Impact of Host Resistance to Tomato Spotted Wilt Orthotospovirus in Peanut Cultivars on Virus Population Genetics and Thrips Fitness
Abstract
:1. Introduction
2. Results
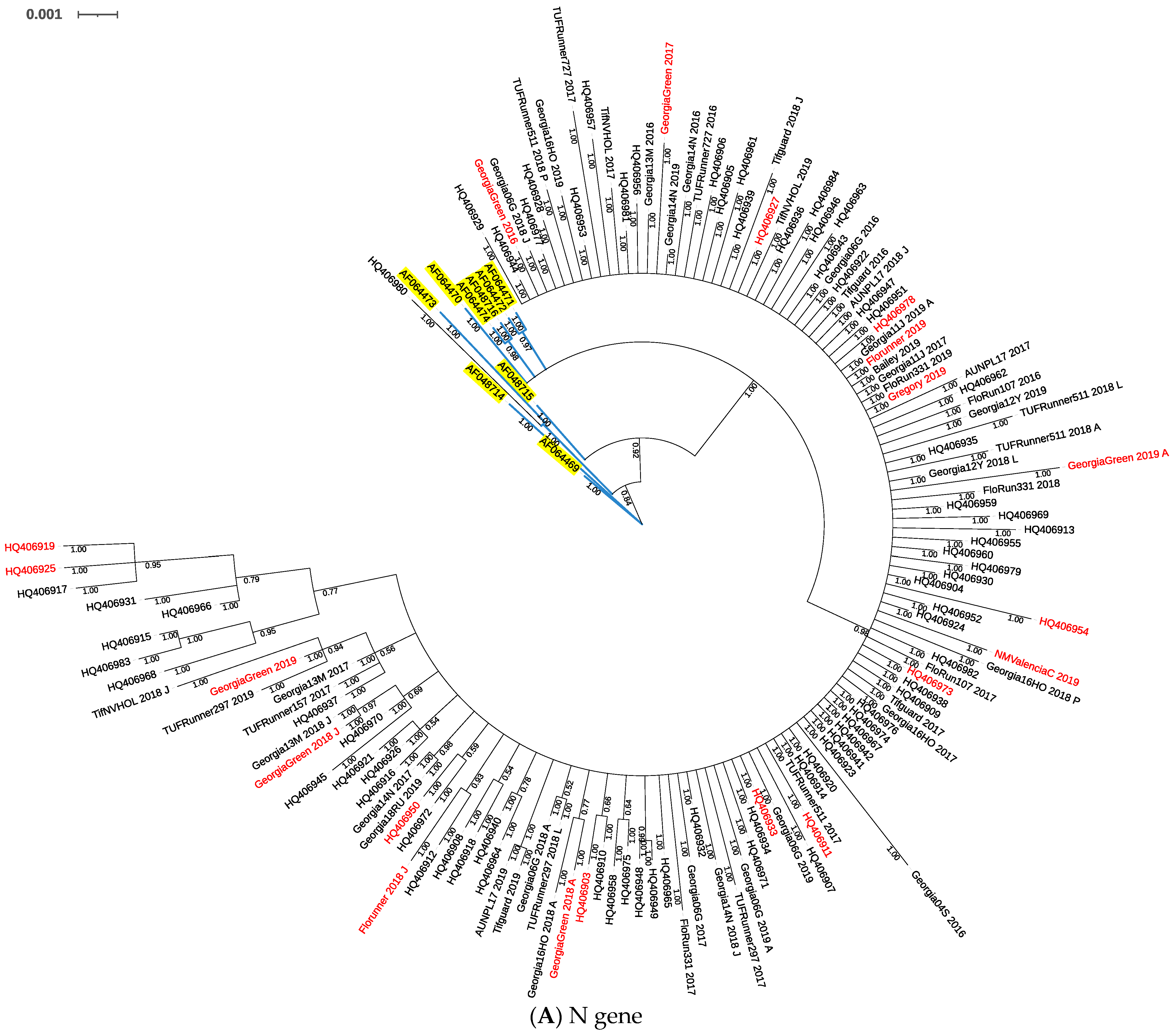
2.1. Phylogeny of TSWV Isolates
2.2. Genetic Diversity, Test of Neutrality, and Identification of Selection
2.3. Genetic Differentiation between Subgroups of TSWV Isolates
2.4. Thrips Feeding Injury and Survival
2.5. Thrips Development, Reproduction, and Oviposition
3. Discussion
4. Materials and Methods
4.1. TSWV Isolates
4.2. RNA Extraction, cDNA Synthesis, PCR, and Sequencing
4.3. TSWV Isolates and Sequence Alignments
4.4. Phylogenetic Analysis and Tree Construction
4.5. Genetic Diversity, Test of Neutrality, and Identification of Selection
4.6. Test of Gene Flow and Population Differentiation
4.7. Thrips Feeding Injury and Survival
4.8. Thrips Development, Reproduction, and Oviposition
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| N | NSs | NSm | Gn/Gc | RdRp | |||||
|---|---|---|---|---|---|---|---|---|---|
| Isolate ID | GenBank Accession | Isolate ID | GenBank Accession | Isolate ID | GenBank Accession | Isolate ID | GenBank Accession | Isolate ID | GenBank Accession |
| FloRun331 2018 | MW519186 | Tifguard 2017 | MW519245 | GeorgiaGreen 2018 A | MW519304 | Georgia14N 2016 | MW519362 | Georgia04S 2016 | MW519421 |
| GeorgiaGreen 2019 A | MW519187 | Florunner 2018 J | MW519246 | FloRun331 2018 L | MW519305 | Georgia12Y 2019 | MW519363 | Tifguard 2018 J | MW519422 |
| Georgia12Y 2018 L | MW519188 | Georgia04S 2016 | MW519247 | GeorgiaGreen 2016 | MW519306 | NMValenciaC 2019 | MW519364 | Florunner 2018 J | MW519423 |
| AUNPL17 2017 | MW519189 | Georgia14N 2017 | MW519248 | Georgia13M 2018 J | MW519307 | FloRun107 2016 | MW519365 | Florunner 2019 | MW519424 |
| Georgia13M 2016 | MW519190 | Georgia18RU 2019 | MW519249 | GeorgiaGreen 2018 J | MW519308 | TUFRunner157 2017 | MW519366 | Georgia06G 2016 | MW519425 |
| Georgia16HO 2019 | MW519191 | GeorgiaGreen 2019 A | MW519250 | Georgia06G 2018 A | MW519309 | Bailey 2019 | MW519367 | GeorgiaGreen 2016 | MW519426 |
| Florunner 2018 J | MW519192 | Georgia12Y 2019 | MW519251 | TUFRunner511 2018 L | MW519310 | Gregory 2019 | MW519368 | TUFRunner297 2018 L | MW519427 |
| Georgia16HO 2017 | MW519193 | Georgia16HO 2017 | MW519252 | NMValenciaC 2019 | MW519311 | Georgia13M 2017 | MW519369 | TUFRunner727 2017 | MW519428 |
| Tifguard 2017 | MW519194 | Georgia16HO 2019 | MW519253 | Georgia06G 2018 J | MW519312 | Georgia06G 2019 A | MW519370 | GeorgiaGreen 2018 A | MW519429 |
| FloRun107 2017 | MW519195 | Georgia06G 2019 | MW519254 | Bailey 2019 | MW519313 | GeorgiaGreen 2016 | MW519371 | Georgia06G 2019 A | MW519430 |
| Georgia06G 2019 | MW519196 | NMValenciaC 2019 | MW519255 | Gregory 2019 | MW519314 | GeorgiaGreen 2018 A | MW519372 | Georgia11J 2017 | MW519431 |
| Georgia14N 2018 J | MW519197 | Georgia14N 2018 J | MW519256 | Georgia06G 2019 A | MW519315 | TifNVHOL 2017 | MW519373 | Georgia16HO 2018 P | MW519432 |
| TUFRunner297 2017 | MW519198 | TUFRunner297 2017 | MW519257 | Georgia14N 2016 | MW519316 | Georgia16HO 2018 A | MW519374 | Tifguard 2016 | MW519433 |
| FloRun107 2016 | MW519199 | Georgia12Y 2018 L | MW519258 | AUNPL17 2017 | MW519317 | Georgia06G 2018 J | MW519375 | TUFRunner511 2018 P | MW519434 |
| Georgia14N 2017 | MW519200 | TUFRunner511 2018 L | MW519259 | Georgia04S 2016 | MW519318 | TUFRunner511 2018 L | MW519376 | FloRun107 2017 | MW519435 |
| Georgia18RU 2019 | MW519201 | Georgia06G 2019 A | MW519260 | AUNPL17 2018 J | MW519319 | Georgia11J 2019 A | MW519377 | GeorgiaGreen 2019 | MW519436 |
| Georgia04S 2016 | MW519202 | Georgia13M 2016 | MW519261 | TifNVHOL 2019 | MW519320 | Georgia13M 2018 J | MW519378 | TUFRunner297 2019 | MW519437 |
| Georgia06G 2017 | MW519203 | TUFRunner511 2017 | MW519262 | TUFRunner511 2018 A | MW519321 | GeorgiaGreen 2018 J | MW519379 | Georgia14N 2016 | MW519438 |
| Georgia06G 2016 | MW519204 | FloRun107 2016 | MW519263 | GeorgiaGreen 2019 | MW519322 | Georgia06G 2017 | MW519380 | Georgia16HO 2018 A | MW519439 |
| AUNPL17 2018 J | MW519205 | TUFRunner511 2018 A | MW519264 | GeorgiaGreen 2017 | MW519323 | Georgia14N 2019 | MW519381 | Georgia14N 2017 | MW519440 |
| TUFRunner331 2019 | MW519206 | AUNPL17 2017 | MW519265 | Georgia13M 2016 | MW519324 | TUFRunner727 2017 | MW519382 | TifNVHOL 2018 J | MW519441 |
| Gregory 2019 | MW519207 | GeorgiaGreen 2016 | MW519266 | TUFRunner297 2019 | MW519325 | FloRun331 2018 L | MW519383 | FloRun331 2018 L | MW519442 |
| Georgia11J 2019 A | MW519208 | Georgia16HO 2018 P | MW519267 | Georgia16HO 2018 P | MW519326 | Georgia06G 2018 A | MW519384 | Georgia12Y 2018 L | MW519443 |
| Bailey 2019 | MW519209 | FloRun331 2018 L | MW519268 | TifNVHOL 2018 J | MW519327 | Georgia04S 2016 | MW519385 | TUFRunner511 2018 L | MW519444 |
| Georgia11J 2017 | MW519210 | AUNPL17 2019 | MW519269 | FloRun107 2017 | MW519328 | Georgia18RU 2019 | MW519386 | Georgia06G 2019 | MW519445 |
| TifNVHOL 2019 | MW519211 | Tifguard 2019 | MW519270 | TUFRunner297 2017 | MW519329 | Georgia06G 2019 | MW519387 | AUNPL17 2017 | MW519446 |
| Tifguard 2016 | MW519212 | Georgia06G 2018 A | MW519271 | Georgia13M 2017 | MW519330 | TUFRunner511 2018 A | MW519388 | TUFRunner297 2017 | MW519447 |
| Georgia13M 2018 J | MW519213 | Georgia06G 2018 J | MW519272 | TUFRunner511 2017 | MW519331 | Georgia06G 2016 | MW519389 | FloRun331 2017 | MW519448 |
| GeorgiaGreen 2018 J | MW519214 | Georgia14N 2019 | MW519273 | FloRun331 2019 | MW519332 | TifNVHOL 2019 | MW519390 | FloRun107 2016 | MW519449 |
| Tifguard 2018 J | MW519215 | Georgia11J 2019 A | MW519274 | AUNPL17 2019 | MW519333 | AUNPL17 2018 J | MW519391 | Georgia12Y 2019 | MW519450 |
| TUFRunner727 2016 | MW519216 | Georgia11J 2017 | MW519275 | Tifguard 2019 | MW519334 | Florunner 2019 | MW519392 | Georgia18RU 2019 | MW519451 |
| Georgia06G 2018 A | MW519217 | Tifguard 2016 | MW519276 | Florunner 2018 J | MW519335 | Georgia14N 2018 J | MW519393 | TifNVHOL 2017 | MW519452 |
| TUFRunner297 2018 L | MW519218 | Florunner 2019 | MW519277 | TUFRunner511 2018 P | MW519336 | Georgia16HO 2018 P | MW519394 | TUFRunner727 2016 | MW519453 |
| AUNPL17 2019 | MW519219 | GeorgiaGreen 2017 | MW519278 | Georgia14N 2018 J | MW519337 | TUFRunner297 2017 | MW519395 | Georgia16HO 2017 | MW519454 |
| Tifguard 2019 | MW519220 | TUFRunner297 2019 | MW519279 | TUFRunner297 2018 L | MW519338 | FloRun107 2017 | MW519396 | TUFRunner157 2017 | MW519455 |
| TUFRunner511 2018 P | MW519221 | TifNVHOL 2018 J | MW519280 | GeorgiaGreen 2019 A | MW519339 | TUFRunner511 2018 P | MW519397 | Georgia06G 2018 J | MW519456 |
| Florunner 2019 | MW519222 | GeorgiaGreen 2019 | MW519281 | Georgia16HO 2019 | MW519340 | AUNPL17 2019 | MW519398 | Georgia16HO 2019 | MW519457 |
| Georgia14N 2016 | MW519223 | Georgia13M 2017 | MW519282 | Tifguard 2016 | MW519341 | Tifguard 2019 | MW519399 | NMValenciaC 2019 | MW519458 |
| Georgia14N 2019 | MW519224 | TUFRunner157 2017 | MW519283 | Florunner 2019 | MW519342 | Georgia13M 2016 | MW519400 | FloRun331 2019 | MW519459 |
| GeorgiaGreen 2017 | MW519225 | Bailey 2019 | MW519284 | Georgia18RU 2019 | MW519343 | TifNVHOL 2018 J | MW519401 | TUFRunner511 2017 | MW519460 |
| Georgia16HO 2018 A | MW519226 | Gregory 2019 | MW519285 | Georgia06G 2016 | MW519344 | TUFRunner297 2019 | MW519402 | Georgia13M 2018 J | MW519461 |
| GeorgiaGreen 2018 A | MW519227 | TifNVHOL 2019 | MW519286 | TUFRunner727 2016 | MW519345 | GeorgiaGreen 2019 | MW519403 | GeorgiaGreen 2018 J | MW519462 |
| TifNVHOL 2017 | MW519228 | Georgia16HO 2018 A | MW519287 | Tifguard 2017 | MW519346 | GeorgiaGreen 2017 | MW519404 | AUNPL17 2019 | MW519463 |
| Georgia06G 2018 J | MW519229 | Tifguard 2018 J | MW519288 | Georgia16HO 2017 | MW519347 | AUNPL17 2017 | MW519405 | Tifguard 2019 | MW519464 |
| Georgia13M 2017 | MW519230 | TUFRunner297 2018 L | MW519289 | FloRun331 2017 | MW519348 | TUFRunner727 2016 | MW519406 | Georgia14N 2018 J | MW519465 |
| TUFRunner157 2017 | MW519231 | TifNVHOL 2017 | MW519290 | Georgia12Y 2018 L | MW519349 | Tifguard 2017 | MW519407 | Georgia14N 2019 | MW519466 |
| TifNVHOL 2018 J | MW519232 | Georgia06G 2016 | MW519291 | Tifguard 2018 J | MW519350 | Georgia16HO 2017 | MW519408 | GeorgiaGreen 2019 A | MW519467 |
| GeorgiaGreen 2019 | MW519233 | FloRun331 2017 | MW519292 | Georgia06G 2019 | MW519351 | FloRun331 2017 | MW519409 | TifNVHOL 2019 | MW519468 |
| TUFRunner297 2019 | MW519234 | TUFRunner727 2016 | MW519293 | Georgia11J 2017 | MW519352 | Georgia11J 2017 | MW519410 | GeorgiaGreen 2017 | MW519469 |
| GeorgiaGreen 2016 | MW519235 | Georgia14N 2016 | MW519294 | Georgia14N 2017 | MW519353 | Georgia14N 2017 | MW519411 | Tifguard 2017 | MW519470 |
| TUFRunner331 2017 | MW519236 | Georgia06G 2017 | MW519295 | FloRun107 2016 | MW519354 | Georgia12Y 2018 L | MW519412 | Georgia06G 2017 | MW519471 |
| TUFRunner727 2017 | MW519237 | FloRun331 2019 | MW519296 | Georgia06G 2017 | MW519355 | Tifguard 2018 J | MW519413 | Georgia13M 2017 | MW519472 |
| Georgia06G 2019 A | MW519238 | TUFRunner511 2018 P | MW519297 | TifNVHOL 2017 | MW519356 | Georgia16HO 2019 | MW519414 | ||
| Georgia16HO 2018 P | MW519239 | AUNPL17 2018 J | MW519298 | Georgia11J 2019 A | MW519357 | Tifguard 2016 | MW519415 | ||
| NMValenciaC 2019 | MW519240 | Georgia13M 2018 J | MW519299 | TUFRunner727 2017 | MW519358 | TUFRunner297 2018 L | MW519416 | ||
| TUFRunner511 2018 L | MW519241 | GeorgiaGreen 2018 J | MW519300 | Georgia16HO 2018 A | MW519359 | GeorgiaGreen 2019 A | MW519417 | ||
| Georgia12Y 2019 | MW519242 | GeorgiaGreen 2018 A | MW519301 | Georgia14N 2019 | MW519360 | FloRun331 2019 | MW519418 | ||
| TUFRunner511 2018 A | MW519243 | TUFRunner727 2017 | MW519302 | TUFRunner157 2017 | MW519361 | Florunner 2018 J | MW519419 | ||
| TUFRunner511 2017 | MW519244 | FloRun107 2017 | MW519303 | TUFRunner511 2017 | MW519420 | ||||
References
- Culbreath, A.K. Tomato Spotted Wilt Virus Epidemic in Flue-Cured Tobacco in Georgia. Plant Dis. 1991, 75, 483. [Google Scholar] [CrossRef]
- Culbreath, A.; Todd, J.; Brown, S. Epidemiology and management of tomato spotted wilt in peanut. Annu. Rev. Phytopathol. 2003, 41, 53–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gitaitis, R.D.; Dowler, C.C.; Chalfant, R.B. Epidemiology of Tomato Spotted Wilt in Pepper and Tomato in Southern Georgia. Plant Dis. 1998, 82, 752–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pappu, H.; Jones, R.; Jain, R. Global status of tospovirus epidemics in diverse cropping systems: Successes achieved and challenges ahead. Virus Res. 2009, 141, 219–236. [Google Scholar] [CrossRef] [PubMed]
- Riley, D.G.; Pappu, H.R. Tactics for management of thrips (Thysanoptera: Thripidae) and tomato spotted wilt virus in tomato. J. Econ. Èntomol. 2004, 97, 1648–1658. [Google Scholar] [CrossRef]
- Srinivasan, R.; Abney, M.; Culbreath, A.; Kemerait, R.; Tubbs, R.; Monfort, W.; Pappu, H. Three decades of managing Tomato spotted wilt virus in peanut in Southeastern United States. Virus Res. 2017, 241, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, R.S.; Philley, G. Spotted wilt of peanut in Texas. Plant Dis. Rep. 1974, 58, 23–25. [Google Scholar]
- Culbreath, A.; Srinivasan, R. Epidemiology of spotted wilt disease of peanut caused by Tomato spotted wilt virus in the southeastern U.S. Virus Res. 2011, 159, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Garcia, L.E.; Brandenburg, R.L.; Bailey, J.E. Incidence of Tomato spotted wilt virus (Bunyaviridae) and Tobacco Thrips in Virginia-Type Peanuts in North Carolina. Plant Dis. 2000, 84, 459–464. [Google Scholar] [CrossRef]
- Hagan, A.K. Tomato Spotted Wilt Virus in Peanut in Alabama. Plant Dis. 1990, 74, 615. [Google Scholar] [CrossRef]
- Black, M.C.; Smith, D.H. Spotted wilt and rust reactions in south Texas among selected peanut genotypes. In Proceedings of the American Peanut Research and Education Society, Orlando, FL, USA, 14–17 July 1987; Volume 19, p. 31. [Google Scholar]
- Culbreath, A.K. Disease Progress of Spotted Wilt in Peanut Cultivars Florunner and Southern Runner. Phytopathology 1992, 82. [Google Scholar] [CrossRef]
- Culbreath, A.K. Disease Progress of Tomato Spotted Wilt Virusin Selected Peanut Cultivars and Advanced Breeding Lines. Plant Dis. 1996, 80, 70–73. [Google Scholar] [CrossRef]
- Moury, B.; Palloix, A.; Selassie, K.G.; Marchoux, G. Hypersensitive resistance to tomato spotted wilt virus in three Capsicum chinense accessions is controlled by a single gene and is overcome by virulent strains. Euphytica 1997, 94, 45–52. [Google Scholar] [CrossRef]
- Thomas-Carroll, M.L.; Jones, R.A.C. Selection, biological properties and fitness of resistance-breaking strains of Tomato spotted wilt virus in pepper. Ann. Appl. Biol. 2003, 142, 235–243. [Google Scholar] [CrossRef]
- Shrestha, A.; Srinivasan, R.; Sundaraj, S.; Culbreath, A.K.; Riley, D.G. Second Generation Peanut Genotypes Resistant to Thrips-Transmitted Tomato Spotted Wilt Virus Exhibit Tolerance Rather Than True Resistance and Differentially Affect Thrips Fitness. J. Econ. Èntomol. 2013, 106, 587–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agarwal, G.; Clevenger, J.; Pandey, M.K.; Wang, H.; Shasidhar, Y.; Chu, Y.; Fountain, J.C.; Choudhary, D.; Culbreath, A.K.; Liu, X.; et al. High-density genetic map using whole-genome resequencing for fine mapping and candidate gene discovery for disease resistance in peanut. Plant Biotechnol. J. 2018, 16, 1954–1967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khera, P.; Pandey, M.K.; Wang, H.; Feng, S.; Qiao, L.; Culbreath, A.K.; Kale, S.; Wang, J.; Holbrook, C.C.; Zhuang, W.; et al. Mapping Quantitative Trait Loci of Resistance to Tomato Spotted Wilt Virus and Leaf Spots in a Recombinant Inbred Line Population of Peanut (Arachis hypogaea L.) from SunOleic 97R and NC94022. PLoS ONE 2016, 11, e0158452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, M.K.; Wang, H.; Khera, P.; Vishwakarma, M.K.; Kale, S.M.; Culbreath, A.K.; Holbrook, C.C.; Wang, X.; Varshney, R.K.; Guo, B. Genetic Dissection of Novel QTLs for Resistance to Leaf Spots and Tomato Spotted Wilt Virus in Peanut (Arachis hypogaea L.). Front. Plant Sci. 2017, 8, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tseng, Y.-C.; Tillman, B.L.; Peng, Z.; Wang, J. Identification of major QTLs underlying tomato spotted wilt virus resistance in peanut cultivar Florida-EP(TM) ‘113’. BMC Genet. 2016, 17, 128. [Google Scholar] [CrossRef] [Green Version]
- Latham, L.J.; Jones, R.A.C. Selection of resistance breaking strains of tomato spotted wilt tospovirus. Ann. Appl. Biol. 1998, 133, 385–402. [Google Scholar] [CrossRef]
- Roggero, P.; Melani, V.; Ciuffo, M.; Tavella, L.; Tedeschi, R.; Stravato, V.M. Two Field Isolates of Tomato Spotted Wilt Tospovirus Overcome the Hypersensitive Response of a Pepper (Capsicum annuum) Hybrid with Resistance Introgressed from C. chinense PI152225. Plant Dis. 1999, 83, 965. [Google Scholar] [CrossRef]
- Roggero, P.; Masenga, V.; Tavella, L. Field Isolates of Tomato spotted wilt virus Overcoming Resistance in Pepper and Their Spread to Other Hosts in Italy. Plant Dis. 2002, 86, 950–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharman, M.; Persley, D.M. Field isolates ofTomato spotted wilt virusovercoming resistance in capsicum in Australia. Australas. Plant Pathol. 2006, 35, 123–128. [Google Scholar] [CrossRef] [Green Version]
- Almási, A.; Nemes, K.; Csömör, Z.; Tóbiás, I.; Palkovics, L.; Salánki, K. A single point mutation in Tomato spotted wilt virus NSs protein is sufficient to overcome Tsw-gene-mediated resistance in pepper. J. Gen. Virol. 2017, 98, 1521–1525. [Google Scholar] [CrossRef] [PubMed]
- López, C.; Aramburu, J.; Galipienso, L.; Soler, S.; Nuez, F.; Rubio, L. Evolutionary analysis of tomato Sw-5 resistance-breaking isolates of Tomato spotted wilt virus. J. Gen. Virol. 2010, 92, 210–215. [Google Scholar] [CrossRef]
- Margaria, P.; Ciuffo, M.; Rosa, C.; Turina, M. Evidence of a tomato spotted wilt virus resistance-breaking strain originated through natural reassortment between two evolutionary-distinct isolates. Virus Res. 2015, 196, 157–161. [Google Scholar] [CrossRef]
- Tentchev, D.; Verdin, E.; Marchal, C.; Jacquet, M.; Aguilar, J.M.; Moury, B. Evolution and structure of Tomato spotted wilt virus populations: Evidence of extensive reassortment and insights into emergence processes. J. Gen. Virol. 2010, 92, 961–973. [Google Scholar] [CrossRef]
- Whitfield, A.E.; Ullman, D.E.; German, T.L. Tospovirus-Thrips Interactions. Annu. Rev. Phytopathol. 2005, 43, 459–489. [Google Scholar] [CrossRef] [PubMed]
- Adkins, S.; Quadt, R.; Choi, T.-J.; Ahlquist, P.; German, T. An RNA-Dependent RNA Polymerase Activity Associated with Virions of Tomato Spotted Wilt Virus, a Plant- and Insect-Infecting Bunyavirus. Virology 1995, 207, 308–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Haan, P.; Kormelink, R.; Resende, R.D.O.; van Poelwijk, F.; Peters, D.; Goldbach, R. Tomato spotted wilt virus L RNA encodes a putative RNA polymerase. J. Gen. Virol. 1991, 72, 2207–2216. [Google Scholar] [CrossRef] [PubMed]
- Kormelink, R.; de Haan, P.; Meurs, C.; Peters, D.; Goldbach, R. The nucleotide sequence of the M RNA segment of tomato spotted wilt virus, a bunyavirus with two ambisense RNA segments. J. Gen. Virol. 1992, 73, 2795–2804. [Google Scholar] [CrossRef] [PubMed]
- Kormelink, R.; Storms, M.; Van Lent, J.; Peters, D.; Goldbach, R. Expression and Subcellular Location of the NSM Protein of Tomato Spotted Wilt Virus (TSWV), a Putative Viral Movement Protein. Virology 1994, 200, 56–65. [Google Scholar] [CrossRef]
- Soellick, T.-R.; Uhrig, J.F.; Bucher, G.L.; Kellmann, J.-W.; Schreier, P.H. The movement protein NSm of tomato spotted wilt tospovirus (TSWV): RNA binding, interaction with the TSWV N protein, and identification of interacting plant proteins. Proc. Natl. Acad. Sci. USA 2000, 97, 2373–2378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storms, M.M.; Kormelink, R.; Peters, D.; Van Lent, J.W.; Goldbach, R.W. The Nonstructural NSm Protein of Tomato Spotted Wilt Virus Induces Tubular Structures in Plant and Insect Cells. Virology 1995, 214, 485–493. [Google Scholar] [CrossRef] [Green Version]
- Bandla, M.D.; Campbell, L.R.; Ullman, D.E.; Sherwood, J.L. Interaction of Tomato Spotted Wilt Tospovirus (TSWV) Glycoproteins with a Thrips Midgut Protein, a Potential Cellular Receptor for TSWV. Phytopathology 1998, 88, 98–104. [Google Scholar] [CrossRef]
- De Haan, P.; Wagemakers, L.; Peters, D.; Goldbach, R. The S RNA Segment of Tomato Spotted Wilt Virus has an Ambisense Character. J. Gen. Virol. 1990, 71, 1001–1007. [Google Scholar] [CrossRef] [PubMed]
- Takeda, A.; Sugiyama, K.; Nagano, H.; Mori, M.; Kaido, M.; Mise, K.; Tsuda, S.; Okuno, T. Identification of a novel RNA silencing suppressor, NSs protein of Tomato spotted wilt virus. FEBS Lett. 2002, 532, 75–79. [Google Scholar] [CrossRef] [Green Version]
- Richmond, K.; Chenault, K.; Sherwood, J.; German, T. Characterization of the Nucleic Acid Binding Properties of Tomato Spotted Wilt Virus Nucleocapsid Protein. Virology 1998, 248, 6–11. [Google Scholar] [CrossRef] [Green Version]
- Sundaraj, S.; Srinivasan, R.; Culbreath, A.K.; Riley, D.G.; Pappu, H.R. Host Plant Resistance Against Tomato spotted wilt virus in Peanut (Arachis hypogaea) and Its Impact on Susceptibility to the Virus, Virus Population Genetics, and Vector Feeding Behavior and Survival. Phytopathology 2014, 104, 202–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garzo, E.; Moreno, A.; Plaza, M.; Fereres, A. Feeding Behavior and Virus-Transmission Ability of Insect Vectors Exposed to Systemic Insecticides. Plants 2020, 9, 895. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, A.L.; Kennedy, G.G. Effect of cyantraniliprole on feeding behavior and virus transmission of Frankliniella fusca and Frankliniella occidentalis (Thysanoptera: Thripidae) on Capsicum annuum. Crop. Prot. 2013, 54, 251–258. [Google Scholar] [CrossRef]
- Maris, P.C.; Joosten, N.N.; Goldbach, R.W.; Peters, D. Restricted Spread of Tomato spotted wilt virus in Thrips-Resistant Pepper. Phytopathology 2003, 93, 1223–1227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maris, P.C.; Joosten, N.N.; Peters, D.; Goldbach, R.W. Thrips Resistance in Pepper and Its Consequences for the Acquisition and Inoculation of Tomato spotted wilt virus by the Western Flower Thrips. Phytopathology 2003, 93, 96–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srinivasan, R.; Abney, M.R.; Lai, P.-C.; Culbreath, A.K.; Tallury, S.; Leal-Bertioli, S.C.M. Resistance to Thrips in Peanut and Implications for Management of Thrips and Thrips-Transmitted Orthotospoviruses in Peanut. Front. Plant Sci. 2018, 9, 1604. [Google Scholar] [CrossRef] [PubMed]
- Pappu, H.; Pappu, S.; Jain, R.; Bertrand, P.; Culbreath, A.; McPherson, R.; Csinos, A. Sequence characteristics of natural populations of tomato spotted wilt tospovirus infecting flue-cured tobacco in Georgia. Virus Genes 1998, 17, 169–177. [Google Scholar] [CrossRef]
- Fabre, F.; Rousseau, E.; Mailleret, L.; Moury, B. Durable strategies to deploy plant resistance in agricultural landscapes. N. Phytol. 2012, 193, 1064–1075. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Sekine, K.-T.; Nishiguchi, M. Breakdown of plant virus resistance: Can we predict and extend the durability of virus resistance? J. Gen. Plant Pathol. 2014, 80, 327–336. [Google Scholar] [CrossRef]
- Little, E.L. Georgia Plant Dis. Loss Estimates; Athens, GA, 2017. 2015. Available online: https://extension.uga.edu/publications/detail.html?number=AP102-8 (accessed on 13 August 2021).
- Little, E.L. Georgia Plant Dis. Loss Estimates; 2019. 2016. Available online: https://extension.uga.edu/publications/detail.html?number=AP102-9 (accessed on 13 August 2021).
- Little, E.L. Georgia Plant Dis. Loss Estimates; 2019. 2017. Available online: https://extension.uga.edu/publications/detail.html?number=AP102-10 (accessed on 13 August 2021).
- Little, E.L. Georgia Plant Dis. Loss Estimates; 2020. 2018. Available online: https://extension.uga.edu/publications/detail.html?number=AP102-11 (accessed on 13 August 2021).
- Tsompana, M.; Abad, J.; Purugganan, M.; Moyer, J.W. The molecular population genetics of the Tomato spotted wilt virus (TSWV) genome. Mol. Ecol. 2004, 14, 53–66. [Google Scholar] [CrossRef]
- Kaye, A.C.; Moyer, J.W.; Parks, E.J.; Carbone, I.; Cubeta, M.A. Population Genetic Analysis of Tomato spotted wilt virus on Peanut in North Carolina and Virginia. Phytopathology 2011, 101, 147–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Storms, M.M.H.; van de Schoot, C.; Prins, M.; Kormelink, R.; van Lent, J.W.M.; Goldbach, R.W. A comparison of two methods of microinjection for assessing altered plasmodesmal gating in tissues expressing viral movement proteins. Plant J. 2002, 13, 131–140. [Google Scholar] [CrossRef]
- Leastro, M.; Pallás, V.; Resende, R.; Sánchez-Navarro, J. The movement proteins (NSm) of distinct tospoviruses peripherally associate with cellular membranes and interact with homologous and heterologous NSm and nucleocapsid proteins. Virology 2015, 478, 39–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, D.; Raikhy, G.; Pappu, H.R. Movement and nucleocapsid proteins coded by two tospovirus species interact through multiple binding regions in mixed infections. Virology 2015, 478, 137–147. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, K.; Qiu, W.P.; Moyer, J.W. Overcoming Host- and Pathogen-Mediated Resistance in Tomato and Tobacco Maps to the M RNA of Tomato spotted wilt virus. Mol. Plant Microbe Interact. 2001, 14, 242–249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peiro, A.; Cañizares, M.C.; Rubio, L.; López, C.; Moriones, E.; Aramburu, J.; Sanchez-Navarro, J.A. The movement protein (NSm) of Tomato spotted wilt virusis the avirulence determinant in the tomatoSw-5gene-based resistance. Mol. Plant Pathol. 2014, 15, 802–813. [Google Scholar] [CrossRef]
- De Ronde, D.; Butterbach, P.; Lohuis, D.; Hedil, M.; van Lent, J.W.M.; Kormelink, R. Tswgene-based resistance is triggered by a functional RNA silencing suppressor protein of the Tomato spotted wilt virus. Mol. Plant Pathol. 2013, 14, 405–415. [Google Scholar] [CrossRef] [PubMed]
- De Ronde, D.; Pasquier, A.; Ying, S.; Butterbach, P.; Lohuis, D.; Kormelink, R. Analysis of Tomato spotted wilt virus NSs protein indicates the importance of the N-terminal domain for avirulence and RNA silencing suppression. Mol. Plant Pathol. 2013, 15, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Margaria, P.; Ciuffo, M.; Pacifico, D.; Turina, M. Evidence That the Nonstructural Protein of Tomato spotted wilt virus Is the Avirulence Determinant in the Interaction with Resistant Pepper Carrying the Tsw Gene. Mol. Plant Microbe Interact. 2007, 20, 547–558. [Google Scholar] [CrossRef] [Green Version]
- Roossinck, M.J. Mechanisms of plant virus evolution. Annu. Rev. Phytopathol. 1997, 35, 191–209. [Google Scholar] [CrossRef] [PubMed]
- García-Arenal, F.; Fraile, A.; Malpica, J.M. Variability and genetic structure of plant virus populations. Annu. Rev. Phytopathol. 2001, 39, 157–186. [Google Scholar] [CrossRef]
- Chare, E.R.; Holmes, E.C. A phylogenetic survey of recombination frequency in plant RNA viruses. Arch. Virol. 2005, 151, 933–946. [Google Scholar] [CrossRef]
- Lian, S.; Lee, J.-S.; Cho, W.K.; Yu, J.; Kim, M.-K.; Choi, H.-S.; Kim, K.-H. Phylogenetic and Recombination Analysis of Tomato Spotted Wilt Virus. PLoS ONE 2013, 8, e63380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, W.; Geske, S.; Hickey, C.; Moyer, J. Tomato Spotted Wilt Tospovirus Genome Reassortment and Genome Segment-Specific Adaptation. Virology 1998, 244, 186–194. [Google Scholar] [CrossRef] [Green Version]
- Qiu, W.; Moyer, J.W. Tomato Spotted Wilt Tospovirus Adapts to the TSWV N Gene-Derived Resistance by Genome Reassortment. Phytopathology 1999, 89, 575–582. [Google Scholar] [CrossRef] [PubMed]
- Maharijaya, A.; Vosman, B.; Verstappen, F.; Steenhuis-Broers, G.; Mumm, R.; Purwito, A.; Visser, R.G.; Voorrips, R.E. Resistance factors in pepper inhibit larval development of thrips (Frankliniella occidentalis). Èntomol. Exp. Appl. 2012, 145, 62–71. [Google Scholar] [CrossRef]
- Mundt, C.C. Durable resistance: A key to sustainable management of pathogens and pests. Infect. Genet. Evol. 2014, 27, 446–455. [Google Scholar] [CrossRef]
- Pilet-Nayel, M.-L.; Moury, B.; Caffier, V.; Montarry, J.; Kerlan, M.-C.; Fournet, S.; Durel, C.-E.; Delourme, R. Quantitative Resistance to Plant Pathogens in Pyramiding Strategies for Durable Crop Protection. Front. Plant Sci. 2017, 8, 1838. [Google Scholar] [CrossRef] [Green Version]
- Catto, M.; Shrestha, A.; Abney, M.; Champagne, D.; Culbreath, A.; Leal-Bertioli, S.; Hunt, B.; Srinivasan, R. Defense-Related Gene Expression Following an Orthotospovirus Infection Is Influenced by Host Resistance in Arachis hypogaea. Viruses 2021, 13, 1303. [Google Scholar] [CrossRef] [PubMed]
- Montarry, J.; Cartier, E.; Jacquemond, M.; Palloix, A.; Moury, B. Virus adaptation to quantitative plant resistance: Erosion or breakdown? J. Evol. Biol. 2012, 25, 2242–2252. [Google Scholar] [CrossRef]
- Tillman, B.L.; Gorbet, D.W. Registration of ‘FloRun “107”’ Peanut. J. Plant Regist. 2015, 9, 162–167. [Google Scholar] [CrossRef]
- Tillman, B.L. Registration of ‘FloRun “331”’ peanut. J. Plant Regist. 2021, 15, 294–299. [Google Scholar] [CrossRef]
- Norden, A.J.; Lipscomb, R.W.; Carver, W.A. Registration of Florunner Peanuts 1 (Reg. No. 2). Crop Sci. 1969, 9, 850. [Google Scholar] [CrossRef]
- Branch, W.D. Registration of ‘Georgia Green’ Peanut. Crop Sci. 1996, 36, 806. [Google Scholar] [CrossRef]
- Branch, W.D. Registration of ‘Georgia-06G’ Peanut. J. Plant Regist. 2007, 1, 120. [Google Scholar] [CrossRef]
- Branch, W.D. Registration of ‘Georgia-12Y’ Peanut. J. Plant Regist. 2013, 7, 151–153. [Google Scholar] [CrossRef]
- Branch, W.D. Registration of ‘Georgia-13M’ Peanut. J. Plant Regist. 2014, 8, 253–256. [Google Scholar] [CrossRef]
- Branch, W.D.; Brenneman, T.B. Registration of ‘Georgia-14N’ Peanut. J. Plant Regist. 2015, 9, 159–161. [Google Scholar] [CrossRef]
- Branch, W.D. Registration of ‘Georgia-16HO’ Peanut. J. Plant Regist. 2017, 11, 231–234. [Google Scholar] [CrossRef]
- Branch, W.D. Registration of ‘Georgia-18RU’ Peanut. J. Plant Regist. 2019, 13, 326–329. [Google Scholar] [CrossRef]
- Holbrook, C.C.; Timper, P.; Culbreath, A.K.; Kvien, C.K. Registration of ‘Tifguard’ Peanut. J. Plant Regist. 2008, 2, 92–94. [Google Scholar] [CrossRef]
- Holbrook, C.C.; Ozias-Akins, P.; Chu, Y.; Culbreath, A.K.; Kvien, C.K.; Brenneman, T.B. Registration of ‘TifNV-High O/L’ Peanut. J. Plant Regist. 2017, 11, 228–230. [Google Scholar] [CrossRef]
- Tillman, B.L. Registration of ‘TUFRunner “297”’ Peanut. J. Plant Regist. 2017, 12, 31–34. [Google Scholar] [CrossRef]
- Tillman, B.L.; Gorbet, D.W. Registration of ‘TUFRunner “511”’ Peanut. J. Plant Regist. 2017, 11, 235–239. [Google Scholar] [CrossRef]
- Branch, W. Registration of ‘Georgia-04S’ Peanut. Crop Sci. 2005, 45, 1653–1654. [Google Scholar] [CrossRef]
- Hsi, D.C. Registration of New Mexico Valencia C Peanut1 (Reg. No. 24). Crop Sci. 1980, 20, 113–114. [Google Scholar] [CrossRef]
- Isleib, T.G.; Milla-Lewis, S.; Pattee, H.E.; Copeland, S.C.; Zuleta, M.C.; Shew, B.B.; Hollowell, J.E.; Sanders, T.H.; Dean, L.O.; Hendrix, K.W.; et al. Registration of ‘Bailey’ Peanut. J. Plant Regist. 2011, 5, 27–39. [Google Scholar] [CrossRef]
- Branch, W.D. Registration of ‘Georgia-11J’ Peanut. J. Plant Regist. 2012, 6, 281–283. [Google Scholar] [CrossRef]
- Isleib, T.G.; Rice, P.W.; Mozingo, R.W.; Pattee, H.E. Registration of ‘Gregory’ Peanut. Crop Sci. 1999, 39, 1526. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2019, 37, 291–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA; Chichester, UK, 1987. [Google Scholar] [CrossRef] [Green Version]
- Watterson, G. On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 1975, 7, 256–276. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.X.; Li, W.-H. Statistical tests of neutrality of mutations. Genetics 1993, 133, 693–709. [Google Scholar] [CrossRef] [PubMed]
- Murrell, B.; Moola, S.; Mabona, A.; Weighill, T.; Sheward, D.; Pond, S.L.K.; Scheffler, K. FUBAR: A Fast, Unconstrained Bayesian AppRoximation for Inferring Selection. Mol. Biol. Evol. 2013, 30, 1196–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hudson, R.R.; Boos, D.D.; Kaplan, N.L. A statistical test for detecting geographic subdivision. Mol. Biol. Evol. 1992, 9, 138–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hudson, R.R.; Slatkin, M.; Maddison, W.P. Estimation of levels of gene flow from DNA sequence data. Genetics 1992, 132, 583–589. [Google Scholar] [CrossRef]
- Marasigan, K.; Toews, M.; Kemerait, R.; Abney, M.R.; Culbreath, A.; Srinivasan, R. Evaluation of Alternatives to Carbamate and Organophosphate Insecticides Against Thrips and Tomato Spotted Wilt Virusin Peanut Production. J. Econ. Èntomol. 2015, 109, 544–557. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, A.; Srinivasan, R.; Riley, D.G.; Culbreath, A.K. Direct and indirect effects of a thrips-transmitted Tospoviruson the preference and fitness of its vector, Frankliniella fusca. Èntomol. Exp. Appl. 2012, 145, 260–271. [Google Scholar] [CrossRef]
- Shrestha, A.; Sundaraj, S.; Culbreath, A.K.; Riley, D.G.; Abney, M.R.; Srinivasan, R. Effects of Thrips Density, Mode of Inoculation, and Plant Age on Tomato Spotted Wilt Virus Transmission in Peanut Plants. Environ. Èntomol. 2015, 44, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Angelella, G.M.; Riley, D.G. Effects of Pine Pollen Supplementation in an Onion Diet on Frankliniella fusca Reproduction. Environ. Èntomol. 2010, 39, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Munger, F. A Method for Rearing Citrus Thrips in the Laboratory. J. Econ. Èntomol. 1942, 35, 373–375. [Google Scholar] [CrossRef]
- Ben-Mahmoud, S.; Smeda, J.R.; Chappell, T.M.; Stafford-Banks, C.; Kaplinsky, C.H.; Anderson, T.; Mutschler, M.A.; Kennedy, G.G.; Ullman, D.E. Acylsugar amount and fatty acid profile differentially suppress oviposition by western flower thrips, Frankliniella occidentalis, on tomato and interspecific hybrid flowers. PLoS ONE 2018, 13, e0201583. [Google Scholar] [CrossRef] [PubMed]







| Gene | Subgroups | N o | S p | π (SD) q | θw r | Tajima’s D s | Fu and Li’s D *, t | Fu and Li’s F *, u |
|---|---|---|---|---|---|---|---|---|
| N | Overall | 150 | 162 | 0.0083 (0.0005) | 0.0373 | −2.550 ***, v | −5.927 ** | −5.232 ** |
| GA-1998 w | 9 | 27 | 0.0102 (0.0021) | 0.0128 | −1.000 | −1.116 | −1.220 | |
| GA-2010 x | 82 | 100 | 0.0075 (0.0006) | 0.0259 | −2.41 ** | −5.315 ** | −4.950 ** | |
| 2010-S x | 10 | 28 | 0.0098 (0.0017) | 0.0127 | −1.111 | −1.012 | −1.170 | |
| 2010-R x | 72 | 89 | 0.0072 (0.0006) | 0.0236 | −2.390 ** | −4.843 ** | −4.634 ** | |
| GA-New y | 59 | 93 | 0.0076 (0.0006) | 0.0258 | −2.484 ** | −4.350 ** | −4.348 ** | |
| New-S y | 10 | 29 | 0.0086 (0.0016) | 0.0132 | −1.6560 | −1.903. | −2.078. | |
| New-R y | 49 | 79 | 0.0075 (0.0006) | 0.0228 | −2.428 ** | −4.113 ** | −4.168 ** | |
| After 2010 z | 141 | 152 | 0.0076 (0.0004) | 0.0354 | −2.574 *** | −5.648 ** | −5.089 ** | |
| After 2010-S z | 20 | 48 | 0.0094 (0.0013) | 0.0174 | −1.8545 * | −2.263 | −2.495 | |
| After 2010-R z | 121 | 137 | 0.0074 (0.0004) | 0.0328 | −2.5791 *** | −5.334 ** | −4.933 ** | |
| NSs | Overall | 59 | 108 | 0.0072 (0.0005) | 0.0268 | −2.586 *** | −4.505 ** | −4.507 ** |
| Susceptible | 10 | 29 | 0.0072 (0.0011) | 0.0118 | −1.853 * | −2.183 * | −2.370 * | |
| Resistant | 49 | 97 | 0.0072 (0.0005) | 0.0250 | −2.573 *** | −4.517 ** | −4.534 ** | |
| NSm | Overall | 58 | 91 | 0.0126 (0.0006) | 0.0292 | −2.047 * | −3.335 * | −3.401 ** |
| Susceptible | 10 | 30 | 0.0126 (0.0011) | 0.0158 | −0.969 | −1.113 | −1.215 | |
| Resistant | 48 | 82 | 0.0126 (0.0007) | 0.0275 | −1.979 * | −3.013 * | −3.142 * | |
| Gn/Gc | Overall | 59 | 147 | 0.0116 (0.0008) | 0.0419 | −2.620 *** | −5.042 ** | −4.910 ** |
| Susceptible | 10 | 34 | 0.0097 (0.0013) | 0.0159 | −1.890 * | −2.241 ** | −2.431 ** | |
| Resistant | 49 | 132 | 0.0119 (0.0009) | 0.0392 | −2.571 *** | −4.910 ** | −4.824 ** | |
| RdRp | Overall | 52 | 226 | 0.0123 (0.0005) | 0.0353 | −2.395 ** | −4.199 ** | −4.202 ** |
| Susceptible | 9 | 65 | 0.0132 (0.0014) | 0.0169 | −1.181 | −1.304 | −1.432 | |
| Resistant | 43 | 202 | 0.0121 (0.0006) | 0.0330 | −2.378 ** | −3.715 ** | −3.851 ** |
| Gene | Codon Site u | α v | Β w | β-α x | Prob [α < β] y | Amino Acid Changes z |
|---|---|---|---|---|---|---|
| N | 7 | 1.508 | 6.379 | 4.871 | 0.902 * | T→I, F |
| 8 | 0.799 | 0.833 | 0.034 | 0.545 | K→M | |
| 10 | 0.531 | 1.180 | 0.648 | 0.738 | S→N | |
| 18 | 0.542 | 0.681 | 0.139 | 0.583 | G→S | |
| 19 | 0.771 | 0.806 | 0.036 | 0.545 | K→I | |
| 40 | 2.687 | 8.793 | 6.105 | 0.828 | G→D, E | |
| 174 | 0.544 | 0.877 | 0.333 | 0.625 | Y→C | |
| 222 | 0.994 | 2.559 | 1.565 | 0.726 | S→C | |
| NSs | 16 | 1.128 | 3.666 | 2.538 | 0.667 | Q→K |
| 53 | 1.049 | 1.253 | 0.204 | 0.582 | S→T | |
| 98 | 1.066 | 1.244 | 0.178 | 0.580 | I→V | |
| 114 | 1.035 | 1.158 | 0.122 | 0.576 | T→A | |
| 152 | 1.144 | 1.174 | 0.030 | 0.567 | E→K | |
| 193 | 0.849 | 1.514 | 0.666 | 0.623 | N→D | |
| 243 | 5.451 | 11.60 | 6.149 | 0.709 | T→I, A | |
| 257 | 1.051 | 1.099 | 0.048 | 0.569 | P→T | |
| 281 | 0.732 | 2.528 | 1.796 | 0.684 | L→M | |
| NSm | 124 | 1.127 | 1.462 | 0.335 | 0.592 | K→R |
| 210 | 1.056 | 5.211 | 4.155 | 0.824 | K→I, T | |
| Gn/Gc | 51 | 0.656 | 0.971 | 0.315 | 0.609 | N→S |
| 82 | 1.018 | 1.138 | 0.120 | 0.561 | Y→S | |
| 135 | 1.401 | 4.190 | 2.789 | 0.753 | S→N | |
| 139 | 0.656 | 0.971 | 0.315 | 0.609 | N→S | |
| 165 | 0.883 | 1.234 | 0.352 | 0.588 | L→V | |
| 223 | 0.619 | 2.307 | 1.688 | 0.790 | D→N | |
| RdRp | 51 | 0.792 | 3.945 | 3.153 | 0.863 | V→I |
| 149 | 1.006 | 6.957 | 5.951 | 0.905 * | K→R | |
| 207 | 0.902 | 11.777 | 10.875 | 0.959 * | Q→R | |
| 208 | 0.967 | 1.051 | 0.085 | 0.545 | N→D | |
| 211 | 0.689 | 0.905 | 0.217 | 0.580 | I→V | |
| 244 | 0.796 | 0.900 | 0.104 | 0.555 | E→K | |
| 280 | 0.699 | 1.067 | 0.368 | 0.596 | N→D | |
| 331 | 2.785 | 5.005 | 2.220 | 0.639 | I→A | |
| 341 | 0.699 | 1.061 | 0.362 | 0.595 | N→S | |
| 368 | 0.865 | 16.328 | 15.463 | 0.992 * | G→R, E, A | |
| 397 | 0.665 | 9.774 | 9.109 | 0.912 * | Q→L, H | |
| 414 | 0.708 | 0.791 | 0.082 | 0.557 | S→T |
| Gene | Comparison r | Ks s | Kst s | p(Ks/Kst) r | Snn t | p (Snn) r | Fst u |
|---|---|---|---|---|---|---|---|
| N | GA-1998 v vs. GA-2010 w | 6.0059 | 0.0782 | <0.001 | 0.9923 | <0.001 | 0.2915 |
| GA-1998 v vs. GA-2016 to 2019 x | 6.1783 | 0.1230 | <0.001 | 1.0000 | <0.001 | 0.3493 | |
| GA-1998 v vs. After 2010 y | 6.0517 | 0.0559 | <0.001 | 0.9953 | <0.001 | 0.3126 | |
| GA-2010 w vs. GA-2016 to 2019 x | 5.8415 | 0.0151 | <0.001 | 0.6490 | <0.001 | 0.0302 | |
| GA-2010 S vs. R z | 5.8045 | −0.0020 | 0.637 | 0.8438 | 0.041 | −0.0081 | |
| GA-2016 to 2019 S vs. R z | 5.9383 | −0.0050 | 0.923 | 0.6825 | 0.776 | −0.0168 | |
| After 2010 S vs. R z | 5.9385 | −0.0013 | 0.722 | 0.7590 | 0.483 | −0.0048 | |
| NSs | S vs. R z | 6.2446 | −0.0052 | 0.982 | 0.6059 | 0.886 | −0.0184 |
| NSm | S vs. R z | 8.4877 | −0.0022 | 0.559 | 0.6997 | 0.614 | −0.0077 |
| GnGc | S vs. R z | 8.7208 | 0.0012 | 0.302 | 0.6439 | 0.779 | 0.0044 |
| RdRp | S vs. R z | 17.3748 | 0.0024 | 0.244 | 0.7212 | 0.606 | 0.0080 |
| Type | Cultivar | Year | Location | Reference | Susceptibility Subgroups z |
|---|---|---|---|---|---|
| Runner | AUNPL-17 | 2017, 2018, 2019 | Tifton | N/A | Resistant |
| Runner | FloRun 107 | 2016, 2017 | Tifton | [74] | Resistant |
| Runner | FloRun 331 | 2017, 2018, 2019 | Tifton | [75] | Resistant |
| Runner | Florunner | 2018, 2019 | Tifton | [76] | Susceptible |
| Runner | Georgia Green | 2016, 2017, 2018, 2019 | Tifton, Attapulgus | [77] | Susceptible |
| Runner | Georgia-06G | 2016, 2017, 2018, 2019 | Tifton, Attapulgus | [78] | Resistant |
| Runner | Georgia-12Y | 2018, 2019 | Tifton | [79] | Resistant |
| Runner | Georgia-13M | 2016, 2017, 2018 | Tifton | [80] | Resistant |
| Runner | Georgia-14N | 2016, 2017, 2018, 2019 | Tifton | [81] | Resistant |
| Runner | Georgia-16HO | 2017, 2018, 2019 | Tifton, Attapulgus | [82] | Resistant |
| Runner | Georgia-18RU | 2019 | Tifton | [83] | Resistant |
| Runner | Tifguard | 2016, 2017, 2018, 2019 | Tifton | [84] | Resistant |
| Runner | TifNV-High O/L | 2017, 2018, 2019 | Tifton | [85] | Resistant |
| Runner | TUFRunner 157 | 2017 | Tifton | N/A | Resistant |
| Runner | TUFRunner 297 | 2017, 2018, 2019 | Tifton | [86] | Resistant |
| Runner | TUFRunner 511 | 2017, 2018 | Tifton, Attapulgus | [87] | Resistant |
| Runner | TUFRunner 727 | 2016, 2017 | Tifton | N/A | Resistant |
| Spanish | Georgia-04S | 2016 | Tifton | [88] | Resistant |
| Valencia | New Mexico Valencia C | 2019 | Tifton | [89] | Susceptible |
| Virginia | Bailey | 2019 | Tifton | [90] | Resistant |
| Virginia | Georgia-11J | 2017, 2019 | Tifton, Attapulgus | [91] | Resistant |
| Virginia | Gregory | 2019 | Tifton | [92] | Susceptible |
| RNA Fragments | Gene | Primer Pairs | Direction | Sequence (5′ -> 3′) | Annealing Temperature (°C) | Amplicon Size (bp) y |
|---|---|---|---|---|---|---|
| L | RdRp | L6885 | F | CTGTCCTCATTGTCGTGCCT | 58 | 1416 |
| L8403 | R | CAACTAACGCCACCCCTGAT | ||||
| M | NSm | M290 | F | ACATCTTCCTTTGGAACCTA | 53 | 672 |
| M962 | R | CCTCTTCTTCTCCAACTGAT | ||||
| GnGc | M2565 | F | ACCAAGCTTCTTCACATCC | 58 | 756 | |
| M3320 | R | TTTATGTTCCAGGCTGTCC | ||||
| S | NSs | S574 | F | GTCTTGTGTCAAAGAGCATACCTATAA | 58 | 869 |
| S1433 | R | TGATCCCGCTTAAATCAAGCT | ||||
| N z | S2057 | F | TTAAGCAAGTTCTGTGAG | 52 | 777 | |
| S2833 | R | ATGTCTAAGGTTAAGCTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, P.-C.; Abney, M.R.; Bag, S.; Culbreath, A.K.; Srinivasan, R. Impact of Host Resistance to Tomato Spotted Wilt Orthotospovirus in Peanut Cultivars on Virus Population Genetics and Thrips Fitness. Pathogens 2021, 10, 1418. https://doi.org/10.3390/pathogens10111418
Lai P-C, Abney MR, Bag S, Culbreath AK, Srinivasan R. Impact of Host Resistance to Tomato Spotted Wilt Orthotospovirus in Peanut Cultivars on Virus Population Genetics and Thrips Fitness. Pathogens. 2021; 10(11):1418. https://doi.org/10.3390/pathogens10111418
Chicago/Turabian StyleLai, Pin-Chu, Mark R. Abney, Sudeep Bag, Albert K. Culbreath, and Rajagopalbabu Srinivasan. 2021. "Impact of Host Resistance to Tomato Spotted Wilt Orthotospovirus in Peanut Cultivars on Virus Population Genetics and Thrips Fitness" Pathogens 10, no. 11: 1418. https://doi.org/10.3390/pathogens10111418







