Antibiotic Resistance, Biofilm Formation, and Presence of Genes Encoding Virulence Factors in Strains Isolated from the Pharmaceutical Production Environment
Abstract
:1. Introduction
2. Results
2.1. P. aeruginosa Dominated among the Isolated Bacterial Strains
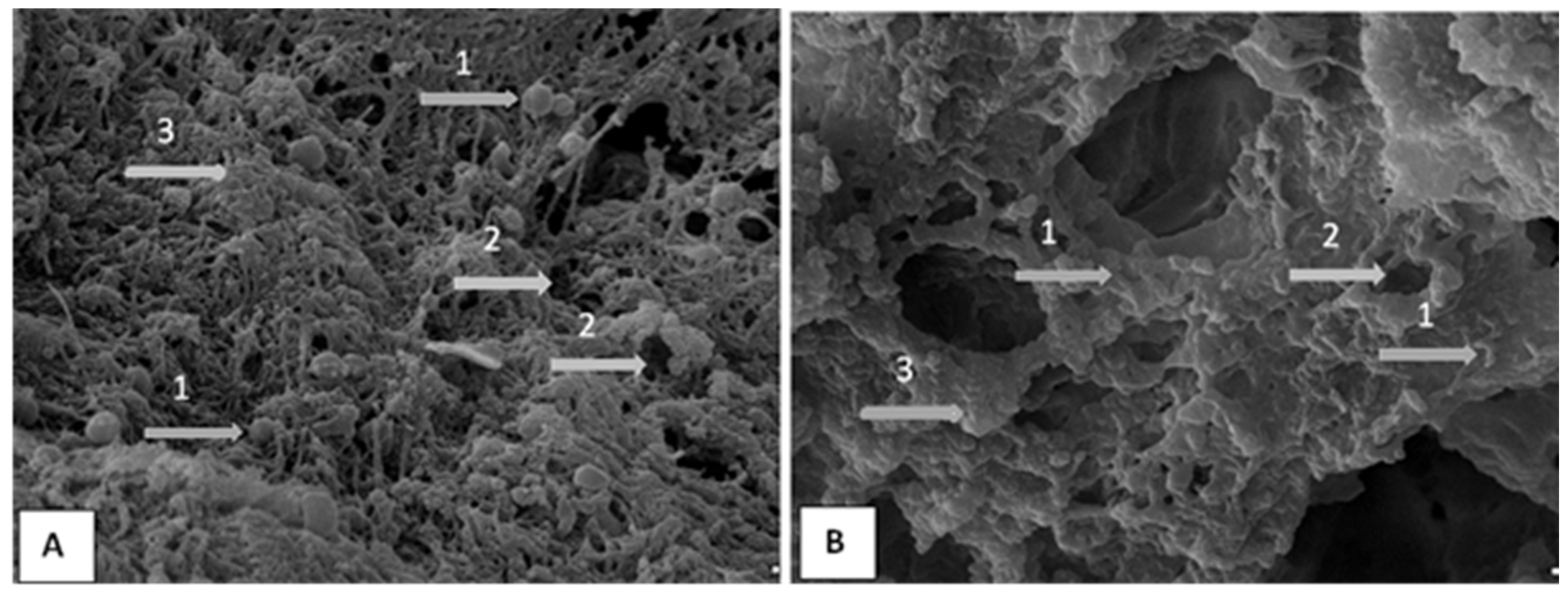
2.2. Majority of P. aeruginosa Strains Identified as Strong Biofilm Producers
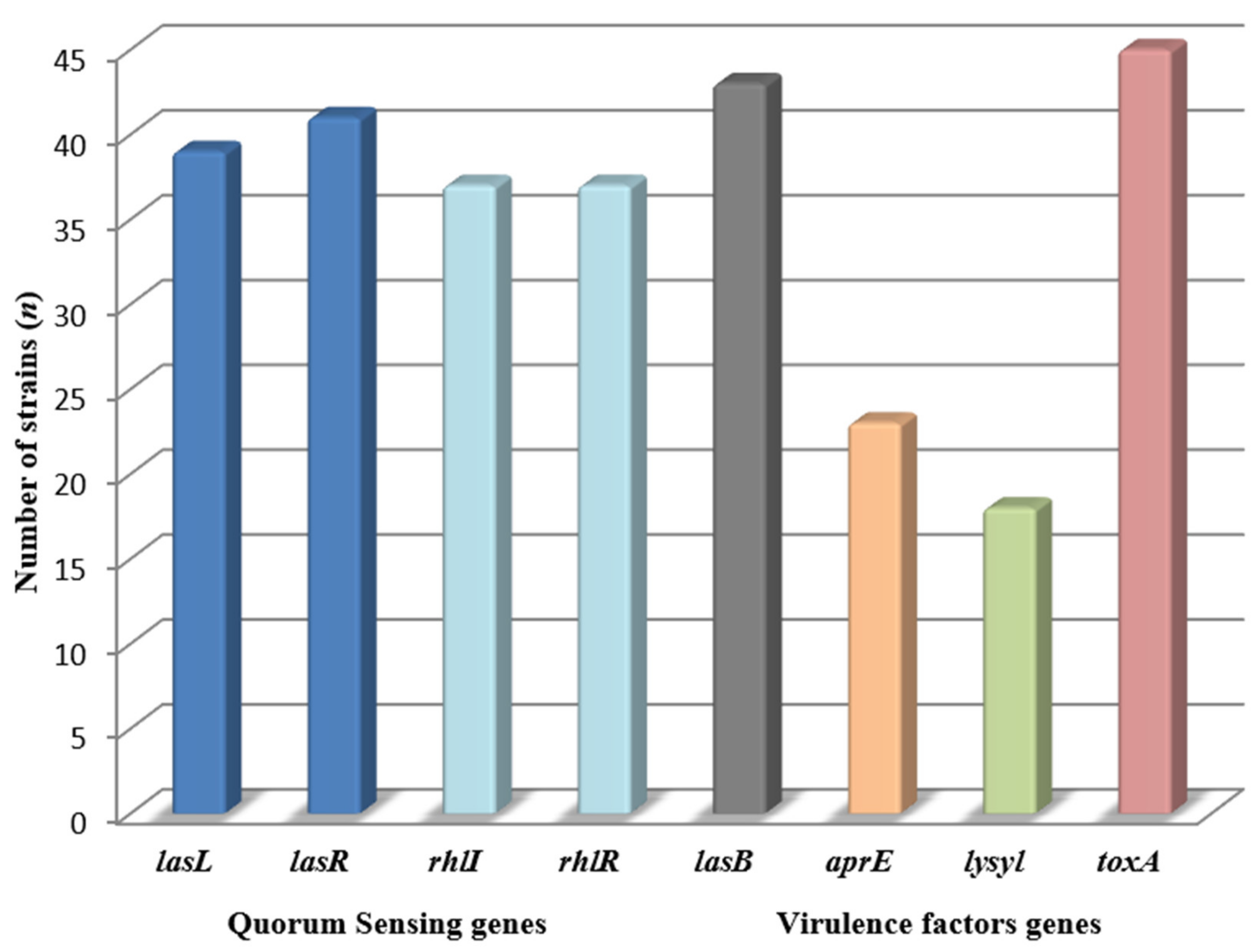
2.3. Genes Involved in the Bacteria Communication and the Virulence Factors Production in P. aeruginosa
2.4. Correlation of the Ability to Create Biofilms and the Presence of Genes Encoding Virulence Factors and QS
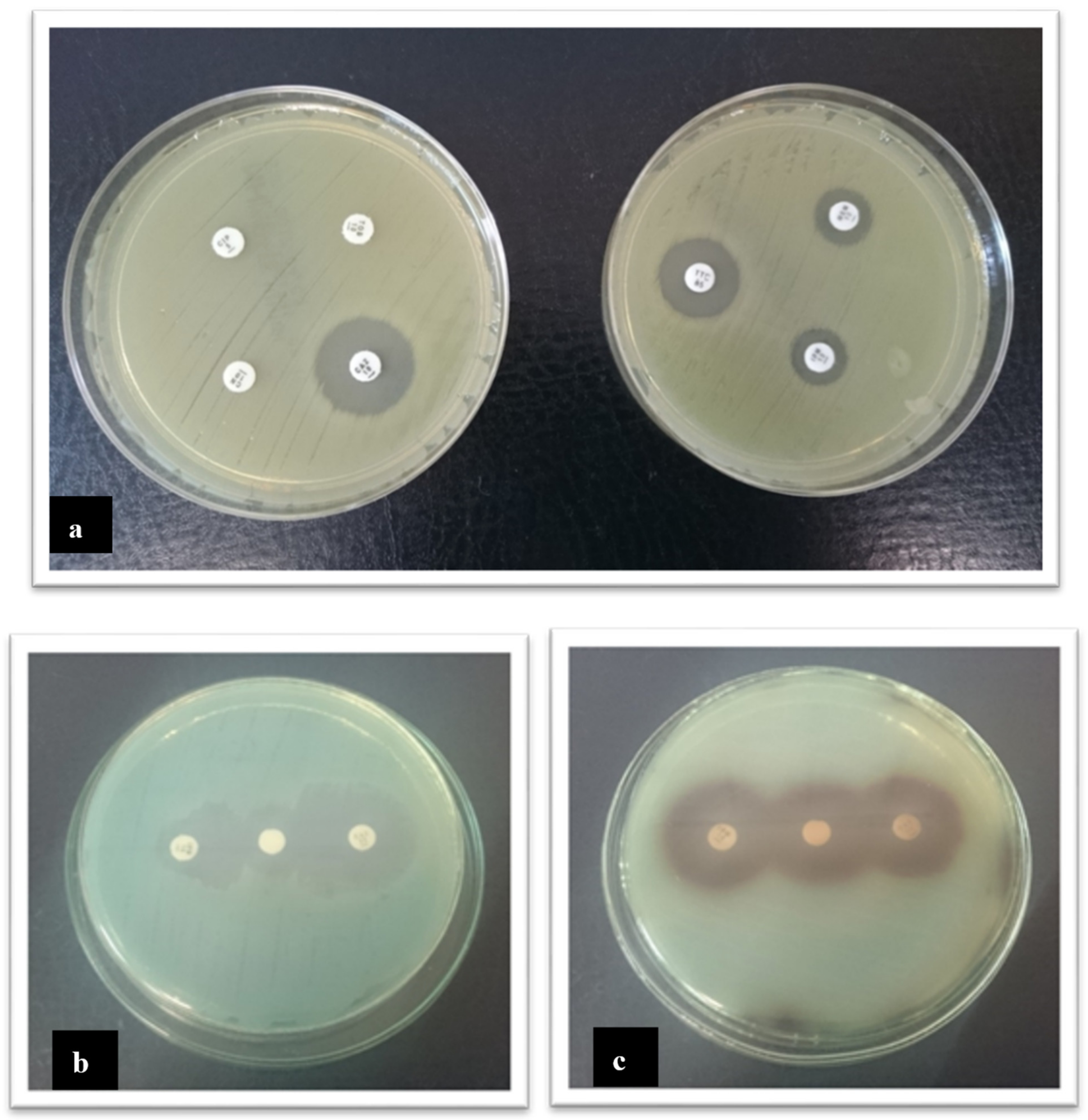
2.5. The Pharmaceutical Production Environment Might Be a Source of Antibiotics Resistant Bacteria
3. Discussion
4. Material and Methods
4.1. Isolation of Bacteria Strains
4.2. Quantitative Assessment of Biofilm Formation in Vitro
4.3. Identification of Genes Taking Part in QS and Encoding Virulence Factors in P. aeruginosa Strains
4.4. Evaluation of Antibiotic Susceptibility of the Isolated Bacterial Strains
4.5. Detection of Mechanisms of Resistance to Antibiotics
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stewart, P.S.; Franklin, M.J. Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 2008, 6, 199–210. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, L.; Rao, X.; Wang, J.; Yu, H.; Jiang, J.; Zhou, W.; Wang, J.; Xiao, Y.; Li, M.; et al. Characterization of lasR-deficient clinical isolates of Pseudomonas aeruginosa. Sci. Rep. 2018, 8, 13344. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Wang, P.-C.; Ma, H.-M.; Chen, S.-Y.; Fu, Y.-H.; Liu, Y.-Y.; Wang, X.; Yu, G.-C.; Huang, T.; Hibbs, D.E.; et al. Design, synthesis and evaluation of halogenated furanone derivatives as quorum sensing inhibitors in Pseudomonas aeruginosa. Eur. J. Pharm. Sci. 2019, 140, 105058. [Google Scholar] [CrossRef] [PubMed]
- Georgi, U.; Lämmel, J.; Datzmann, T.; Schmitt, J.; Decker, S. Do drug-related safety warnings have the expected impact on drug therapy? A systematic review. Pharmacoepidemiol. Drug Saf. 2020, 29, 229–251. [Google Scholar] [CrossRef]
- Ochoa, S.A.; Cruz-Córdova, A.; Rodea, G.E.; Cazares-Dominguez, V.; Escalona, G.; Arellano-Galindo, J.; Hernandez-Castro, R.; Reyes-Lopez, A.; Xicohtencatl-Corteees, J. Phenotypic characterization of multidrug-resistant Pseudomonas aeruginosa strains isolated from pediatric patients associated to biofilm formation. Microbiol. Res. 2015, 172, 68–78. [Google Scholar] [CrossRef] [PubMed]
- Andrejko, M.; Zdybicka-Barabas, A.; Janczarek, M.; Cytryńska, M. Three Pseudomonas aeruginosa strains with different protease profiles. Acta Biochim. Pol. 2013, 60, 83–90. [Google Scholar] [CrossRef]
- Khan, F.; Manivasagan, P.; Pham, D.T.N.; Oh, J.; Kim, S.K.; Kim, Y.M. Antibiofilm and antivirulence properties of chitosan-polypyrrole nanocomposites to Pseudomonas aeruginosa. Microb. Pathog. 2019, 128, 363–373. [Google Scholar] [CrossRef]
- Saleh, M.M.; Abbas, H.A.; Askoura, M.M. Repositioning secnidazole as a novel virulence factors attenuating agent in Pseudomonas aeruginosa. Microb. Pathog. 2019, 127, 31–38. [Google Scholar] [CrossRef]
- Vandeplassche, E.; Sass, A.; Lemarcq, A.; Dandekar, A.A.; Coenye, T.; Crabbé, A. In vitro evolution of Pseudomonas aeruginosa AA2 biofilms in the presence of cystic fibrosis lung microbiome members. Sci. Rep. 2019, 9, 12859. [Google Scholar] [CrossRef] [Green Version]
- Samira, H.; Fereshteh, E. Biofilm formation and β-Lactamase production in Burn isolates of Pseudomonas aeruginosa. Jundishapur J. Microbiol. 2015, 8, 15514. [Google Scholar] [CrossRef] [Green Version]
- Cipolla, D.; Blanchard, J.; Gonda, I. Development of Liposomal Ciprofloxacin to Treat Lung Infections. Pharmaceutics 2016, 8, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbara, S.; Pitsch, A.; Jochmans, S.; Hodjat, K.; Cherrier, P.; Monchi, M.; Vinsonneau, C.; Diamantis, S. Impact of a multimodal strategy combining a new standard of care and restriction of carbapenems, fluoroquinolones and cephalosporins on antibiotic consumption and resistance of Pseudomonas aeruginosa in a French intensive care unit. Int. J. Antimicrob. Agents 2019, 53, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Somboro, A.M.; Sekyere, O.J.; Amoako, D.G.; Essack, S.Y.; Bester, L.A. Diversity and Proliferation of Metallo-β-Lactamases: A Clarion Call for Clinically Effective Metallo-β-Lactamase Inhibitors. Appl. Environ. Microbiol. 2018, 84, e00698-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masadeh, M.M.; Alzoubi, K.H.; Ahmed, W.S.; Magaji, A.S. In Vitro Comparison of Antibacterial and Antibiofilm Activities of Selected Fluoroquinolones against Pseudomonas aeruginosa and Methicillin-Resistant Staphylococcus aureus. Pathogens 2019, 8, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Centre for Disease Prevention and Control. Available online: https://www.ecdc.europa.eu/en (accessed on 16 April 2020).
- Długaszewska, J.; Ratajczak, M.; Kamińska, D.; Gajecka, M. Are dietary supplements containing plant-derived ingredients safe microbiologically? Saudi Pharm. J. 2019, 27, 240–245. [Google Scholar] [CrossRef]
- EMA Annual Report 2018 Published. Available online: https://www.ema.europa.eu/en/news/ema-annual-report-2018-published (accessed on 12 April 2020).
- Enforcement Reports. Available online: https://www.fda.gov/safety/recalls-market-withdrawals-safetyalerts/enforcement-reports (accessed on 7 May 2020).
- Jimenez, L. Microbial diversity in pharmaceutical product recalls and environments. PDA J. Pharm. Sci. Technol. 2007, 61, 383–399. [Google Scholar]
- Tresoldi, A.T.; Padoveze, M.C.; Trabasso, P.; Veiga, J.F.S.; Marba, S.T.M.; Nowakonski, A.; Moretti Branchini, M.L. Enterobacter cloacae sepsis outbreak in a newborn unit caused by contaminated total parenteral nutrition solution. Am. J. Infect. Control. 2002, 8, 258–261. [Google Scholar] [CrossRef]
- Watson, J.T.; Roderick, C.; Jones, A.M.; Siston, J.R.; Fernandez, K.M.; Beck, E.; Sokalski, S.; Bette, J.; Matthew, J.; Srinivasan, A.; et al. Outbreak of Catheter-Associated Klebsiella oxytoca and Enterobacter cloacae Bloodstream Infections in an Oncology Chemotherapy Center. Arch. Intern. Med. 2005, 165, 2639–2643. [Google Scholar] [CrossRef] [Green Version]
- Długaszewska, J.; Muszyński, Z. Biological purity of the medicinal product manufacturing environment. Farm. Pol. 2009, 65, 323–326. [Google Scholar]
- Dozens of Drug Products Recalled due to Microbial Contamination. Available online: https://www.pharmacytimes.com/resource-centers/veterinary-pharmacy/dozens-of-drug-products-recalled-due-to-microbial-contamination (accessed on 24 April 2020).
- Tršan, M.; Seme, K.; Srčič, S. The environmental monitoring in hospital pharmacy cleanroom and microbiota catalogue preparation. Saudi Pharm. J. 2019, 27, 455–462. [Google Scholar] [CrossRef]
- Karatuna, O.; Yagci, A. Analysis of quorum sensing-dependent virulence factor production and its relationship with antimicrobial susceptibility in Pseudomonas aeruginosa respiratory isolates. Clin. Microbiol. Infect. 2010, 16, 1770–1775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Rev. Microbiol. 2012, 8, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Claessen, D.; Rozen, D.E.; Kuipers, O.P.; Sogaard-Andersen, L.; Van Wezel, G.P. Bacterial solutions to multicellularity: A tale of biofilms, filaments and fruiting bodies. Nat. Rev. Microbiol. 2014, 12, 115–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchand, S.; De Block, J.; De Jonghe, V.; Coorevits, A.; Heyndrickx, M.; Herman, L. Biofilm formation in milk production and processing environments; influence on milk quality and safety. Compr Rev. Food Sci. Food Saf. 2012, 11, 133–147. [Google Scholar] [CrossRef]
- Harmsen, M.; Yang, L.; Pamp, S.J.; Tolker-Nielsen, T. An update on Pseudomonas aeruginosa biofilm formation, tolerance, and dispersal. FEMS Immunol. Med. Microbiol. 2012, 59, 253–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadhim, D.; Ali, M.R. Prevalence study of quorum sensing groups among clinical isolates of Pseudomonas aeruginosa. Int. Curr. Microbiol. Appl. Sci. 2014, 3, 204–2015. [Google Scholar]
- Senturk, S.; Ulusoy, S.; Bosgelmez-Tinaz, G.; Yagci, A. Quorum sensing and virulence of Pseudomonas aeruginosa during urinary tract infections. J. Infect. Dev. Ctries 2012, 6, 501–507. [Google Scholar] [CrossRef] [Green Version]
- Wolska, K.; Szweda, P. Genetic features of clinical P. aeruginosa strains. Polish J. Microbiol. 2009, 58, 255–260. [Google Scholar]
- Mittal, R.; Sharma, S.; Chhibber, S.; Harjai, K. Contribution of quorum-sensing systems to virulence of Pseudomonas aeruginosa in an experimental pyelonephritis model. J. Microbiol. Immunol. Infect. 2006, 39, 302–309. [Google Scholar]
- Sabharwal, N.; Dhall, S.; Chhibber, S.; Harjai, K. Molecular detection of virulence genes as markers in Pseudomonas aeruginosa isolated from urinary tract infections. Int. Mol. Epidemiol. Genet. 2014, 5, 125–134. [Google Scholar]
- Cornaglia, G.; Giamarellouu, H.; Rossolini, G.M. Metallo-β-lactamases: A last frontier for β-lactams? Lancet Infect. Dis. 2011, 11, 381–393. [Google Scholar] [CrossRef]
- Demirdjian, S.; Sanchez, H.; Hopkins, D.; Berwin, B. Motility-Independent Formation of Antibiotic-Tolerant Pseudomonas aeruginosa Aggregates. Appl. Environ. Microbiol. 2019, 85, e00844-19. [Google Scholar] [CrossRef] [Green Version]
- Commission Directive (UE) 2017/1572 of 15 September 2017, Supplementing Directive 2001/83/EC of the European Parliament and of the Council as Regards the Principles and Guidelines of Good Manufacturing Practice for Medicinal Products for Human Use. Available online: https://eur-lex.europa.eu/eli/dir/2017/1572/oj (accessed on 18 June 2020).
- Sanganyado, E.; Gwenzi, W. Antibiotic resistasnce in drinking water systems: Occurrence, removal, and human health risks. Sci. Total Environ. 2019, 669, 785–797. [Google Scholar] [CrossRef] [PubMed]
- EDQM. Methods of Analysis. In European Pharmacopoeia; European Directorate for the Quality of Medicines & Health Care (EDQM): Strasbourg, France, 2016; pp. 195–234, 579–580. [Google Scholar]
- Długaszewska, J.; Antczak, M.; Kaczmarek, I.; Jankowiak, R.; Buszkiewicz, M.; Herkowiak, M.; Michalak, K.; Kukuła, H.; Ratajczak, M. In vitro biofilm formation and antibiotic susceptibility of Pseudomonas aeruginosa isolated from airways of patient with cystic fibrosis. J. Med. Sci. 2016, 85, 245–253. [Google Scholar] [CrossRef] [Green Version]
- European Committee on Antimicrobial Susceptibility Testing Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 8.1. Available online: http://www.eucast.org (accessed on 21 January 2020).

| Cefoxitin | Ceftazidime | Piperacylina | Ticarcilin/ Clavulanate | Imipenem | Meropenem | Gentamicin | Tobramycin | Clindamycin | Erythromycin | Ciprofloxacin | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P. aeruginosa (n = 46) | R (%) | - | 6.5 | - | 2.2 | 4.3 | 2.2 | 13.0 | 6.5 | - | - | 2.2 |
| S (%) | - | 93.5 | - | 97.8 | 93.5 | 95.6 | 87.0 | 93.5 | - | - | 95.6 | |
| Enterobacterales (n = 14) | R (%) | - | 21.4 | 0.0 | 7.1 | - | - | 21.4 | 35.7 | - | - | 0.0 |
| S (%) | - | 78.6 | 100 | 92.9 | - | - | 78.6 | 64.3 | - | - | 100 | |
| Staphylococcus spp. (n = 7) | R (%) | 0.0 | - | - | - | - | - | - | - | 0.0 | 0.0 | - |
| S (%) | 100 | - | - | - | - | - | - | - | 100 | 100 | - | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ratajczak, M.; Kaminska, D.; Dlugaszewska, J.; Gajecka, M. Antibiotic Resistance, Biofilm Formation, and Presence of Genes Encoding Virulence Factors in Strains Isolated from the Pharmaceutical Production Environment. Pathogens 2021, 10, 130. https://doi.org/10.3390/pathogens10020130
Ratajczak M, Kaminska D, Dlugaszewska J, Gajecka M. Antibiotic Resistance, Biofilm Formation, and Presence of Genes Encoding Virulence Factors in Strains Isolated from the Pharmaceutical Production Environment. Pathogens. 2021; 10(2):130. https://doi.org/10.3390/pathogens10020130
Chicago/Turabian StyleRatajczak, Magdalena, Dorota Kaminska, Jolanta Dlugaszewska, and Marzena Gajecka. 2021. "Antibiotic Resistance, Biofilm Formation, and Presence of Genes Encoding Virulence Factors in Strains Isolated from the Pharmaceutical Production Environment" Pathogens 10, no. 2: 130. https://doi.org/10.3390/pathogens10020130






