marzi, A.; Murphy, A.A.; Feldmann, F.; Parkins, C.J.; Haddock, E.; Hanley, P.W.; Emery, M.J.; Engelmann, F.; Messaoudi, I.; Feldmann, H.; et al. Cytomegalovirus-based vaccine expressing Ebola virus glycoprotein protects nonhuman primates from Ebola virus infection. Sci. Rep. 2016, 6, 21674. [Google Scholar] [CrossRef] [PubMed]
- Marzi, A.; Robertson, S.J.; Haddock, E.; Feldmann, F.; Hanley, P.W.; Scott, D.P.; Strong, J.E.; Kobinger, G.; Best, S.M.; Feldmann, H. EBOLA VACCINE. VSV-EBOV rapidly protects macaques against infection with the 2014/15 Ebola virus outbreak strain. Science 2015, 349, 739–742. [Google Scholar] [CrossRef] [PubMed]
- Wong, G.; Audet, J.; Fernando, L.; Fausther-Bovendo, H.; Alimonti, J.B.; Kobinger, G.P.; Qiu, X. Immunization with vesicular stomatitis virus vaccine expressing the Ebola glycoprotein provides sustained long-term protection in rodents. Vaccine 2014, 32, 5722–5729. [Google Scholar] [CrossRef]
- Warren, T.K.; Whitehouse, C.A.; Wells, J.; Welch, L.; Heald, A.E.; Charleston, J.S.; Sazani, P.; Iversen, P.L.; Bavari, S. A single phosphorodiamidate morpholino oligomer targeting VP24 protects rhesus monkeys against lethal Ebola virus infection. mBio 2015, 6, e02344-14. [Google Scholar] [CrossRef]
- Srivastava, P.N.; Jain, R.; Dubey, S.D.; Bhatnagar, S.; Ahmad, N. Prediction of epitope-based peptides for vaccine development from coat proteins GP2 and VP24 of Ebola virus using immunoinformatics. Int. J. Pept. Res. Ther. 2016, 22, 119–133. [Google Scholar] [CrossRef]
- FDA. Available online: https://www.fda.gov/news-events/press-announcements/first-fda-approved-vaccine-prevention-ebola-virus-disease-marking-critical-milestone-public-health (accessed on 7 May 2020).
- Jain, S.; Khaiboullina, S.F.; Baranwal, M. Immunological Perspective for Ebola Virus Infection and Various Treatment Measures Taken to Fight the Disease. Pathogens 2020, 9, 850. [Google Scholar] [CrossRef]
- Chung, E.H. Vaccine allergies. Clin. Exp. Vaccine Res. 2014, 3, 50–57. [Google Scholar] [CrossRef]
- Skwarczynski, M.; Toth, I. Peptide-based synthetic vaccines. Chem. Sci. 2016, 7, 842–854. [Google Scholar] [CrossRef] [PubMed]
- Andersen, P.; Doherty, T.M. The success and failure of BCG—Implications for a novel tuberculosis vaccine. Nat. Rev. Microbiol. 2005, 3, 656–662. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).