Acinetobacter baumannii Strains Deficient in the Clp Chaperone-Protease Genes Have Reduced Virulence in a Murine Model of Pneumonia
Abstract
:1. Introduction
2. Results
2.1. PCR Analysis Was Performed to Validate the Clp Chaperone-Protease Gene Mutations in A. Baumannii Strain AB5075.
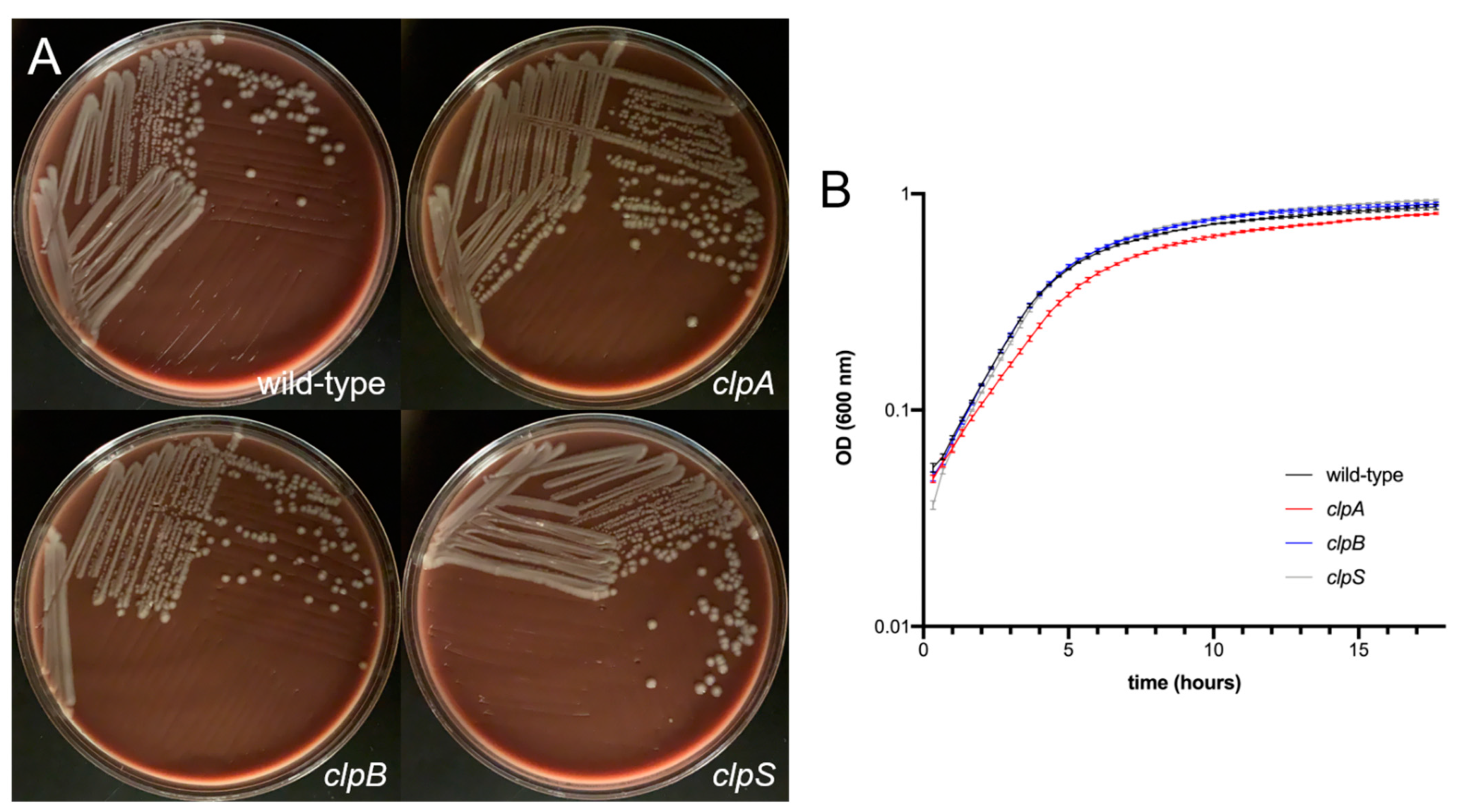
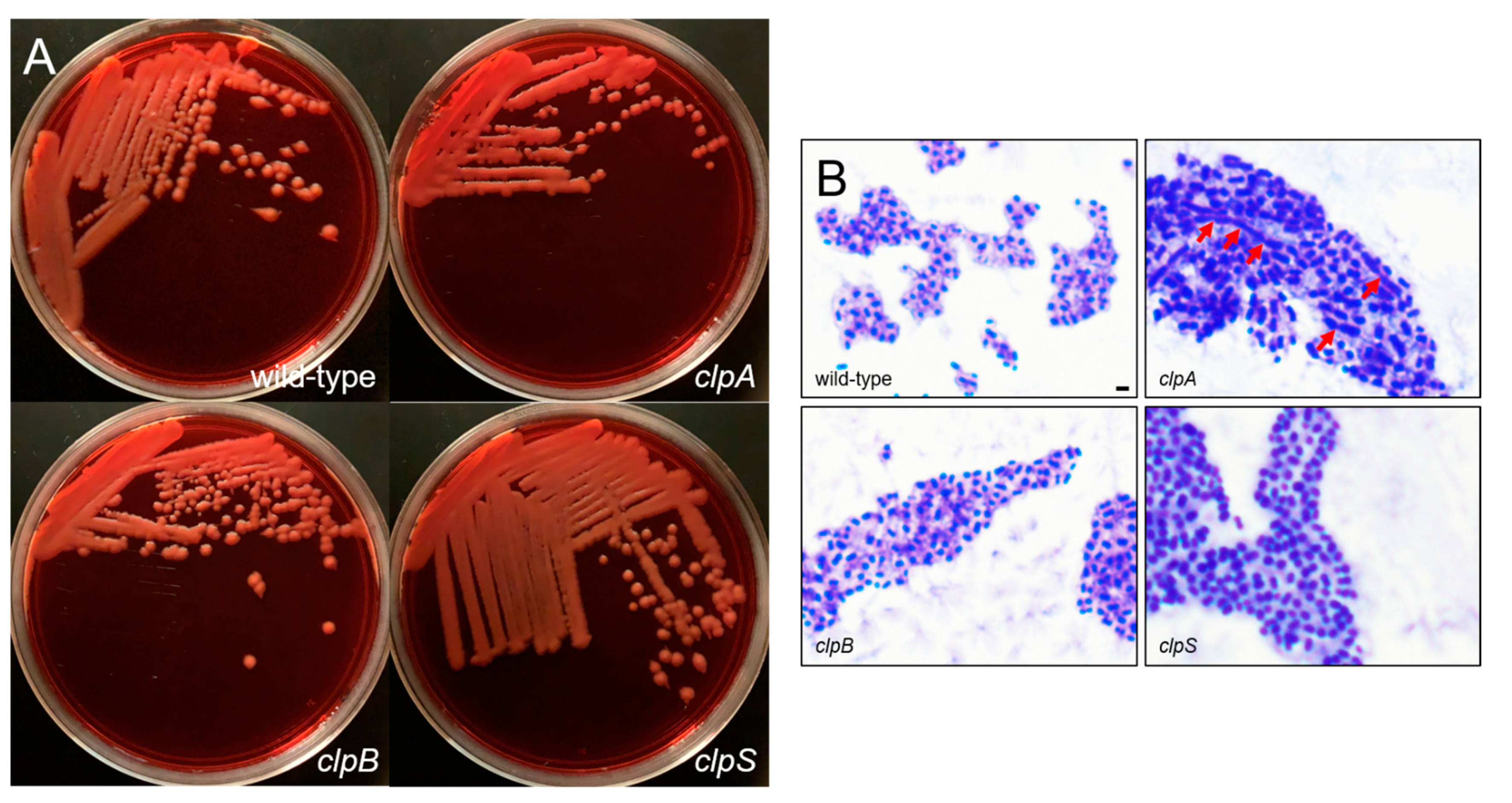
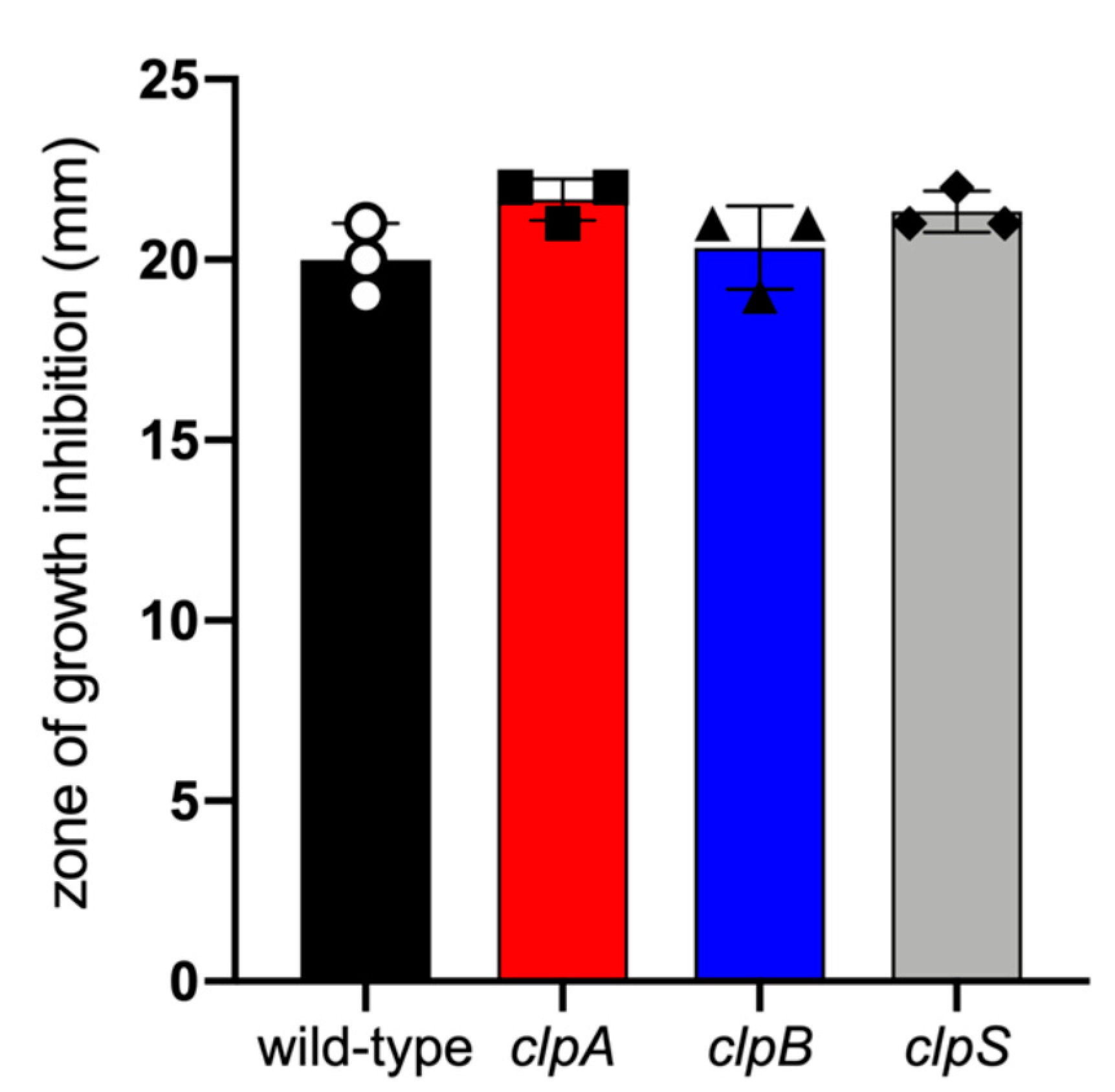
2.2. Deletion of the Clp Chaperone-Protease Genes Does Not Affect Microbial Growth, Encapsulation and Sensitivity to Oxidative Stress
2.3. A. baumannii Requires the Clp Chaperone-Protease Genes for Virulence and to Cause Disease In Vivo
2.4. The Clp Chaperone-Protease Genes Are Necessary for A. baumannii Survival Inside of J774.16 Macrophage-Like Cells
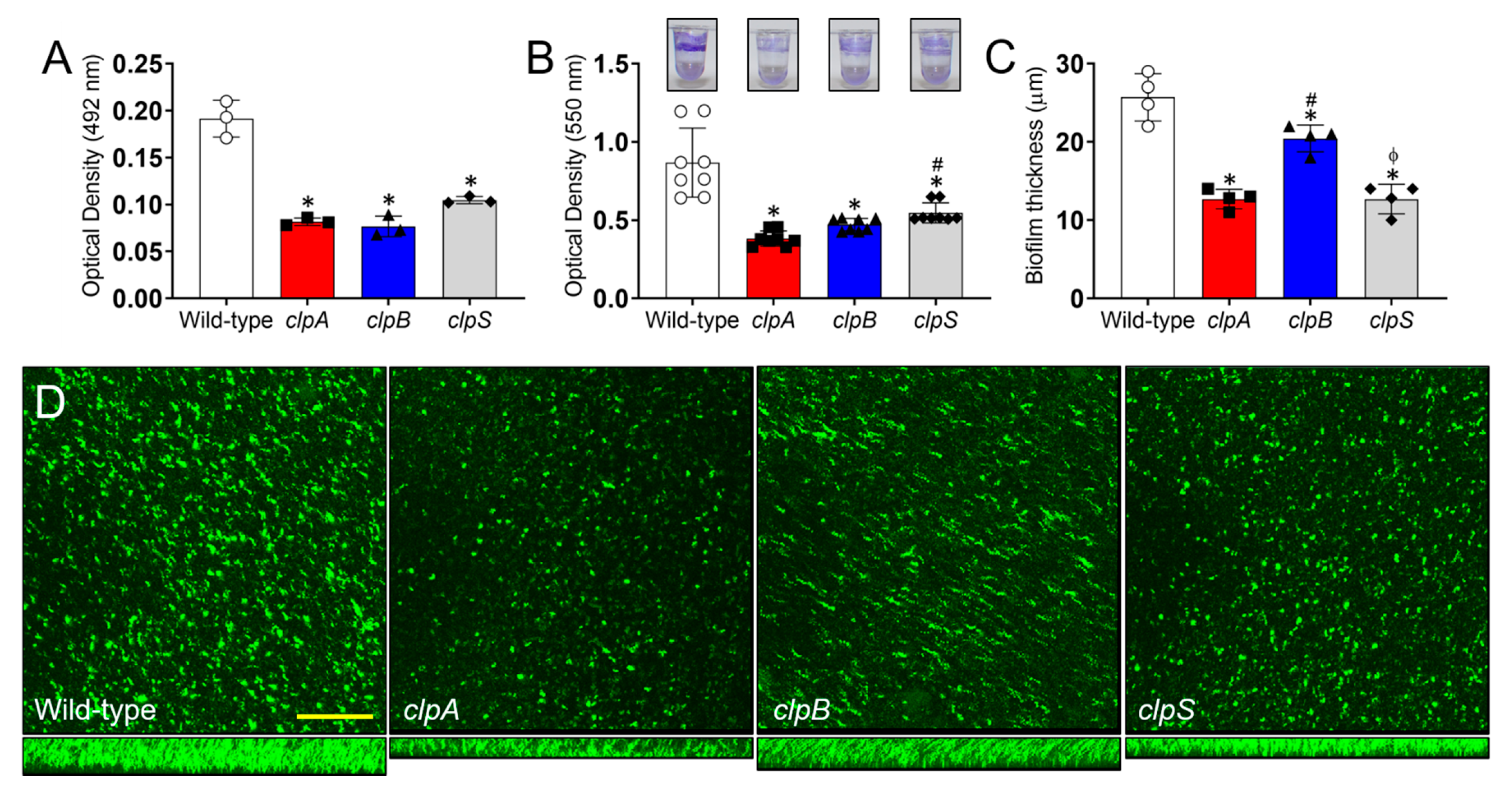
2.5. The Clp Chaperone-Protease Genes Are Crucial for A. baumannii Biofilm Formation In Vitro
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Acinetobacter baumannii
5.2. PCR Analysis of clp Genes
5.3. Growth Curves
5.4. Capsule Staining
5.5. Sensitivity to Hydrogen Peroxide
5.6. Bacterial Infection
5.7. CFU Count Determinations in Tissues
5.8. J774.16 Macrophage-Like Cells
5.9. Phagocytosis and Killing Assay
5.10. Biofilm Formation
5.11. Measurement of Biofilm Metabolic Activity by XTT Reduction Assay
5.12. Crystal Violet Assay
5.13. Confocal Microscopy
5.14. Statistical Analysis
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lin, M.F.; Lan, C.Y. Antimicrobial resistance in Acinetobacter baumannii: From bench to bedside. World J. Clin. Cases 2014, 2, 787–814. [Google Scholar] [CrossRef] [PubMed]
- Fournier, P.E.; Richet, H. The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin. Infect. Dis. 2006, 42, 692–699. [Google Scholar] [CrossRef] [Green Version]
- Mortensen, B.L.; Rathi, S.; Chazin, W.J.; Skaar, E.P. Acinetobacter baumannii response to host-mediated zinc limitation requires the transcriptional regulator Zur. J. Bacteriol. 2014, 196, 2616–2626. [Google Scholar] [CrossRef] [Green Version]
- Clark, R.B. Imipenem resistance among Acinetobacter baumannii: Association with reduced expression of a 33-36 kDa outer membrane protein. J. Antimicrob. Chemother. 1996, 38, 245–251. [Google Scholar] [CrossRef] [Green Version]
- Lambert, T.; Gerbaud, G.; Courvalin, P. Transferable amikacin resistance in Acinetobacter spp. due to a new type of 3’-aminoglycoside phosphotransferase. Antimicrob. Agents Chemother. 1988, 32, 15–19. [Google Scholar] [CrossRef] [Green Version]
- US CDC. Healthcare-Associated Infections. Acinetobater in Healthcare. Available online: https://www.cdc.gov/hai/organisms/acinetobacter.html (accessed on 4 December 2020).
- Dijkshoorn, L.; Nemec, A.; Seifert, H. An increasing threat in hospitals: Multidrug-resistant Acinetobacter baumannii. Nat. Rev. Microbiol. 2007, 5, 939–951. [Google Scholar] [CrossRef] [PubMed]
- de Breij, A.; Dijkshoorn, L.; Lagendijk, E.; van der Meer, J.; Koster, A.; Bloemberg, G.; Wolterbeek, R.; van den Broek, P.; Nibbering, P. Do biofilm formation and interactions with human cells explain the clinical success of Acinetobacter baumannii? PLoS ONE 2010, 5, e10732. [Google Scholar] [CrossRef]
- Choi, C.H.; Lee, E.Y.; Lee, Y.C.; Park, T.I.; Kim, H.J.; Hyun, S.H.; Kim, S.A.; Lee, S.K.; Lee, J.C. Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell. Microbiol. 2005, 7, 1127–1138. [Google Scholar] [CrossRef]
- Loehfelm, T.W.; Luke, N.R.; Campagnari, A.A. Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J. Bacteriol. 2008, 190, 1036–1044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.W.; Koh, Y.M.; Kim, J.; Lee, J.C.; Lee, Y.C.; Seol, S.Y.; Cho, D.T.; Kim, J. Capacity of multidrug-resistant clinical isolates of Acinetobacter baumannii to form biofilm and adhere to epithelial cell surfaces. Clin. Microbiol. Infect. 2008, 14, 49–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shlaes, D.M.; Bradford, P.A. Antibiotics-From There to Where?: How the antibiotic miracle is threatened by resistance and a broken market and what we can do about it. Pathog. Immun. 2018, 3, 19–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, F.C.; Dexter, C.; Kostoulias, X.; Uddin, M.I.; Peleg, A.Y. The Mechanisms of Disease Caused by Acinetobacter baumannii. Front. Microbiol. 2019, 10, 1601. [Google Scholar] [CrossRef] [Green Version]
- Choi, C.H.; Hyun, S.H.; Lee, J.Y.; Lee, J.S.; Lee, Y.S.; Kim, S.A.; Chae, J.P.; Yoo, S.M.; Lee, J.C. Acinetobacter baumannii outer membrane protein A targets the nucleus and induces cytotoxicity. Cell. Microbiol. 2008, 10, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Weber, B.S.; Harding, C.M.; Feldman, M.F. Pathogenic Acinetobacter: From the Cell Surface to Infinity and Beyond. J. Bacteriol. 2015, 198, 880–887. [Google Scholar] [CrossRef] [Green Version]
- Moffatt, J.H.; Harper, M.; Mansell, A.; Crane, B.; Fitzsimons, T.C.; Nation, R.L.; Li, J.; Adler, B.; Boyce, J.D. Lipopolysaccharide-deficient Acinetobacter baumannii shows altered signaling through host Toll-like receptors and increased susceptibility to the host antimicrobial peptide LL-37. Infect. Immun. 2013, 81, 684–689. [Google Scholar] [CrossRef] [Green Version]
- Boll, J.M.; Tucker, A.T.; Klein, D.R.; Beltran, A.M.; Brodbelt, J.S.; Davies, B.W.; Trent, M.S. Reinforcing Lipid A Acylation on the Cell Surface of Acinetobacter baumannii Promotes Cationic Antimicrobial Peptide Resistance and Desiccation Survival. mBio 2015, 6, e00478-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chin, C.Y.; Tipton, K.A.; Farokhyfar, M.; Burd, E.M.; Weiss, D.S.; Rather, P.N. A high-frequency phenotypic switch links bacterial virulence and environmental survival in Acinetobacter baumannii. Nat. Microbiol. 2018, 3, 563–569. [Google Scholar] [CrossRef]
- Geisinger, E.; Isberg, R.R. Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii. PLoS Pathog 2015, 11, e1004691. [Google Scholar] [CrossRef] [Green Version]
- Fiester, S.E.; Arivett, B.A.; Schmidt, R.E.; Beckett, A.C.; Ticak, T.; Carrier, M.V.; Ghosh, R.; Ohneck, E.J.; Metz, M.L.; Sellin Jeffries, M.K.; et al. Iron-Regulated Phospholipase C Activity Contributes to the Cytolytic Activity and Virulence of Acinetobacter baumannii. PLoS ONE 2016, 11, e0167068. [Google Scholar] [CrossRef] [Green Version]
- Bentancor, L.V.; Camacho-Peiro, A.; Bozkurt-Guzel, C.; Pier, G.B.; Maira-Litran, T. Identification of Ata, a multifunctional trimeric autotransporter of Acinetobacter baumannii. J. Bacteriol. 2012, 194, 3950–3960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eijkelkamp, B.A.; Stroeher, U.H.; Hassan, K.A.; Paulsen, I.T.; Brown, M.H. Comparative analysis of surface-exposed virulence factors of Acinetobacter baumannii. BMC Genom. 2014, 15, 1020. [Google Scholar] [CrossRef] [Green Version]
- Coyne, S.; Rosenfeld, N.; Lambert, T.; Courvalin, P.; Perichon, B. Overexpression of resistance-nodulation-cell division pump AdeFGH confers multidrug resistance in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2010, 54, 4389–4393. [Google Scholar] [CrossRef] [Green Version]
- Damier-Piolle, L.; Magnet, S.; Bremont, S.; Lambert, T.; Courvalin, P. AdeIJK, a resistance-nodulation-cell division pump effluxing multiple antibiotics in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2008, 52, 557–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magnet, S.; Courvalin, P.; Lambert, T. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob. Agents Chemother. 2001, 45, 3375–3380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Ozer, E.A.; Mandel, M.J.; Hauser, A.R. Genome-wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. mBio 2014, 5, e01163-14. [Google Scholar] [CrossRef] [Green Version]
- Frees, D.; Chastanet, A.; Qazi, S.; Sorensen, K.; Hill, P.; Msadek, T.; Ingmer, H. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 2004, 54, 1445–1462. [Google Scholar] [CrossRef] [PubMed]
- Webb, C.; Moreno, M.; Wilmes-Riesenberg, M.; Curtiss, R., 3rd; Foster, J.W. Effects of DksA and ClpP protease on sigma S production and virulence in Salmonella typhimurium. Mol. Microbiol. 1999, 34, 112–123. [Google Scholar] [CrossRef]
- Jackson, M.W.; Silva-Herzog, E.; Plano, G.V. The ATP-dependent ClpXP and Lon proteases regulate expression of the Yersinia pestis type III secretion system via regulated proteolysis of YmoA, a small histone-like protein. Mol. Microbiol. 2004, 54, 1364–1378. [Google Scholar] [CrossRef]
- Gallagher, L.A.; Ramage, E.; Weiss, E.J.; Radey, M.; Hayden, H.S.; Held, K.G.; Huse, H.K.; Zurawski, D.V.; Brittnacher, M.J.; Manoil, C. Resources for Genetic and Genomic Analysis of Emerging Pathogen Acinetobacter baumannii. J. Bacteriol. 2015, 197, 2027–2035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, J.; Ruan, W.; Sun, J.; Wang, F.; Yan, W. Functional Characterization of c-di-GMP Signaling-Related Genes in the Probiotic Lactobacillus acidophilus. Front. Microbiol. 2018, 9, 1935. [Google Scholar] [CrossRef] [Green Version]
- Olsen, A.; Jonsson, A.; Normark, S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 1989, 338, 652–655. [Google Scholar] [CrossRef]
- Capestany, C.A.; Tribble, G.D.; Maeda, K.; Demuth, D.R.; Lamont, R.J. Role of the Clp system in stress tolerance, biofilm formation, and intracellular invasion in Porphyromonas gingivalis. J. Bacteriol. 2008, 190, 1436–1446. [Google Scholar] [CrossRef] [Green Version]
- Orsinger-Jacobsen, S.J.; Patel, S.S.; Vellozzi, E.M.; Gialanella, P.; Nimrichter, L.; Miranda, K.; Martinez, L.R. Use of a stainless steel washer platform to study Acinetobacter baumannii adhesion and biofilm formation on abiotic surfaces. Microbiology 2013, 159, 2594–2604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, L.; Wang, H.; Zhang, R.; Li, H. The regulation of Porphyromonas gingivalis biofilm formation by ClpP. Biochem. Biophys. Res. Commun. 2019, 509, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Lo, H.H.; Liao, C.T.; Li, C.E.; Chiang, Y.C.; Hsiao, Y.M. The clpX gene plays an important role in bacterial attachment, stress tolerance, and virulence in Xanthomonas campestris pv. campestris. Arch. Microbiol. 2020, 202, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Mawla, G.D.; Hall, B.M.; Carcamo-Oyarce, G.; Grant, R.A.; Zhang, J.J.; Kardon, J.R.; Ribbeck, K.; Sauer, R.T.; Baker, T.A. ClpP1P2 peptidase activity promotes biofilm formation in Pseudomonas aeruginosa. Mol. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Kahne, S.C.; Darwin, K.H. Structural determinants of regulated proteolysis in pathogenic bacteria by ClpP and the proteasome. Curr. Opin. Struct. Biol. 2020, 67, 120–126. [Google Scholar] [CrossRef]
- Moreno-Cinos, C.; Goossens, K.; Salado, I.G.; Van Der Veken, P.; De Winter, H.; Augustyns, K. ClpP Protease, a Promising Antimicrobial Target. Int. J. Mol. Sci. 2019, 20, 2232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olivares, A.O.; Baker, T.A.; Sauer, R.T. Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat. Rev. Microbiol. 2016, 14, 33–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Corro, J.H.; Palmer, C.D.; Ojha, A.K. Progression from remodeling to hibernation of ribosomes in zinc-starved mycobacteria. Proc. Natl. Acad. Sci. USA 2020, 117, 19528–19537. [Google Scholar] [CrossRef] [PubMed]
- Panasenko, O.O.; Bezrukov, F.; Komarynets, O.; Renzoni, A. YjbH Solubility Controls Spx in Staphylococcus aureus: Implication for MazEF Toxin-Antitoxin System Regulation. Front. Microbiol. 2020, 11, 113. [Google Scholar] [CrossRef]
- Pennetzdorfer, N.; Lembke, M.; Pressler, K.; Matson, J.S.; Reidl, J.; Schild, S. Regulated Proteolysis in Vibrio cholerae Allowing Rapid Adaptation to Stress Conditions. Front. Cell. Infect. Microbiol. 2019, 9, 214. [Google Scholar] [CrossRef] [PubMed]
- Kock, H.; Gerth, U.; Hecker, M. MurAA, catalysing the first committed step in peptidoglycan biosynthesis, is a target of Clp-dependent proteolysis in Bacillus subtilis. Mol. Microbiol. 2004, 51, 1087–1102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frees, D.; Gerth, U.; Ingmer, H. Clp chaperones and proteases are central in stress survival, virulence and antibiotic resistance of Staphylococcus aureus. Int. J. Med. Microbiol. 2014, 304, 142–149. [Google Scholar] [CrossRef]
- Roy, R.; You, R.I.; Lin, M.D.; Lin, N.T. Mutation of the Carboxy-Terminal Processing Protease in Acinetobacter baumannii Affects Motility, Leads to Loss of Membrane Integrity, and Reduces Virulence. Pathogens 2020, 9, 322. [Google Scholar] [CrossRef]
- Ching, C.; Yang, B.; Onwubueke, C.; Lazinski, D.; Camilli, A.; Godoy, V.G. Lon Protease Has Multifaceted Biological Functions in Acinetobacter baumannii. J. Bacteriol. 2019, 201, e00536-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chastanet, A.; Derre, I.; Nair, S.; Msadek, T. clpB, a novel member of the Listeria monocytogenes CtsR regulon, is involved in virulence but not in general stress tolerance. J. Bacteriol. 2004, 186, 1165–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, P.; Singh, L.K.; Kumari, S.; Hakiem, O.R.; Batra, J.K. ClpB is an essential stress regulator of Mycobacterium tuberculosis and endows survival advantage to dormant bacilli. Int. J. Med. Microbiol. 2020, 310, 151402. [Google Scholar] [CrossRef]
- Sangpuii, L.; Dixit, S.K.; Kumawat, M.; Apoorva, S.; Kumar, M.; Kappala, D.; Goswami, T.K.; Mahawar, M. Comparative roles of clpA and clpB in the survival of Salmonella Typhimurium under stress and virulence in poultry. Sci. Rep. 2018, 8, 4481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, T.; Kedzierska-Mieszkowska, S.; Liu, H.; Cheng, C.; Ganta, R.R.; Zolkiewski, M. Aggregate-reactivation activity of the molecular chaperone ClpB from Ehrlichia chaffeensis. PLoS ONE 2013, 8, e62454. [Google Scholar] [CrossRef] [Green Version]
- Brodmann, M.; Dreier, R.F.; Broz, P.; Basler, M. Francisella requires dynamic type VI secretion system and ClpB to deliver effectors for phagosomal escape. Nat. Commun. 2017, 8, 15853. [Google Scholar] [CrossRef]
- Ojha, A.; Anand, M.; Bhatt, A.; Kremer, L.; Jacobs, W.R., Jr.; Hatfull, G.F. GroEL1: A dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 2005, 123, 861–873. [Google Scholar] [CrossRef] [Green Version]
- Arita-Morioka, K.; Yamanaka, K.; Mizunoe, Y.; Ogura, T.; Sugimoto, S. Novel strategy for biofilm inhibition by using small molecules targeting molecular chaperone DnaK. Antimicrob. Agents Chemother. 2015, 59, 633–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubern, J.F.; Lagendijk, E.L.; Lugtenberg, B.J.; Bloemberg, G.V. The heat shock genes dnaK, dnaJ, and grpE are involved in regulation of putisolvin biosynthesis in Pseudomonas putida PCL1445. J. Bacteriol. 2005, 187, 5967–5976. [Google Scholar] [CrossRef] [Green Version]
- Tipton, K.A.; Dimitrova, D.; Rather, P.N. Phase-Variable Control of Multiple Phenotypes in Acinetobacter baumannii Strain AB5075. J. Bacteriol. 2015, 197, 2593–2599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tipton, K.A.; Chin, C.Y.; Farokhyfar, M.; Weiss, D.S.; Rather, P.N. Role of Capsule in Resistance to Disinfectants, Host Antimicrobials, and Desiccation in Acinetobacter baumannii. Antimicrob. Agents Chemother. 2018, 62, e01188-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobs, A.C.; Thompson, M.G.; Black, C.C.; Kessler, J.L.; Clark, L.P.; McQueary, C.N.; Gancz, H.Y.; Corey, B.W.; Moon, J.K.; Si, Y.; et al. AB5075, a Highly Virulent Isolate of Acinetobacter baumannii, as a Model Strain for the Evaluation of Pathogenesis and Antimicrobial Treatments. mBio 2014, 5, e01076-14. [Google Scholar] [CrossRef] [Green Version]
- Asplund, M.B.; Coelho, C.; Cordero, R.J.; Martinez, L.R. Alcohol impairs J774.16 macrophage-like cell antimicrobial functions in Acinetobacter baumannii infection. Virulence 2013, 4, 467–472. [Google Scholar] [CrossRef] [Green Version]
- Moffat, J.F.; Tompkins, L.S. A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect. Immun. 1992, 60, 296–301. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belisario, J.C.; Lee, H.H.; Luknauth, H.; Rigel, N.W.; Martinez, L.R. Acinetobacter baumannii Strains Deficient in the Clp Chaperone-Protease Genes Have Reduced Virulence in a Murine Model of Pneumonia. Pathogens 2021, 10, 204. https://doi.org/10.3390/pathogens10020204
Belisario JC, Lee HH, Luknauth H, Rigel NW, Martinez LR. Acinetobacter baumannii Strains Deficient in the Clp Chaperone-Protease Genes Have Reduced Virulence in a Murine Model of Pneumonia. Pathogens. 2021; 10(2):204. https://doi.org/10.3390/pathogens10020204
Chicago/Turabian StyleBelisario, J Christian, Hiu Ham Lee, Harshani Luknauth, Nathan W. Rigel, and Luis R. Martinez. 2021. "Acinetobacter baumannii Strains Deficient in the Clp Chaperone-Protease Genes Have Reduced Virulence in a Murine Model of Pneumonia" Pathogens 10, no. 2: 204. https://doi.org/10.3390/pathogens10020204
APA StyleBelisario, J. C., Lee, H. H., Luknauth, H., Rigel, N. W., & Martinez, L. R. (2021). Acinetobacter baumannii Strains Deficient in the Clp Chaperone-Protease Genes Have Reduced Virulence in a Murine Model of Pneumonia. Pathogens, 10(2), 204. https://doi.org/10.3390/pathogens10020204




